Abstract
Interaction between the iron transporter protein transferrin (Tf) and its receptor at the cell surface is fundamental for most living organisms. Tf receptor (TfR) binds iron-loaded Tf (holo-Tf) and transports it to endosomes, where acidic pH favors iron release. Iron-free Tf (apo-Tf) is then brought back to the cell surface and dissociates from TfR. Here we investigated the Tf-TfR interaction at the single-molecule level under different conditions encountered during the Tf cycle. An atomic force microscope tip functionalized with holo-Tf or apo-Tf was used to probe TfR. We tested both purified TfR anchored to a mica substrate and in situ TfR at the surface of living cells. Dynamic force measurements showed similar results for TfR on mica or at the cell surface but revealed striking differences between holo-Tf-TfR and apo-Tf-TfR interactions. First, the forces necessary to unbind holo-Tf and TfR are always stronger compared to the apo-Tf–TfR interaction. Second, dissociation of holo-Tf-TfR complex involves overcoming two energy barriers, whereas the apo-Tf-TfR unbinding pathway comprises only one energy barrier. These results agree with a model that proposes differences in the contact points between holo-Tf-TfR and apo-Tf-TfR interactions.
INTRODUCTION
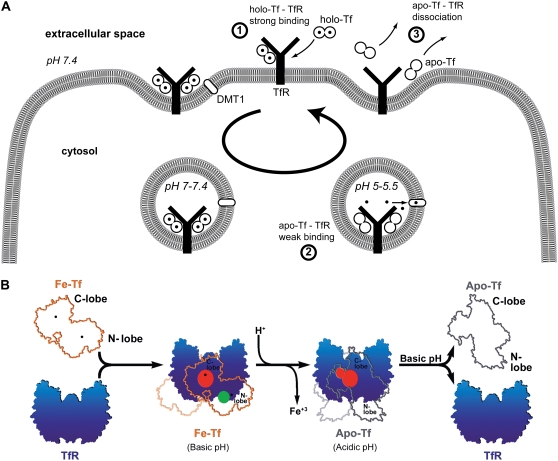
Iron is very important for life, intervening as a cofactor in various biological functions, such as respiration or DNA replication. However, free Fe3+ is both toxic for living cells and insoluble. Serum transferrin (Tf) plays a crucial role as iron transporter to safely supply cells through its interaction with the transferrin receptor (TfR) (1–4). After binding of iron-loaded Tf (holo-Tf) with TfR, holo-Tf-TfR complex is internalized to endosomes where an acidic pH triggers conformational changes and iron release. Iron is then transported in the cytosol through the divalent metal transporter 1. Iron free Tf (apo-Tf) remains bound to TfR in endosomes and dissociates when the complex is brought back at the slightly basic pH of the surface (5,6), which allows apo-Tf to fetch new iron ions for another cycle.
TfR is a 190-kDa homodimeric transmembrane receptor expressed at the surface of most mammalian cells. Each TfR monomer contains 671 amino acid residues and comprises a large extracellular ectodomain, a single-pass transmembrane domain, and a short intracellular domain (2,4). The extracellular portion is divided into three domains called apical, protease-like, and helical domains (7). Tf is a 79-kDa protein containing 679 amino acid residues (2,3) and organized in two homologous subunits known as the C-lobe and the N-lobe (1). Both lobes are able to bear one Fe3+ ion and Tf exists therefore as diferric (holo-Tf), monoferric, or iron-free type (apo-Tf). Diferric Tf binds TfR at the cell surface with an affinity 30 times higher compared to monoferric Tf (2). In acidic endosomes, iron release is facilitated by a conformational change of Tf lobes (8,9) and modulated by interlobe interactions and TfR presence (4,10).
Crystal structures of holo-Tf, apo-Tf, and TfR have been successfully obtained individually (7,8,11), but are lacking for the Tf-TfR complex. However, alternative techniques have yielded significant advances in understanding the mechanisms and structure of Tf-TfR interaction. Cheng et al. have used cryoelectron microscopy to show that holo-Tf binds TfR through its two lobes (12). The C-lobe interacts with the TfR helical domain, whereas the N-lobe contacts the TfR stalk region. These findings have been confirmed by mutational analysis (9) and by radiolytic footprinting coupled with mass spectroscopy (13). In contrast, Giannetti et al. have proposed, based on a mutational analysis, that apo-Tf binds TfR at acidic pH through its C-lobe only (9). However, this result does not fully agree with an electron-density map study that proposes the same interaction for holo-Tf–TfR as for apo-Tf–TfR (14).
To better understand the Tf-TfR complex and possible differences between holo-Tf-TfR and apo-Tf-TfR interactions, we used atomic force microscopy (AFM). In recent years, AFM has become a powerful tool, allowing measurement of interactions between single molecules under nearly physiological conditions (15–17). For these measurements, a specific ligand is cross-linked to a small tip, mounted at the end of a flexible cantilever. This tip is then used to probe either a purified receptor attached to an artificial surface or a native receptor expressed at the cell surface. The tip is first brought in contact with the surface to allow ligand-receptor recognition. Tip retraction then induces stretching of the molecules followed by forced dissociation of the ligand-receptor complex. This technique has already permitted us to quantify unbinding forces of numerous ligand-receptor pairs, either on an artificial surface (18–22) or at the surface of living cells (23–26).
The unbinding pathway of a ligand-receptor complex is frequently treated according to the Bell model (27), where the transition from bound to unbound state requires escaping from a potential well and overcoming one or several activation energy barriers (28,29). When an external force F is applied to the ligand-receptor complex, as it occurs during AFM experiments, energy barriers are lowered and the dissociation rate constant k(F) increases as follows:
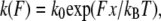 |
(1) |
where 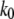 is the dissociation rate constant in the absence of external force, x is the width of the dominating energy barrier along the direction of the applied force,
is the dissociation rate constant in the absence of external force, x is the width of the dominating energy barrier along the direction of the applied force, 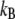 is Boltzmann's constant and T is temperature. In addition, if the external force applied increases at a constant rate
is Boltzmann's constant and T is temperature. In addition, if the external force applied increases at a constant rate 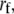 the most probable unbinding force
the most probable unbinding force 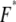 is given by (29)
is given by (29)
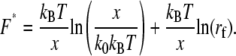 |
(2) |
Therefore, the most probable unbinding force, 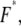 is not an absolute value for a given ligand-receptor pair but depends on the loading rate applied. Measuring the dependence of the unbinding force on the applied loading rate (dynamic force spectroscopy) gives access to parameters x and k0, which characterize energy barriers (16,29). The unbinding pathway of various receptor-ligand complexes has thus been successfully studied (30–34).
is not an absolute value for a given ligand-receptor pair but depends on the loading rate applied. Measuring the dependence of the unbinding force on the applied loading rate (dynamic force spectroscopy) gives access to parameters x and k0, which characterize energy barriers (16,29). The unbinding pathway of various receptor-ligand complexes has thus been successfully studied (30–34).
In this study, we investigated human Tf-TfR interaction at single-molecule level and explored its energy landscape. TfR interactions were probed with holo-Tf (diferric) and with apo-Tf. We tested both purified full-length TfR cross-linked onto a mica surface and native TfR endogenously expressed at the surface of living cells. For experiments on mica, we used full-length TfR rather than TfR ectodomain to reproduce a possible effect of the stalk on the interaction with Tf (12). Moreover, the two cysteine residues contained in TfR intramembraneous domain have appeared as useful anchoring points for cross-linkers.
MATERIALS AND METHODS
Proteins
Human plasma holo-Tf (iron content 1.56 μg/mg) and apo-Tf (iron content 0 μg/g) were purchased from USB (Cleveland, OH) with a >99% purity certified. From iron contents and Tf molecular mass (79 kDa), we assumed that holo-Tf molecules were bearing two iron ions, whereas apo-Tf molecules were iron-free. The proteins were diluted in 10 mM phosphate-buffered saline (PBS) to a final concentration of ∼0.25 mg/ml, estimated by absorbance at 280 nm (35). Full-length TfR purified from human placenta was purchased from Alpha Diagnostic (San Antonio, TX) and HyTest (Turku, Finland). TfR was diluted in PBS to a final concentration of ∼0.2 mg/ml, estimated by absorbance at 280 nm (36).
Tip and mica substrate preparation
Cantilever spring constants (OMCL-TR400PSA, Olympus, Tokyo, Japan; nominal value 0.02 N/m) were determined by thermal noise analysis (37), with uncertainty of ∼10%. Coupling of Tf and TfR to AFM Si3N4 tips and mica substrates (Pelco, Redding, CA) was done using a heterobifunctional polyethylene glycol (PEG) linker (38–41) in a three-step binding protocol, as schematized in Fig. 1. Tips and freshly cleaved mica substrates were cleaned under ultraviolet light and exposed for 2 h to APTES vapors in a 2-liter desiccator filled with argon and containing 30 μl of (3-aminopropyl)triethoxysilane and 10 μl of N,N-diisopropylethylamine (Sigma-Aldrich, Tokyo, Japan) (42,43). Tips and mica substrates were then kept for up to 3 days in an argon-filled atmosphere until use.
FIGURE 1.
The three-step functionalizing protocol of AFM tips and samples. (A) First, Si3N4 tips are aminosilanized by exposure to APTES vapors. Second, a heterobifunctional PEG linker is anchored to amino-group bearing tips through its NHS end. Third, Tf is attached to the PEG linker free end via a maleimide-cysteine bond. (B) The same method is used to cross-link purified full-length TfR to mica.
Amino-group bearing tips and mica were incubated for 60 min with 1 mg/ml of N-hydroxy-succinimide ester-PEG-maleimide (NHS-PEG-MAL, 3400 Da, Nektar Therapeutics, Huntsville, AL) in PBS. They were then washed several times with PBS to remove unanchored linker molecules. From the molecular weight, we estimated the PEG linker length to be ∼32 nm (41).
The final binding step was achieved by a reaction between the linker maleimide end and cysteine residues naturally present in Tf and TfR. A few minutes before deposition on mica substrate, 10 mM 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonic acid (CHAPS, Siyaku.Com, Osaka, Japan) was added to solubilize TfR (6,44). CHAPS was reported to form micelles that prevent TfR aggregation without altering Tf binding (6). Tips were incubated with Tf and mica substrates with TfR-CHAPS for 30 min. Finally, tips and samples were abundantly washed with buffer (at pH 7.4 for holo-Tf, at pH 5.3 for apo-Tf) to remove unbound molecules and CHAPS micelles from substrates.
AFM measurement on mica substrate
Force distance cycles were recorded with a MultiMode AFM Nanoscope III (Digital Instruments, Santa Barbara, CA) working in Force Volume mode. Force curves were captured at 256 different positions evenly spread over a surface of 1 μm2 on the sample. To limit the force exerted by the tip on the sample, a relative trigger of 10–15 nm was applied to the cantilever deflection. Holo-Tf-TfR experiments were done in PBS at pH 7.4 and apo-Tf-TfR experiments were done at pH 5.3 (PBS adjusted with HCl). For binding-competitor experiments, 0.1 μM TfR solubilized in 10 mM CHAPS (final concentrations) was added to the measuring buffer. Dynamic force spectroscopy experiments were performed by varying the retraction speed from 75 to 6000 nm/s.
Cell culture and AFM measurement
HeLa cells were cultivated at 37°C under 5% CO2 atmosphere in Dulbecco's minimum essential medium supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% heat-inactivated fetal bovine serum (Gibco, Auckland, New Zealand). One or two days before experiments, cells were plated in 60-mm-diameter dishes.
For AFM measurements, cells were washed three times with PBS at pH 7.4, and immediately placed on the stage of a Bioscope (Digital Instruments) mounted on an inverted optical microscope (Olympus). Force curves were recorded on different points at the cell surface using a relative trigger of 20–40 nm on the cantilever deflection. Dynamic force spectroscopy experiments were performed by varying the retraction speed from 120 to 7000 nm/s.
Data analysis
Force curves were analyzed off-line with a fuzzy-logic algorithm (45). Most probable unbinding forces were obtained from the force histograms of the unbinding events. Histograms were fitted with a Gaussian curve multiplied by a window function to account for the limited force sensitivity (46,47). The window function was designed to match the low force range and kept the same for all histograms. The most probable unbinding force was taken as the mean of the Gaussian component. A detailed description of the fitting method can be found in Supplementary Material. The loading rate was obtained from the slope on the force curve just before unbinding occurred (20,38). Uncertainties of the mean unbinding force and loading rate were calculated by adding the standard error of the mean of the distribution and the cantilever spring constant uncertainty. Mean unbinding lengths were obtained by fitting a Gaussian curve to unbinding-length histograms and uncertainty was given by the standard error of the mean.
RESULTS
Holo-Tf interaction with purified transferrin receptor
To investigate interactions between Tf and TfR at the single-molecule level, we used AFM working in force-volume mode. Holo-Tf molecules bearing two Fe3+ ions were cross-linked to the tip via a heterobifunctional PEG linker and full-length purified TfR was similarly cross-linked to a flat mica substrate, as schematized in Fig. 1. AFM force curves were then recorded in PBS at pH 7.4 (Fig. 2 A). Slightly basic media were often used to mimic the extracellular environment in which the holo-Tf-TfR interaction should occur (6,48). Although some retraction curves displayed no particular features (Fig. 2 A, upper curve), ∼25% presented a downward deflection abruptly ended by a force jump (Fig. 2 A, lower curves). Most likely, this typical pattern resulted from a binding-unbinding event between holo-Tf on the tip and TfR on the mica. Using a PEG linker ensured that holo-Tf-TfR unbinding occurred while the tip and sample were several tenths of nanometers apart. In contrast, events occurring immediately after tip-sample contact (Fig. 2 A, second curve from top) represented nonspecific tip-surface adhesion and were systematically discarded from our analysis.
FIGURE 2.
Specific interaction between holo-Tf and purified TfR at pH 7.4 in PBS. (A) Typical retraction force curves recorded with a holo-Tf coated tip on a TfR functionalized mica. Vertical axis indicates cantilever deflections as a function of the cantilever-sample retraction distance. The first curve from the top shows no particular features, and the second curve from the top shows an unspecific tip-surface adhesion event. The other curves display specific unbinding events between holo-Tf and TfR, which can clearly be differentiated form unspecific adhesion. The vertical force jump of events allows access to the holo-Tf-TfR unbinding force. The unbinding length of an event is measured by the distance separating the force jump from the tip-sample contact point (represented by the kink on the left of the curve). (B) Force histogram of 389 unbinding events obtained after analysis of 1500 force curves. The mean unbinding force is 56 ± 7 pN, for a mean loading rate of 4.6 nN/s. The gray line is a fitting curve. (C) Unbinding-length distribution for the events plotted in B, showing a mean unbinding length of 44 ± 2 nm. The gray line is a Gaussian fit.
About 1500 force curves were recorded over different spots on the substrate and several hundreds of specific events were collected, analyzed, and plotted in a force histogram (Fig. 2 B). A clear peak arose from this force distribution and yielded a mean unbinding force of 56 ± 7 pN (measurement performed at a mean loading rate of 4.6 nN/s). The distribution of unbinding length (tip-sample distance at unbinding time) for these events showed a clear peak centered at 44 ± 2 nm (Fig. 2 C). Additional measurements performed under similar conditions on different days with new tips and samples essentially provided the same force distributions and values as shown in Fig. 2, B and C (data not shown here), which demonstrated the good reproducibility of our results.
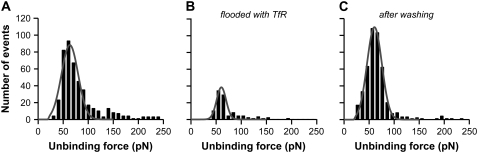
To verify the specificity of the unbinding events measured here, we performed control experiments in which binding was inhibited. First, 1500 force curves were recorded in PBS at pH 7.4, as explained above, yielding an unbinding probability of 31.6% and a mean unbinding force of 63 ± 8 pN (Fig. 3 A; measurements performed at a mean loading rate of 5.5 nN/s). Free TfR (0.1 μM) was then added to the measuring buffer and 1500 new force curves were recorded with the same functionalized tip and mica (Fig. 3 B). The number of events clearly decreased and the unbinding probability fell to 6.8%. Most likely, this diminution resulted from the competition taking place between free TfR in solution and TfR immobilized on mica for the binding to holo-Tf. However, the mean unbinding force obtained from the reduced histogram remained similar to the initial measurement (61 ± 8 pN). This finding strongly suggests that the events persisting despite inhibition resulted from recognition between holo-Tf on the tip and TfR on the mica. Finally, both tip and mica were washed with fresh PBS and 1500 force curves were recorded again in PBS (Fig. 3 C). The unbinding probability increased to 28.6%, showing a recovery close to the initial level (31.6%), and the mean unbinding force remained unchanged (61 ± 8 pN). The same experiment was repeated on different days with new tips and samples, resulting in the same inhibition and recovery pattern.
FIGURE 3.
Holo-Tf-TfR binding competitor experiments. (A) Force histogram of holo-Tf interaction with purified TfR at pH 7.4 in PBS. (B) As in A, after tip blocking with free TfR molecules (0.1 μM). (C) As in B, after washing with fresh PBS. Unbinding probabilities are 31.6%, 6.8%, and 28.6%, and unbinding forces 63 ± 8 pN, 61 ± 8 pN and 61 ± 8 pN for A–C, respectively. The mean loading rate applied has been 5.5 nN/s. Gray lines are fitting curves.
Apo-Tf interaction with purified transferrin receptor
After binding of holo-Tf with TfR, the complex is internalized to endosomes where acidic pH favors conformational changes and iron release. The iron-free apo-Tf remains bound to TfR in endosomes and the complex is recycled at the cell surface where it dissociates (2,4). To further investigate Tf-TfR interactions taking place during the Tf cycle, we performed AFM measurements between apo-Tf (anchored to the tip) and purified TfR (attached to the mica) at endosomal pH of 5.3. First, 1500 force curves were recorded (Supplementary Material, Fig. 1 A), showing specific unbinding events with a probability of 15.3%. Off-line analysis resulted in a force histogram presenting a clear peak (Fig. 4 A), characterized by a mean unbinding force of 44 ± 5 pN (for a mean loading rate of 3.5 nN/s) and a mean unbinding length of 38 ± 2 nm (Supplementary Material, Fig. 1 B).
FIGURE 4.
pH dependence of the apo-Tf-TfR interaction. (A) Force histogram of apo-Tf interaction with purified TfR at pH 5.3, obtained from 235 unbinding events. The mean unbinding force is 44 ± 5 pN and the mean unbinding probability is 15.3%. (B) As in A, after placing the whole system at pH 7.4. The mean unbinding probability has decreased to 3.1% and the number of unbinding events to 48. (C) As in B, after restoring the system to pH 5.3. The mean unbinding force is 41 ± 6 pN, the mean unbinding probability 15.5% and the number of unbinding events 238. The mean loading rate applied has been 3.3 nN/s. Gray lines are fitting curves.
To mimic the conditions encountered by the Tf-TfR complex as it is recycled from endosomes back to the cell surface, we then exchanged the acidic buffer with a buffer at pH 7.4. Keeping the same functionalized tip and sample, another set of 1500 force curves was recorded and analyzed. The number of events detected at pH 7.4 dramatically decreased compared with acidic pH, with a mean unbinding probability falling to 3.1%. Moreover, no force peak was distinguishable on the corresponding histogram (Fig. 4 B). These results strongly suggest that no specific interaction took place between apo-Tf and TfR at pH 7.4, in agreement with a previous study (6). Finally, the neutral buffer was removed and replaced by an acidic buffer to restore initial conditions. Again, we recorded 1500 force curves with the same tip and mica (Fig. 4 C). The number of events recorded clearly increased and the unbinding probability reached 15.5%, which was almost identical to the initial level (15.3%). The mean unbinding force (41 ± 6 pN) and unbinding length (37 ± 2 nm) were close to the initial values measured (44 ± 5 pN and 38 ± 2 nm, respectively), suggesting a recovery of the apo-Tf-TfR interaction. In addition, this apparent recovery demonstrated that the absence of specific interaction between apo-Tf and TfR at pH 7.4 was a relevant fact and stemmed neither from the pulling of apo-Tf off the tip nor from the sample degradation. However, a clear broadening of the force distribution was observed when pH was brought back to 5.3 compared to the initial situation (compare the width of the force histograms on Fig. 4, A and C). This finding suggested that the exposure to pH 7.4, followed by the recovery of acidic pH, induced a higher variability of the apo-Tf-TfR interaction at the single-molecule level.
Dynamic force spectroscopy of transferrin-transferrin receptor complexes
To investigate unbinding kinetics of the Tf-TfR complex, we measured the mean unbinding force between Tf and purified TfR at various loading rates ranging from 0.5 to 70 nN/s. At each loading rate tested, a minimum of 1000 force curves was recorded and analyzed. The holo-Tf-TfR interaction was measured at pH 7.4 (Fig. 5 A) and apo-Tf-TfR at pH 5.3 (Fig. 5 B). In both cases, a shift toward high unbinding forces was clearly observed with increasing loading rates, although it was more pronounced for holo-Tf-TfR than for apo-Tf-TfR experiments. Dynamic force spectroscopy of the Tf-TfR interaction was obtained by plotting the mean unbinding force as a function of the logarithm of the loading rate (Fig. 5 C). Two striking differences appeared between the force spectra of holo-Tf-TfR (Fig. 5 C, solid circles) and apo-Tf-TfR (Fig. 5 C, open circles). First, two different loading-rate regimes were clearly distinguishable on the holo-Tf-TfR force spectrum, whereas only one regime was detected for the apo-Tf-TfR complex. Second, at the same loading rate, apo-Tf-TfR unbinding forces were always weaker than holo-Tf-TfR unbinding forces.
FIGURE 5.
Loading-rate dependence of the Tf-TfR unbinding force. (A) Force histograms of holo-Tf-TfR interaction measured at pH 7.4 under different loading rates. The mean unbinding force (F), the mean loading rate applied (rf), and the number of unbinding events analyzed (N) are given for each histogram. An obvious shift toward higher unbinding forces is visible with increasing loading rates (bottom to top). (B) Force histograms of apo-Tf-TfR interaction measured at pH 5.3. Gray lines are fitting curves. (C) Dynamic force spectroscopy of the holo-Tf-TfR interaction at pH 7.4 (solid circles) and of the apo-Tf-TfR interaction at pH 5.3 (open circles). The unbinding force is plotted as a function of the loading-rate logarithm. Two regimes are visible for holo-Tf-TfR interaction but only one appears for apo-Tf-TfR interaction. Solid lines represent fitting curves derived from Eq. 2. Error bars were calculated as explained in Materials and Methods.
As predicted by Eq. 2, within each regime the mean unbinding force was found to depend logarithmically on the loading rate. In the low regime starting at 0.5 nN/s, holo-Tf-TfR unbinding forces increased with loading rate up to 4.5 nN/s, where an abrupt change of slope marked the transition between the two regimes. Beyond 4.5 nN/s, the force increase was clearly steeper. In contrast apo-Tf-TfR unbinding forces increased at a constant rate along the whole range of loading rate tested. According to Bell-Evans model (27–29), our measurements were consistent with unbinding energy pathways that involved two energy barriers for the holo-Tf-TfR complex and only one barrier for the apo-Tf–TfR complex (see Discussion). Fitting Eq. 2 with the loading-rate regimes displayed in Fig. 5 C provided the Bell model parameters for these barriers (Table 1).
TABLE 1.
Bell model parameters of the Tf-TfR complex
| On tip | Substrate | pH | Loading rate (nN/s) | k0 (s−1) | x (Å) |
|---|---|---|---|---|---|
| holo-Tf | TfR on mica | 7.4 | 0.5–4.5 | 0.0050 ± 0.0033 | 9.3 ± 1.5 |
| holo-Tf | TfR on mica | 7.4 | 4.5–70 | 22.9 ± 0.4 | 1.5 ± 0.1 |
| apo-Tf | TfR on mica | 5.3 | 0.5–40 | 0.25 ± 0.08 | 8.1 ± 1.0 |
| holo-Tf | TfR on cell | 7.4 | 0.5–4.5 | 0.0034 ± 0.0071 | 10.6 ± 6.5 |
| holo-Tf | TfR on cell | 7.4 | 4.5–40 | 19.1 ± 1.4 | 1.9 ± 0.3 |
Transferrin interaction with native transferrin receptor at cell surface
To compare the interaction measured between Tf and purified TfR with the interaction occurring at the cell surface between Tf and native TfR, we replaced the functionalized mica substrate with cultivated HeLa cells endogenously expressing TfR. The cells were placed in PBS at pH 7.4 and probed with a holo-Tf functionalized tip (Fig. 6 A). About 1000 force curves were recorded and 21.2% of them presented unbinding events (Fig. 6 B). These events were analyzed off-line and plotted in a force histogram (Fig. 6 C), yielding a mean unbinding force of 56 ± 7 pN for a mean loading rate of 2.5 nN/s. In contrast, when the holo-Tf functionalized tip was replaced with an apo-Tf functionalized tip, the unbinding probability was only 2.2%. The resulting force histogram did not show any interaction peak (Fig. 6 D), suggesting that no specific interaction took place between apo-Tf and TfR at the cell surface at pH 7.4.
FIGURE 6.
Measurements of Tf-TfR interaction at the surface of living HeLa cells. (A) A tip functionalized with holo-Tf (at the end of the cantilever, not visible since the picture is taken from above) is used to probe the cell surface. Scale bar, 20 μm. (B) Retraction force curves recorded at the cell surface in PBS at pH 7.4. From the top, the first curve is a reference curve showing no unbinding events. The other curves display single unbinding events between holo-Tf and native TfR. (C) Force histogram of 242 unbinding events obtained after analysis of 1000 force curves recorded with a holo-Tf functionalized tip at the surface of a living cell. The mean unbinding force is 56 ± 7 pN, for a mean loading rate of 2.5 nN/s. The gray line is a fitting curve. (D) As in C, but with an apo-Tf functionalized tip. The number of unbinding events is 22. (E) Dynamic force spectroscopy of holo-Tf interaction with native TfR at the surface of living cells (open squares). The unbinding force is plotted as a function of the loading-rate logarithm. Two regimes are visible; solid lines represent fitting curves derived from Eq. 2. Dynamic force spectroscopy of purified TfR interaction with holo-Tf at pH 7.4 (solid circles) and with apo-Tf at pH 5.3 (open circles) is shown for comparison. Error bars were calculated as described in Materials and Methods.
Finally, we investigated the loading-rate dependence of the holo-Tf-TfR unbinding force at the cell surface. A minimum of 1000 force curves was recorded for each loading rate tested. The mean unbinding force obtained from the force histogram was plotted against the mean loading rate applied (Fig. 6 E, open squares). The holo-Tf-TfR interaction in living cells presented two loading-rate regimes, similar to the holo-Tf-TfR interaction on mica (Fig. 6 E, solid circles (shown for comparison)). The Bell model parameters characterizing these two regimes were obtained by fitting Eq. 2 to Fig. 6 E (Table 1).
DISCUSSION
In this study, we investigated interactions between Tf and TfR at the single-molecule level and performed dynamic force spectroscopy measurements. First, we tested the interaction of holo-Tf with purified TfR deposited on a mica substrate and showed the reproducibility of our measurements (Fig. 2 A). In addition, experiments involving binding competitors demonstrated the specificity of these measurements (Fig. 3). Indeed, the dramatic but reversible decrease of unbinding events measured in the presence of free TfR molecules indicated the blocking of recognition between holo-Tf on the tip and TfR anchored on mica. Inhibition was not absolute, however, and several unbinding events persisted under blocking conditions. Previous experiments involving binding competitors often showed similar pertaining events (22,31,41,49). Persistence of these events despite blocking might have stemmed from a forced binding due to the contact between tip and sample.
Unbinding-length analysis of the holo-Tf-TfR interaction on mica showed that no events were detected beyond an extension of ∼75 nm (Fig. 2 C). This distance is consistent with our estimated maximal unbinding length of 73 nm (2 × 32 nm for the PEG linkers and 9 nm estimated for Tf-TfR complex from Cheng et al. (12)). The most probable unbinding length was between 40 and 50 nm, which suggested either that unbinding occurred while the PEG linker was not fully stretched or that the linker was anchored on the tip side rather than at the apex. Similar findings have been reported previously in experiments using the same type of cross-linker (41,50).
In another set of experiments, we showed that the specific interaction taking place between apo-Tf and purified TfR at pH 5.3 was abolished at pH 7.4, but could be recovered when pH was brought back to acidic (Fig. 4). Our results demonstrate, therefore, that variations of Tf-TfR interaction occur along the Tf cycle, as explained in Fig. 7. First holo-Tf binds TfR at the cell surface at pH 7.4 (unbinding force of 54 pN for a loading rate of 3.3 nN/s). Second, the complex is internalized to acidic endosomes, where conformational changes trigger iron release. Apo-Tf remains loosely bound to TfR (unbinding force of 44 pN for a loading rate of 3.5 nN/s). Third, apo-Tf-TfR complex is brought back at the cell surface, where it encounters slightly basic pH and dissociates (no specific interaction was measured between apo-Tf and TfR at pH 7.4). These findings agree with a study showing an overall affinity of TfR 15 times stronger for holo-Tf at neutral pH than for apo-Tf at acidic pH (6) and support the hypothesis of pH-dependent conformational changes affecting Tf, TfR, or both (6,9).
FIGURE 7.
Models of Tf-TfR interactions along the Tf cycle. (A) Variations of unbinding forces along the cycle. First, holo-Tf strongly binds TfR at the cell surface (unbinding force of 54 pN at 3.3 nN/s). Second, the complex is internalized and transported to acidic endosomes. Conformational changes trigger iron release (solid circles) from holo-Tf, which becomes apo-Tf and remains weakly bound to TfR (unbinding force of 43 pN at 3.5 nN/s). Third, the complex is brought back at the cell surface and apo-Tf dissociates from TfR. (B) The model adapted from Giannetti et al. for the binding of holo-Tf and apo-Tf to TfR (9). At the cell surface pH, holo-Tf (here named Fe-Tf, orange outline with iron ions marked as black dots) binds TfR (blue) through its C-lobe (red binding site) and through its N-lobe (green binding site). At endosomal pH, iron is released and conformational changes occur. Apo-Tf (gray outline) binds TfR through its C-lobe only (red binding site). Upon return to the cell surface, apo-Tf dissociates from TfR. The two binding sites of holo-Tf might explain the two barriers of the holo-Tf-TfR energy landscape (see Fig. 8), whereas the absence of binding between the apo-Tf N-lobe and TfR might explain the absence of an inner barrier in the apo-Tf-TfR energy landscape.
We then exchanged the purified TfR on mica for native TfR expressed at the surface of living HeLa cells. We found that the forces needed to unbind holo-Tf from TfR at the cell surface or on mica were very close (56 ± 7 pN and 54 ± 7 pN, respectively, for a loading rate of ∼3 nN/s). In addition, dynamic force spectroscopy measurements of holo-Tf-TfR interaction showed two similar loading-rate regimes at the cell surface and on mica (Fig. 6 E). Finally, no specific interaction was detected between apo-Tf and native TfR at the cell surface (Fig. 6 D), which agreed with the result obtained with purified TfR on mica (Fig. 4 B). Therefore, these findings indicated that the interaction studied on mica with purified TfR was very similar to the interaction taking place at the cell surface. Furthermore, they suggested that the forces binding Tf and TfR do not depend on the molecular environment surrounding TfR in vivo. This conclusion is probably not true for all receptors, however. Indeed, a recent study showed that a cytoskeleton anchorage might modify the shape of unbinding events recorded between a receptor and its ligand (51). Since native TfR is not linked to cytoskeleton (52), our conclusion does not disagree with this study.
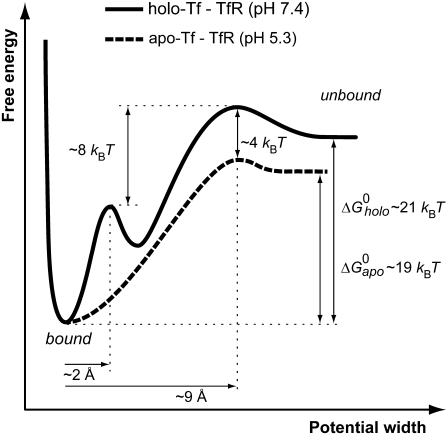
In terms of the Bell-Evans model (27,28), our dynamic force spectroscopy measurements (Figs. 5 C and 6 E, and Table 1) indicate that the dissociation of holo-Tf-TfR complex involves overcoming two energy barriers, whereas apo-Tf-TfR complex dissociation involves overcoming only one, as schematized in Fig. 8. The transition states of holo-Tf-TfR barriers are situated, respectively, at ∼1.5–1.9 Å and ∼9.3–10.6 Å from equilibrium, whereas the transition state of the apo-Tf-TfR single barrier is situated at ∼8.1 Å from equilibrium (Table 1). The positions of apo-Tf-TfR single barrier and holo-Tf-TfR outer barrier are close (Fig. 5 C; note that in the low range of loading rates, the slopes are almost parallel), which suggests a correspondence between them. The energy difference between these two barriers was estimated by 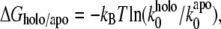 where
where 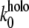 and
and 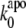 are the Bell model parameters
are the Bell model parameters 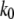 for the holo-Tf-TfR outer barrier and the apo-Tf-TfR single barrier, respectively. This analysis showed that holo-Tf-TfR outer barrier is ∼4 kBT higher than apo-Tf-TfR barrier (Fig. 8) and revealed that apo-Tf-TfR dissociation requires less energy than holo-Tf-TfR dissociation. The absence of an inner barrier for apo-Tf-TfR complex made it significantly easier to dissociate at high loading rates compared with holo-Tf-TfR complex (51 ± 9 pN at 27.4 nN/s compared with 106 ± 14 pN at 29.2 nN/s).
for the holo-Tf-TfR outer barrier and the apo-Tf-TfR single barrier, respectively. This analysis showed that holo-Tf-TfR outer barrier is ∼4 kBT higher than apo-Tf-TfR barrier (Fig. 8) and revealed that apo-Tf-TfR dissociation requires less energy than holo-Tf-TfR dissociation. The absence of an inner barrier for apo-Tf-TfR complex made it significantly easier to dissociate at high loading rates compared with holo-Tf-TfR complex (51 ± 9 pN at 27.4 nN/s compared with 106 ± 14 pN at 29.2 nN/s).
FIGURE 8.
Energy landscape of Tf-TfR complex. The dissociation of holo-Tf-TfR complex at pH 7.4 involves overcoming two activation barriers (solid line), whereas the unbinding pathway of apo-Tf-TfR complex at pH 5.3 has only one barrier (dashed line). Positions of the barriers and energy differences between them were obtained from the Bell model parameters given in Table 1. Estimations of 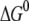 were calculated from the dissociation constants (see text for details).
were calculated from the dissociation constants (see text for details).
The energy difference 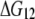 between the two transition states of holo-Tf–TfR activation barriers (Fig. 8) was estimated by
between the two transition states of holo-Tf–TfR activation barriers (Fig. 8) was estimated by 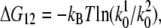 where
where 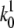 and
and 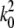 are the Bell model parameters
are the Bell model parameters 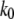 for the inner and outer barrier respectively (Table 1). This calculation showed that the outer activation barrier is ∼8
for the inner and outer barrier respectively (Table 1). This calculation showed that the outer activation barrier is ∼8 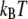 higher than the inner barrier for unforced dissociation. The energy difference
higher than the inner barrier for unforced dissociation. The energy difference 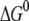 between bound and unbound state (Fig. 8) was calculated as
between bound and unbound state (Fig. 8) was calculated as 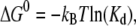 where
where 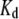 is the dissociation constant. Applying the values reported by Giannetti et al. (9) yielded
is the dissociation constant. Applying the values reported by Giannetti et al. (9) yielded 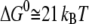 for holo-Tf-TfR complex and
for holo-Tf-TfR complex and 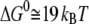 for apo-Tf-TfR complex.
for apo-Tf-TfR complex.
Applying an external force to Tf–TfR complex distorted its energy landscape and lowered the activation barriers (28,29). In the case of holo-Tf-TfR complex, the outer barrier was dominating unbinding kinetics at low loading rates (<4.5 nN/s), but at loading rates >4.5 nN/s, the outer activation barrier was lowered under the inner barrier, which became dominant for dissociation. Assuming that the outer barrier is also prevalent for unforced dissociation, the corresponding Bell parameter, 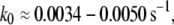 should correspond to the natural dissociation rate k0. A value of
should correspond to the natural dissociation rate k0. A value of 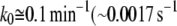 ) has previously been reported (53,54), which is in the range of our result, although smaller. The difference may come from the nonlinear loading induced by the PEG cross-linker. Indeed, in a recent study, Ray et al. claimed that the Bell-Evans model yields underestimation of the barrier width and overestimation of the dissociation rate, when the elasticity of the polymeric tether is ignored (46). We applied to our data the algorithm proposed by Ray et al. to correct these systematic errors. Although the barrier width seemed to show a very small error (∼2%), our k0 value might, in contrast, have been overestimated by 20–25%, which may then partially explain the difference from the published value.
) has previously been reported (53,54), which is in the range of our result, although smaller. The difference may come from the nonlinear loading induced by the PEG cross-linker. Indeed, in a recent study, Ray et al. claimed that the Bell-Evans model yields underestimation of the barrier width and overestimation of the dissociation rate, when the elasticity of the polymeric tether is ignored (46). We applied to our data the algorithm proposed by Ray et al. to correct these systematic errors. Although the barrier width seemed to show a very small error (∼2%), our k0 value might, in contrast, have been overestimated by 20–25%, which may then partially explain the difference from the published value.
Our measurements revealed striking differences between holo-Tf-TfR and apo-Tf-TfR dynamic force spectra (Figs. 5 C and 6 E), which reflected clearly distinct energy landscapes (Fig. 8). Two different models have previously been proposed for Tf-TfR interaction, in which the contact points between TfR and holo-Tf or apo-Tf are different. On one side, Giannetti et al. have proposed from a mutational analysis that holo-Tf binds TfR through its two lobes, whereas apo-Tf binds TfR through its C-lobe only (9), as schematized in Fig. 7 B. On the other side, Cheng et al. have concluded, from electron density mapping, that holo-Tf and apo-Tf bind TfR similarly via their two lobes (12,14). Therefore, the clear differences observed here between holo-Tf-TfR and apo-Tf-TfR interactions seem in better agreement with the model proposing different binding points for holo-Tf-TfR and apo-Tf-TfR interactions (Fig. 7 B). In addition, this model might provide a structural interpretation for the energy barriers postulated here. Indeed, the two barriers of holo-Tf-TfR interaction might stem from the two binding sites of holo-Tf (C-lobe and N-lobe), whereas the single barrier of apo-Tf-TfR interaction might originate from the single binding site of apo-Tf (C-lobe). The common binding site of holo-Tf and apo-Tf (Fig. 7 B, red) would then explain the similarity between the holo-Tf-TfR outer barrier and the apo-Tf-TfR single barrier. Finally, the absence of an inner barrier for apo-Tf-TfR complex would reflect the absence of interaction between apo-Tf N-lobe and TfR. These hypotheses would imply that the two lobes of holo-Tf unbind from TfR at different rates, which seems possible given the different responsiveness of the lobes to complex formation with TfR. Zak and Aisen showed that the C-lobe by itself is able to bind TfR, whereas the N-lobe is not (55). In addition, they showed that 76% of the binding energy of the Tf-TfR complex is due to the C-lobe. However, we cannot exclude different origins for these energy barriers, especially since the crystal structure of Tf-TfR complex is not known yet.
In conclusion, we have shown that the interaction between Tf and TfR strongly depends on pH and on iron load of Tf. Measurements performed with purified TfR anchored on mica and with native TfR at the cell surface are in good agreement. Dissociation of holo-Tf-TfR complex at pH 7.4 involves overcoming two energy barriers, whereas only one barrier characterizes apo-Tf-TfR dissociation at pH 5.3. These results seem to agree with the model proposing a binding of holo-Tf to TfR via both of its lobes and a binding of apo-Tf to TfR via its C-lobe only (9).
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Acknowledgments
We thank Rehana Afrin and Hiroshi Sekiguchi for critical reading of the manuscript.
A.Y. acknowledges the Japan Society for the Promotion of Science (JSPS) for supporting him through a postdoctoral fellowship. This work was financially supported by a JSPS Grant-in-Aid for Creative Scientific Research to A.I. (No. 19GS0418).
Editor: Peter Hinterdorfer.
References
- 1.Baker, H. M., B. F. Anderson, and E. N. Baker. 2003. Dealing with iron: common structural principles in proteins that transport iron and heme. Proc. Natl. Acad. Sci. USA. 100:3579–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daniels, T. R., T. Delgado, J. A. Rodriguez, G. Helguera, and M. L. Penichet. 2006. The transferrin receptor part I: biology and targeting with cytotoxic antibodies for the treatment of cancer. Clin. Immunol. 121:144–158. [DOI] [PubMed] [Google Scholar]
- 3.Gomme, P. T., K. B. McCann, and J. Bertolini. 2005. Transferrin: structure, function and potential therapeutic actions. Drug Discov. Today. 10:267–273. [DOI] [PubMed] [Google Scholar]
- 4.Aisen, P. 2004. Transferrin receptor 1. Int. J. Biochem. Cell Biol. 36:2137–2143. [DOI] [PubMed] [Google Scholar]
- 5.Lamb, J. E., F. Ray, J. H. Ward, J. P. Kushner, and J. Kaplan. 1983. Internalization and subcellular localization of transferrin and transferrin receptors in HeLa cells. J. Biol. Chem. 258:8751–8758. [PubMed] [Google Scholar]
- 6.Hemadi, M., P. H. Kahn, G. Miquel, and J. M. El Hage Chahine. 2004. Transferrin's mechanism of interaction with receptor 1. Biochemistry. 43:1736–1745. [DOI] [PubMed] [Google Scholar]
- 7.Lawrence, C. M., S. Ray, M. Babyonyshev, R. Galluser, D. W. Borhani, and S. C. Harrison. 1999. Crystal structure of the ectodomain of human transferrin receptor. Science. 286:779–782. [DOI] [PubMed] [Google Scholar]
- 8.MacGillivray, R. T., S. A. Moore, J. Chen, B. F. Anderson, H. Baker, Y. Luo, M. Bewley, C. A. Smith, M. E. Murphy, Y. Wang, A. B. Mason, R. C. Woodworth, G. D. Brayer, and E. N. Baker. 1998. Two high-resolution crystal structures of the recombinant N-lobe of human transferrin reveal a structural change implicated in iron release. Biochemistry. 37:7919–7928. [DOI] [PubMed] [Google Scholar]
- 9.Giannetti, A. M., P. M. Snow, O. Zak, and P. J. Bjorkman. 2003. Mechanism for multiple ligand recognition by the human transferrin receptor. PLoS Biol. 1:E51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zak, O., and P. Aisen. 2003. Iron release from transferrin, its C-lobe, and their complexes with transferrin receptor: presence of N-lobe accelerates release from C-lobe at endosomal pH. Biochemistry. 42:12330–12334. [DOI] [PubMed] [Google Scholar]
- 11.Wally, J., P. J. Halbrooks, C. Vonrhein, M. A. Rould, S. J. Everse, A. B. Mason, and S. K. Buchanan. 2006. The crystal structure of iron-free human serum transferrin provides insight into inter-lobe communication and receptor binding. J. Biol. Chem. 281:24934–24944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng, Y., O. Zak, P. Aisen, S. C. Harrison, and T. Walz. 2004. Structure of the human transferrin receptor-transferrin complex. Cell. 116:565–576. [DOI] [PubMed] [Google Scholar]
- 13.Xu, G., R. Liu, O. Zak, P. Aisen, and M. R. Chance. 2005. Structural allostery and binding of the transferrin-receptor complex. Mol. Cell. Proteomics. 4:1959–1967. [DOI] [PubMed] [Google Scholar]
- 14.Cheng, Y., E. Wolf, M. Larvie, O. Zak, P. Aisen, N. Grigorieff, S. C. Harrison, and T. Walz. 2006. Single particle reconstructions of the transferrin-transferrin receptor complex obtained with different specimen preparation techniques. J. Mol. Biol. 355:1048–1065. [DOI] [PubMed] [Google Scholar]
- 15.Hinterdorfer, P., and Y. F. Dufrene. 2006. Detection and localization of single molecular recognition events using atomic force microscopy. Nat. Methods. 3:347–355. [DOI] [PubMed] [Google Scholar]
- 16.Lee, C. K., Y. M. Wang, L. S. Huang, and S. Lin. 2006. Atomic force microscopy: determination of unbinding force, off rate and energy barrier for protein-ligand interaction. Micron. 38:446–461. [DOI] [PubMed] [Google Scholar]
- 17.Allison, D. P., P. Hinterdorfer, and W. Han. 2002. Biomolecular force measurements and the atomic force microscope. Curr. Opin. Biotechnol. 13:47–51. [DOI] [PubMed] [Google Scholar]
- 18.Yersin, A., H. Hirling, P. Steiner, S. Magnin, R. Regazzi, B. Huni, P. Huguenot, P. De los Rios, G. Dietler, S. Catsicas, and S. Kasas. 2003. Interactions between synaptic vesicle fusion proteins explored by atomic force microscopy. Proc. Natl. Acad. Sci. USA. 100:8736–8741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwesinger, F., R. Ros, T. Strunz, D. Anselmetti, H. J. Guntherodt, A. Honegger, L. Jermutus, L. Tiefenauer, and A. Pluckthun. 2000. Unbinding forces of single antibody-antigen complexes correlate with their thermal dissociation rates. Proc. Natl. Acad. Sci. USA. 97:9972–9977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dettmann, W., M. Grandbois, S. Andre, M. Benoit, A. K. Wehle, H. Kaltner, H. J. Gabius, and H. E. Gaub. 2000. Differences in zero-force and force-driven kinetics of ligand dissociation from β-galactoside-specific proteins (plant and animal lectins, immunoglobulin G) monitored by plasmon resonance and dynamic single molecule force microscopy. Arch. Biochem. Biophys. 383:157–170. [DOI] [PubMed] [Google Scholar]
- 21.Baumgartner, W., P. Hinterdorfer, W. Ness, A. Raab, D. Vestweber, H. Schindler, and D. Drenckhahn. 2000. Cadherin interaction probed by atomic force microscopy. Proc. Natl. Acad. Sci. USA. 97:4005–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartels, F. W., B. Baumgarth, D. Anselmetti, R. Ros, and A. Becker. 2003. Specific binding of the regulatory protein ExpG to promoter regions of the galactoglucan biosynthesis gene cluster of Sinorhizobium meliloti–a combined molecular biology and force spectroscopy investigation. J. Struct. Biol. 143:145–152. [DOI] [PubMed] [Google Scholar]
- 23.Bustanji, Y., C. R. Arciola, M. Conti, E. Mandello, L. Montanaro, and B. Samori. 2003. Dynamics of the interaction between a fibronectin molecule and a living bacterium under mechanical force. Proc. Natl. Acad. Sci. USA. 100:13292–13297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, S. S., and G. Banting. 2002. Characterisation of the lumenal domain of TGN38 and effects of elevated expression of TGN38 on glycoprotein secretion. Eur. J. Cell Biol. 81:609–621. [DOI] [PubMed] [Google Scholar]
- 25.Pfister, G., C. M. Stroh, H. Perschinka, M. Kind, M. Knoflach, P. Hinterdorfer, and G. Wick. 2005. Detection of HSP60 on the membrane surface of stressed human endothelial cells by atomic force and confocal microscopy. J. Cell Sci. 118:1587–1594. [DOI] [PubMed] [Google Scholar]
- 26.Yersin, A., H. Hirling, S. Kasas, C. Roduit, K. Kulangara, G. Dietler, F. Lafont, S. Catsicas, and P. Steiner. 2007. Elastic properties of the cell surface and trafficking of single AMPA receptors in living hippocampal neurons. Biophys. J. 92:4482–4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bell, G. I. 1978. Models for the specific adhesion of cells to cells. Science. 200:618–627. [DOI] [PubMed] [Google Scholar]
- 28.Merkel, R., P. Nassoy, A. Leung, K. Ritchie, and E. Evans. 1999. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature. 397:50–53. [DOI] [PubMed] [Google Scholar]
- 29.Evans, E., and K. Ritchie. 1997. Dynamic strength of molecular adhesion bonds. Biophys. J. 72:1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, F., S. D. Redick, H. P. Erickson, and V. T. Moy. 2003. Force measurements of the α5β1 integrin-fibronectin interaction. Biophys. J. 84:1252–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonanni, B., A. S. Kamruzzahan, A. R. Bizzarri, C. Rankl, H. J. Gruber, P. Hinterdorfer, and S. Cannistraro. 2005. Single molecule recognition between cytochrome C 551 and gold-immobilized azurin by force spectroscopy. Biophys. J. 89:2783–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kokkoli, E., S. E. Ochsenhirt, and M. Tirrell. 2004. Collective and single-molecule interactions of α(5)β(1) integrins. Langmuir. 20:2397–2404. [DOI] [PubMed] [Google Scholar]
- 33.Nevo, R., C. Stroh, F. Kienberger, D. Kaftan, V. Brumfeld, M. Elbaum, Z. Reich, and P. Hinterdorfer. 2003. A molecular switch between alternative conformational states in the complex of Ran and importin β1. Nat. Struct. Biol. 10:553–557. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, X., E. Wojcikiewicz, and V. T. Moy. 2002. Force spectroscopy of the leukocyte function-associated antigen-1/intercellular adhesion molecule-1 interaction. Biophys. J. 83:2270–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ross, D. C., T. J. Egan, and L. R. Purves. 1995. Periodate modification of human serum transferrin Fe(III)-binding sites. Inhibition of carbonate insertion into Fe(III)- and Cu(II)-chelator-transferrin ternary complexes. J. Biol. Chem. 270:12404–12410. [DOI] [PubMed] [Google Scholar]
- 36.Bali, P. K., O. Zak, and P. Aisen. 1991. A new role for the transferrin receptor in the release of iron from transferrin. Biochemistry. 30:324–328. [DOI] [PubMed] [Google Scholar]
- 37.Hutter, J. L., and J. Bechhoefer. 1993. Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 64:1868–1873. [Google Scholar]
- 38.Hukkanen, E. J., J. A. Wieland, A. Gewirth, D. E. Leckband, and R. D. Braatz. 2005. Multiple-bond kinetics from single-molecule pulling experiments: evidence for multiple NCAM bonds. Biophys. J. 89:3434–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baumgartner, W., N. Golenhofen, N. Grundhofer, J. Wiegand, and D. Drenckhahn. 2003. Ca2+ dependency of N-cadherin function probed by laser tweezer and atomic force microscopy. J. Neurosci. 23:11008–11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kienberger, F., G. Kada, H. Mueller, and P. Hinterdorfer. 2005. Single molecule studies of antibody-antigen interaction strength versus intra-molecular antigen stability. J. Mol. Biol. 347:597–606. [DOI] [PubMed] [Google Scholar]
- 41.Hinterdorfer, P., W. Baumgartner, H. J. Gruber, K. Schilcher, and H. Schindler. 1996. Detection and localization of individual antibody-antigen recognition events by atomic force microscopy. Proc. Natl. Acad. Sci. USA. 93:3477–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stroh, C., H. Wang, R. Bash, B. Ashcroft, J. Nelson, H. Gruber, D. Lohr, S. M. Lindsay, and P. Hinterdorfer. 2004. Single-molecule recognition imaging microscopy. Proc. Natl. Acad. Sci. USA. 101:12503–12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riener, C. K., C. M. Stroh, A. Ebner, C. Klampfl, A. A. Gall, C. Romanin, Y. L. Lyubchenko, P. Hinterdorfer, and H. J. Gruber. 2003. Simple test system for single molecule recognition force microscopy. Anal. Chim. Acta. 479:59–75. [Google Scholar]
- 44.Fuchs, H., R. Gessner, R. Tauber, and R. Ghosh. 1995. Functional reconstitution of the human placental transferrin receptor into phospholipid bilayers leads to long tubular structures proceeding from the vesicle surface. Biochemistry. 34:6196–6207. [DOI] [PubMed] [Google Scholar]
- 45.Kasas, S., B. M. Riederer, S. Catsicas, B. Cappella, and G. Dietler. 2000. Fuzzy logic algorithm to extract specific interaction forces from atomic force microscopy data. Rev. Sci. Instrum. 71:2082–2086. [Google Scholar]
- 46.Ray, C., J. R. Brown, and B. B. Akhremitchev. 2007. Correction of systematic errors in single-molecule force spectroscopy with polymeric tethers by atomic force microscopy. J. Phys. Chem. 111:1963–1974. [DOI] [PubMed] [Google Scholar]
- 47.Ray, C., and B. B. Akhremitchev. 2005. Conformational heterogeneity of surface-grafted amyloidogenic fragments of alpha-synuclein dimers detected by atomic force microscopy. J. Am. Chem. Soc. 127:14739–14744. [DOI] [PubMed] [Google Scholar]
- 48.Halbrooks, P. J., A. M. Giannetti, J. S. Klein, P. J. Bjorkman, J. R. Larouche, V. C. Smith, R. T. MacGillivray, S. J. Everse, and A. B. Mason. 2005. Composition of pH-sensitive triad in C-lobe of human serum transferrin. Comparison to sequences of ovotransferrin and lactoferrin provides insight into functional differences in iron release. Biochemistry. 44:15451–15460. [DOI] [PubMed] [Google Scholar]
- 49.Ros, R., F. Schwesinger, D. Anselmetti, M. Kubon, R. Schafer, A. Pluckthun, and L. Tiefenauer. 1998. Antigen binding forces of individually addressed single-chain Fv antibody molecules. Proc. Natl. Acad. Sci. USA. 95:7402–7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ratto, T. V., K. C. Langry, R. E. Rudd, R. L. Balhorn, M. J. Allen, and M. W. McElfresh. 2004. Force spectroscopy of the double-tethered concanavalin-A mannose bond. Biophys. J. 86:2430–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Afrin, R., and A. Ikai. 2006. Force profiles of protein pulling with or without cytoskeletal links studied by AFM. Biochem. Biophys. Res. Commun. 348:238–244. [DOI] [PubMed] [Google Scholar]
- 52.Huang, H., J. Sylvan, M. Jonas, R. Barresi, P. T. So, K. P. Campbell, and R. T. Lee. 2005. Cell stiffness and receptors: evidence for cytoskeletal subnetworks. Am. J. Physiol. 288:C72–C80. [DOI] [PubMed] [Google Scholar]
- 53.Graziadei, I., R. Kaserbacher, H. Braunsteiner, and W. Vogel. 1993. The hepatic acute-phase proteins α1-antitrypsin and α2-macroglobulin inhibit binding of transferrin to its receptor. Biochem. J. 290:109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ciechanover, A., A. L. Schwartz, A. Dautry-Varsat, and H. F. Lodish. 1983. Kinetics of internalization and recycling of transferrin and the transferrin receptor in a human hepatoma cell line. Effect of lysosomotropic agents. J. Biol. Chem. 258:9681–9689. [PubMed] [Google Scholar]
- 55.Zak, O., and P. Aisen. 2002. A new method for obtaining human transferrin C–lobe in the native conformation: preparation and properties. Biochemistry. 41:1647–1653. [DOI] [PubMed] [Google Scholar]