Abstract
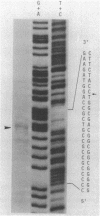
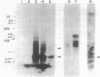
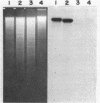
The nucleotide sequence of afsB from Streptomyces coelicolor A3(2), a pleiotropic gene which positively controls the biosynthesis of A-factor and the pigmented antibiotics actinorhodin and undecylprodigiosin in S. coelicolor A3(2) and "Streptomyces lividans," was determined. The determinant of the afsB gene, which includes the putative AfsB protein consisting of 243 amino acids, was mapped functionally by tests for A-factor and pigment production in "S. lividans" and S. coelicolor A3(2) after introduction of recombinant plasmids containing various restriction endonuclease fragments on the vector plasmids pIJ41 and pIJ702. The putative AfsB protein contains two regions separated by 167 residues which resemble conserved domains of known DNA-binding proteins. High-resolution nuclease S1 protection mapping revealed that the afsB mRNA, approximately 1,300 base pairs (bp) long, which was determined by Northern blot hybridization, had its start point 340 bp upstream of the putative methionine start codon. The Northern hybridization experiment also suggested that the afsB gene was constitutively transcribed throughout growth. Also shown by the Northern hybridization was the presence of an unidentified gene with an extraordinary amount of 880-bp mRNA located downstream from afsB. Dot hybridization with the brown pigment production genes, possibly involved in polyketide biosynthesis, as the probe suggested that the afsB gene did not stimulate transcription of the pigment production genes. In Southern blot DNA-DNA hybridization analysis with the afsB sequence as the probe, sequences exhibiting various degrees of homology were found in several Streptomyces spp. A DNA sequence showing strong homology to the afsB in Streptomyces griseus FT-1, a high streptomycin producer, behaved like an extrachromosomal element, homologous to the afsA gene, a structural gene for A-factor biosynthesis.
Full text
PDF

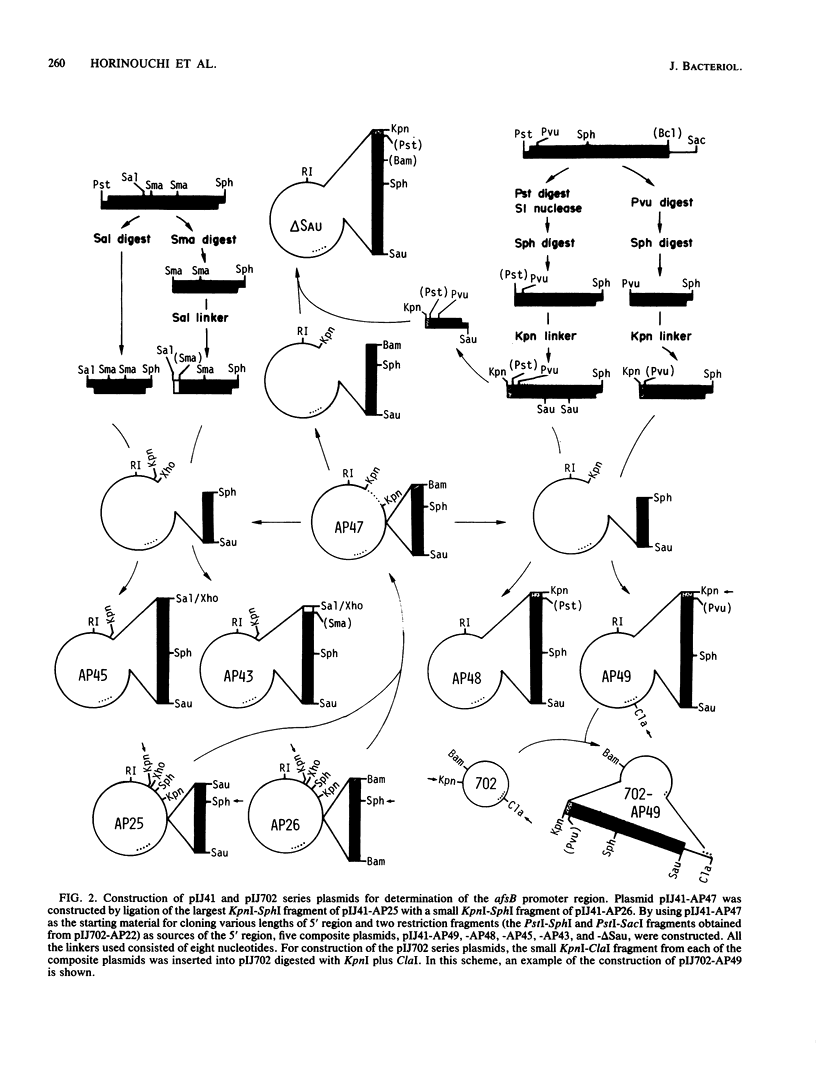

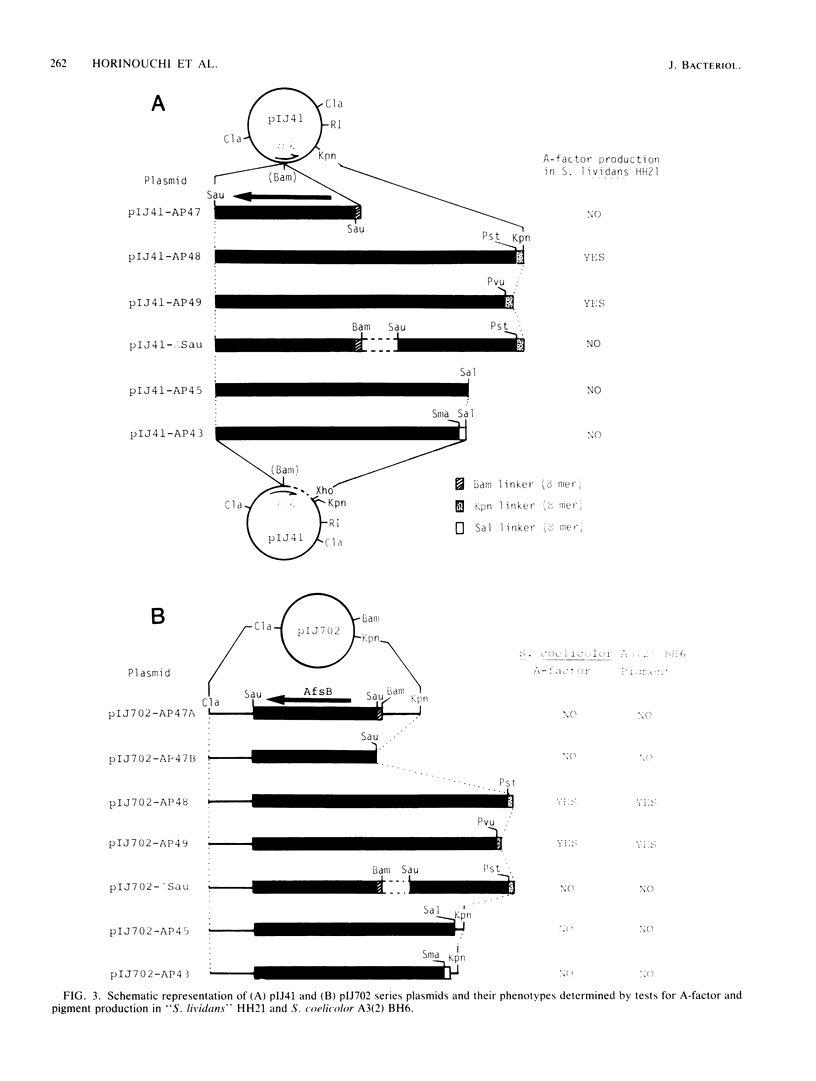
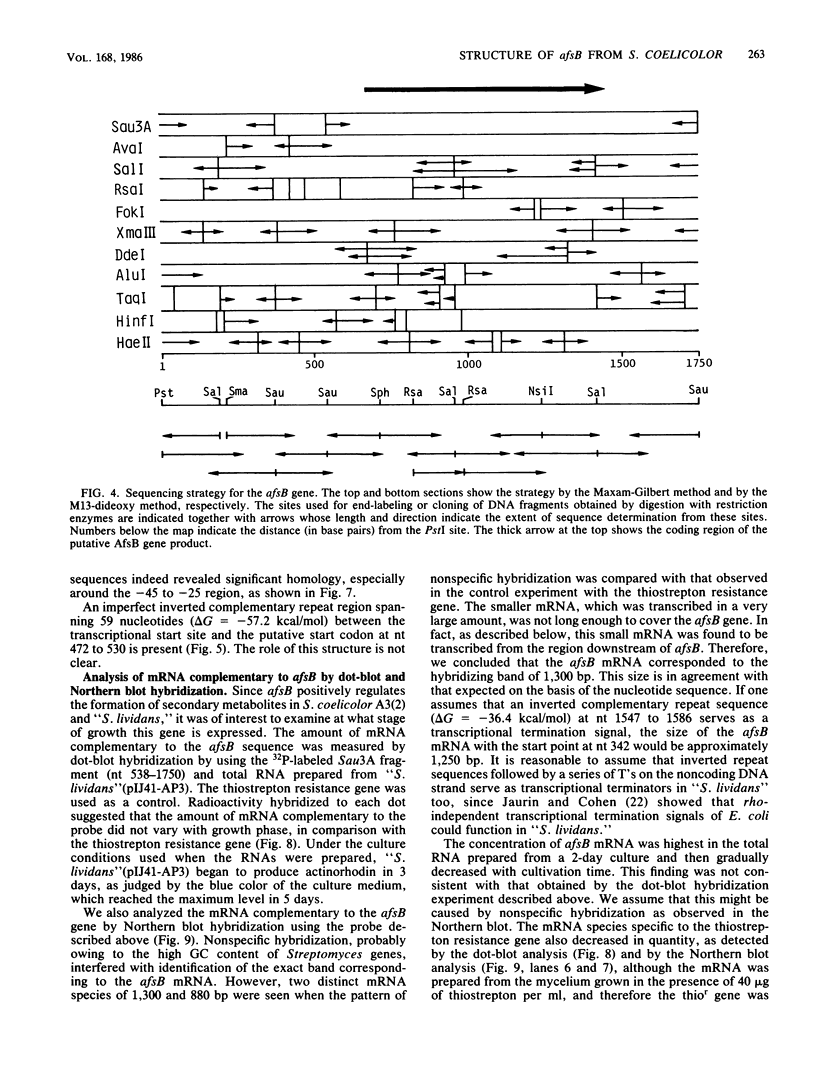


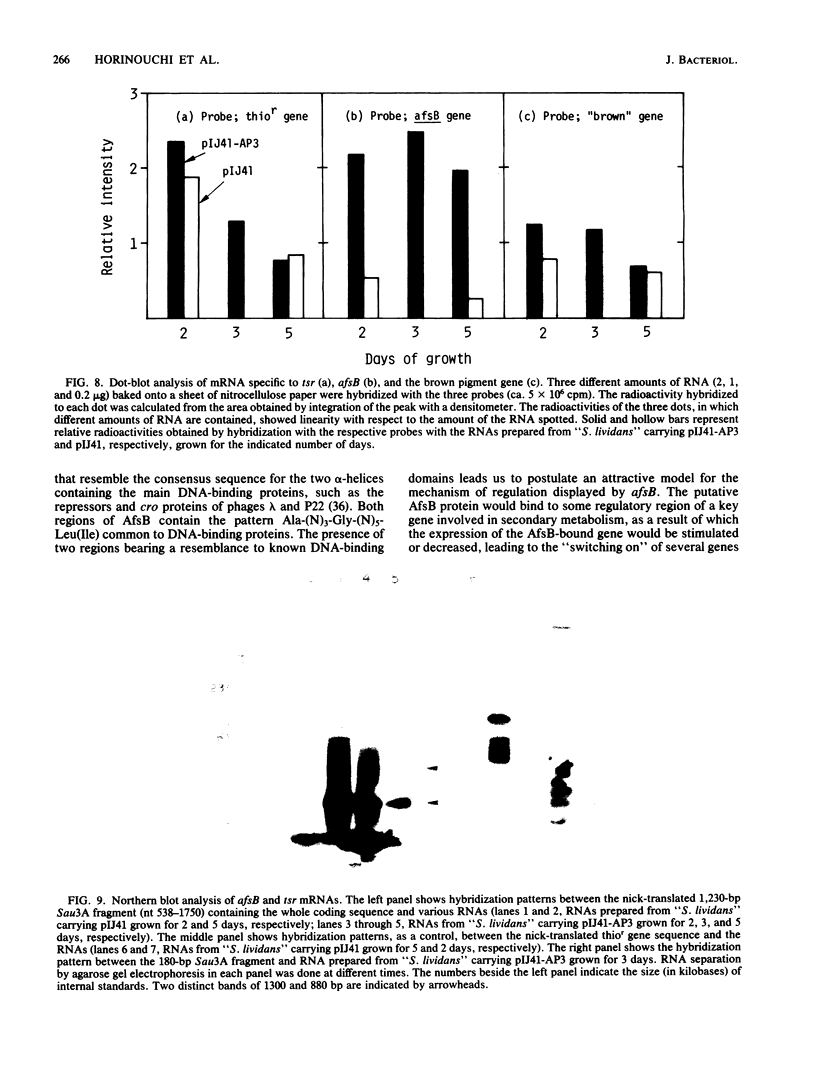


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aguilar A., Hopwood D. A. Determination of methylenomycin A synthesis by the pSV1 plasmid from Streptomyces violaceus-ruber SANK 95570. J Gen Microbiol. 1982 Aug;128(8):1893–1901. doi: 10.1099/00221287-128-8-1893. [DOI] [PubMed] [Google Scholar]
- Barnes W. M., Bevan M., Son P. H. Kilo-sequencing: creation of an ordered nest of asymmetric deletions across a large target sequence carried on phage M13. Methods Enzymol. 1983;101:98–122. doi: 10.1016/0076-6879(83)01008-3. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Cohen S. N. Gene expression in Streptomyces: construction and application of promoter-probe plasmid vectors in Streptomyces lividans. Mol Gen Genet. 1982;187(2):265–277. doi: 10.1007/BF00331128. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Findlay P. R., Johnson M. W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984 Oct;30(1-3):157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- Burton Z., Burgess R. R., Lin J., Moore D., Holder S., Gross C. A. The nucleotide sequence of the cloned rpoD gene for the RNA polymerase sigma subunit from E coli K12. Nucleic Acids Res. 1981 Jun 25;9(12):2889–2903. doi: 10.1093/nar/9.12.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M., Brzustowicz L., Hannett N., Pero J. Bacteriophage SPO1 genes 33 and 34. Location and primary structure of genes encoding regulatory subunits of Bacillus subtilis RNA polymerase. J Mol Biol. 1984 Dec 15;180(3):533–547. doi: 10.1016/0022-2836(84)90026-3. [DOI] [PubMed] [Google Scholar]
- Gitt M. A., Wang L. F., Doi R. H. A strong sequence homology exists between the major RNA polymerase sigma factors of Bacillus subtilis and Escherichia coli. J Biol Chem. 1985 Jun 25;260(12):7178–7185. [PubMed] [Google Scholar]
- Gold L., Pribnow D., Schneider T., Shinedling S., Singer B. S., Stormo G. Translational initiation in prokaryotes. Annu Rev Microbiol. 1981;35:365–403. doi: 10.1146/annurev.mi.35.100181.002053. [DOI] [PubMed] [Google Scholar]
- Hara O., Beppu T. Mutants blocked in streptomycin production in Streptomyces griseus - the role of A-factor. J Antibiot (Tokyo) 1982 Mar;35(3):349–358. doi: 10.7164/antibiotics.35.349. [DOI] [PubMed] [Google Scholar]
- Hara O., Horinouchi S., Uozumi T., Beppu T. Genetic analysis of A-factor synthesis in Streptomyces coelicolor A3(2) and Streptomyces griseus. J Gen Microbiol. 1983 Sep;129(9):2939–2944. doi: 10.1099/00221287-129-9-2939. [DOI] [PubMed] [Google Scholar]
- Hirschman J., Wong P. K., Sei K., Keener J., Kustu S. Products of nitrogen regulatory genes ntrA and ntrC of enteric bacteria activate glnA transcription in vitro: evidence that the ntrA product is a sigma factor. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7525–7529. doi: 10.1073/pnas.82.22.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood D. A., Kieser T., Wright H. M., Bibb M. J. Plasmids, recombination and chromosome mapping in Streptomyces lividans 66. J Gen Microbiol. 1983 Jul;129(7):2257–2269. doi: 10.1099/00221287-129-7-2257. [DOI] [PubMed] [Google Scholar]
- Hopwood D. A., Wright H. M. CDA is a new chromosomally-determined antibiotic from Streptomyces coelicolor A3(2). J Gen Microbiol. 1983 Dec;129(12):3575–3579. doi: 10.1099/00221287-129-12-3575. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Beppu T. Construction and application of a promoter-probe plasmid that allows chromogenic identification in Streptomyces lividans. J Bacteriol. 1985 Apr;162(1):406–412. doi: 10.1128/jb.162.1.406-412.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Hara O., Beppu T. Cloning of a pleiotropic gene that positively controls biosynthesis of A-factor, actinorhodin, and prodigiosin in Streptomyces coelicolor A3(2) and Streptomyces lividans. J Bacteriol. 1983 Sep;155(3):1238–1248. doi: 10.1128/jb.155.3.1238-1248.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Kumada Y., Beppu T. Unstable genetic determinant of A-factor biosynthesis in streptomycin-producing organisms: cloning and characterization. J Bacteriol. 1984 May;158(2):481–487. doi: 10.1128/jb.158.2.481-487.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaurin B., Cohen S. N. Streptomyces lividans RNA polymerase recognizes and uses Escherichia coli transcriptional signals. Gene. 1984 Apr;28(1):83–91. doi: 10.1016/0378-1119(84)90090-8. [DOI] [PubMed] [Google Scholar]
- Kirby R., Hopwood D. A. Genetic determination of methylenomycin synthesis by the SCP1 plasmid of Streptomyces coelicolor A3(2). J Gen Microbiol. 1977 Jan;98(1):239–252. doi: 10.1099/00221287-98-1-239. [DOI] [PubMed] [Google Scholar]
- Lakey J. H., Lea E. J., Rudd B. A., Wright H. M., Hopwood D. A. A new channel-forming antibiotic from Streptomyces coelicolor A3(2) which requires calcium for its activity. J Gen Microbiol. 1983 Dec;129(12):3565–3573. doi: 10.1099/00221287-129-12-3565. [DOI] [PubMed] [Google Scholar]
- Landick R., Vaughn V., Lau E. T., VanBogelen R. A., Erickson J. W., Neidhardt F. C. Nucleotide sequence of the heat shock regulatory gene of E. coli suggests its protein product may be a transcription factor. Cell. 1984 Aug;38(1):175–182. doi: 10.1016/0092-8674(84)90538-5. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Merrick M. J., Gibbins J. R. The nucleotide sequence of the nitrogen-regulation gene ntrA of Klebsiella pneumoniae and comparison with conserved features in bacterial RNA polymerase sigma factors. Nucleic Acids Res. 1985 Nov 11;13(21):7607–7620. doi: 10.1093/nar/13.21.7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Novotny J. Matrix program to analyze primary structure homology. Nucleic Acids Res. 1982 Jan 11;10(1):127–131. doi: 10.1093/nar/10.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosbash M., Blank D., Fahrner K., Hereford L., Ricciardi R., Roberts B., Ruby S., Woolford J. R-looping and structural gene indentification of recombinant DNA. Methods Enzymol. 1979;68:454–469. doi: 10.1016/0076-6879(79)68035-7. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Rudd B. A., Hopwood D. A. A pigmented mycelial antibiotic in Streptomyces coelicolor: control by a chromosomal gene cluster. J Gen Microbiol. 1980 Aug;119(2):333–340. doi: 10.1099/00221287-119-2-333. [DOI] [PubMed] [Google Scholar]
- Rudd B. A., Hopwood D. A. Genetics of actinorhodin biosynthesis by Streptomyces coelicolor A3(2). J Gen Microbiol. 1979 Sep;114(1):35–43. doi: 10.1099/00221287-114-1-35. [DOI] [PubMed] [Google Scholar]
- Sauer R. T., Yocum R. R., Doolittle R. F., Lewis M., Pabo C. O. Homology among DNA-binding proteins suggests use of a conserved super-secondary structure. Nature. 1982 Jul 29;298(5873):447–451. doi: 10.1038/298447a0. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stragier P., Bouvier J., Bonamy C., Szulmajster J. A developmental gene product of Bacillus subtilis homologous to the sigma factor of Escherichia coli. Nature. 1984 Nov 22;312(5992):376–378. doi: 10.1038/312376a0. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C. J., Gray G. S. Nucleotide sequence of a streptomycete aminoglycoside phosphotransferase gene and its relationship to phosphotransferases encoded by resistance plasmids. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5190–5194. doi: 10.1073/pnas.80.17.5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trach K. A., Chapman J. W., Piggot P. J., Hoch J. A. Deduced product of the stage 0 sporulation gene spo0F shares homology with the Spo0A, OmpR, and SfrA proteins. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7260–7264. doi: 10.1073/pnas.82.21.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright L. F., Hopwood D. A. Identification of the antibiotic determined by the SCP1 plasmid of Streptomyces coelicolor A3(2). J Gen Microbiol. 1976 Jul;95(1):96–106. doi: 10.1099/00221287-95-1-96. [DOI] [PubMed] [Google Scholar]
- Wurtzel E. T., Chou M. Y., Inouye M. Osmoregulation of gene expression. I. DNA sequence of the ompR gene of the ompB operon of Escherichia coli and characterization of its gene product. J Biol Chem. 1982 Nov 25;257(22):13685–13691. [PubMed] [Google Scholar]
- Yura T., Tobe T., Ito K., Osawa T. Heat shock regulatory gene (htpR) of Escherichia coli is required for growth at high temperature but is dispensable at low temperature. Proc Natl Acad Sci U S A. 1984 Nov;81(21):6803–6807. doi: 10.1073/pnas.81.21.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]