Abstract
We have rigorously defined 2,820 orthologous mRNA and protein sequence pairs from rats, mice, and humans. Evolutionary rate analyses indicate that mammalian genes are evolving 17–30% more slowly than previous textbook values. Data are presented on the average properties of mRNA and protein sequences, on variations in sequence conservation in coding and noncoding regions, and on the absolute and relative frequencies of repetitive elements and splice sites in untranslated regions of mRNAs. Our data set contains 1,880 unique human/rodent sequence pairs that represent about 2–4% of all mammalian genes. Of the 1,880 human orthologs, 70% are present on a new gene map of the human genome, thus providing a valuable resource for cross-referencing human and rodent genomes. In addition to comparative mapping, these results have practical applications in the interpretation of noncoding sequence conservation between syntenic regions of human and mouse genomic sequence, and in the design and calibration of gene expression arrays.
Genome science and technology have brought us to the brink of being able to describe the genetic blueprint and molecular evolutionary history of the human species. But we will not be able to fully interpret these data in isolation. This is one of the reasons why the Human Genome Project has, from its inception, included the study of so-called “model organisms” whose biology, experimental advantages, and smaller, simpler genomes have provided not only important biological insights but also stepping stones for technology development.
The completion of genomic sequences for multiple prokaryotes and yeast has provided a wealth of information, and the value of comparative analysis of coding sequences from distantly related organisms (e.g., yeast and human) is beyond dispute (1, 2). Nevertheless there are limitations to functional inferences based on interspecies comparison of anciently diverged coding sequences (3). Furthermore, noncoding regions are generally not amenable to comparative analyses across such vast evolutionary distances because sequence divergence is simply too great (4). Thus it is necessary to study more closely related organisms to detect and interpret the conservation of regulatory, noncoding sequences (5).
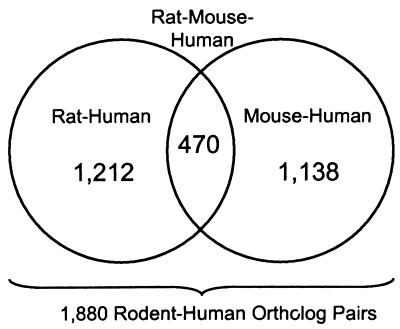
The mouse is the premier organism for studying mammalian genetics and development, and the rat has been extensively used for physiological and pharmacological studies. Mouse and rat genome projects (involving genetic and physical mapping and expressed gene surveys) are underway (6, 7). Cross-referencing molecular genetic data from rodents with human genome maps and sequences has many important applications, and a critical component is the identification of orthologous (8) genes. All too often, however, researchers mistake “homologous” for “orthologous.” Thus we have worked to define ortholog sets by using a rigorous phylogenetic approach (9) and also have devised a “triplet test” that yields confidence values for our conclusions of orthology. This work has resulted in 1,212 human/rat orthologs, 1,138 human/mouse orthologs and 470 orthologs shared by all three species (Fig. 1). These data sets contain 1,880 nonredundant human–rodent ortholog pairs and constitute the largest collection (by an order of magnitude) of transcribed sequences ever subjected to evolutionary distance analysis. Statistical distributions of sequence conservation in translated and untranslated regions are described and will be useful: (i) for identifying orthologs among human and rodent expressed sequence tag (EST) collections; (ii) for interpreting the relative significance of sequence conservation in nontranscribed genomic sequences; (iii) for developing and cross-referencing gene-based physical maps of mammalian genomes; and (iv) for calibrating hybridization specificity in gene expression arrays. Data on size distributions of mRNAs will also be useful in planning efforts to construct cDNA libraries that are optimal for the conversion of ESTs to full-length cDNA sequences.
Figure 1.
Data sets of orthologous sequence pairs.
MATERIALS AND METHODS
Selection of Sequence Pairs.
Orthologous rat and human sequences were selected as described previously (10) with the exception that release 19 of the HOVERGEN database (9) was used. Of the 4,705 protein families in HOVERGEN 19, 1,213 corresponded to full-length protein sequences available for both rat and human species. The cognate mRNAs for these proteins were retrieved from GenBank (11), always choosing the longest available sequence when several alternatives were present. This rat/human data set was compared with a mouse/human data set (10) to identify the overlapping subset of genes.
In total, there were 1,212 orthologous rat/human mRNA pairs and 470 orthologous mouse/rat mRNA pairs analyzed in this study (Fig. 1). A number of these mRNAs had either very short or unknown 5′ and 3′ untranslated regions (UTRs), and all 5′ UTRs shorter than 20 nucleotides and all 3′ UTRs shorter than 40 nucleotides were excluded from analysis. Consequently, 850 5′ UTRs and 1,028 3′ UTRs from rat/human data, and 292 5′ UTRs and 364 3′ UTRs from mouse/rat data set met these minimum-length criteria.
Sequence Alignment.
The desired mRNA sequences were extracted directly from GenBank by using the dump_cds program (J. Zhang, unpublished). This program extracts different regions of an mRNA to separate text files based on annotation in the GenBank features table. This procedure ensured that the most recent data were always used. The alignments of both nucleotide and protein sequences were computed by using the gap program,‡ which utilizes a global optimal alignment algorithm and fixed penalty for long gaps. Coding sequences (CDSs) used in substitution rate calculations were aligned by using the protein alignments as templates. Because the gap program does not penalize terminal gaps, each protein alignment was visually inspected and such errors in the alignments were corrected.
Synonymous and Nonsynonymous Substitution Distances.
The evolutionary distance, K, between two homologous sequences is estimated in terms of the number of base substitutions, but corrections are necessary to control for multiple and revertant mutation events (4). Distances were computed by method 1 of Ina (12), which includes a correction for multiple substitutions at single sites based on the two-parameter model of Kimura (13). Evolutionary distances are expressed in terms of the number of base substitutions per site. For coding regions, substitutions may be further classified as occurring at synonymous (silent) and nonsynonymous sites, and the corresponding distances are referred to as Ks and Ka, respectively. Substitution distances were calculated for three sets of sequence pairs: rat/human and rat/mouse (this study) and mouse/human based on data in Makałowski et al. (10). Distances, K, may be converted to rates, r, by using the equation r = K/(2T), where T is the divergence time between the two species (4).
RESULTS
Nucleotide and protein sequences were aligned as described in Materials and Methods. Gaps were excluded from all identity calculations. Results are summarized in Table 1. A large table containing GenBank sequence accession numbers (acc. nos.), alignment lengths, sequence identity values, and mutation distances for all rat/human and rat/mouse sequence pairs used in this study is available as an electronic supplement on the World Wide Web at http://www.ncbi.nlm.nih.gov/Makalowski/PNAS. Statistical properties of the data sets are discussed below.
Table 1.
Summary of sequence properties for 2,820 aligned orthologous human–rodent mRNAs and protein sequences
| Property | Rat–human
|
Mouse–human
|
Mouse–rat
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean (SD) | Median | Range | Mean (SD) | Median | Range | Mean (SD) | Median | Range | |
| 5′ UTR | |||||||||
| % Identity | 68.4 (13.0) | 66.7 | 36.6–100 | 69.7 (12.9) | 67 | 40.7–100 | 84.5 (12.9) | 87.1 | 41.7–100 |
| Aligned length, bp | 98 (96) | 65 | 20–879 | 124 (129) | 94 | 20–1521 | 97 (99) | 64 | 20–752 |
| Mutation distance K | 0.486 (0.260) | 0.453 | 0.00–1.595 | 0.493 (0.273) | 0.458 | 0.00–1.559 | 0.212 (0.224) | 0.142 | 0.00–1.237 |
| Mutation rate (×10−9) | 2.9 (1.6) | 2.8 | 0.00–10.0 | 2.75 (1.7) | 2.86 | 0.0–9.7 | 67 (4.7) | 7.5 | 0.0–41.0 |
| CDS | |||||||||
| % Identity (protein) | 88.0 (11.8) | 91.3 | 40.3–100 | 86.4 (12.3) | 89 | 41.1–100 | 94.5 (6.3) | 96.6 | 62.4–100 |
| % Identity (DNA) | 85.9 (6.0) | 87 | 58.3–98.4 | 85.2 (6.5) | 86.2 | 60.7–97.6 | 93.8 (3.2) | 94.3 | 75.4–98.9 |
| Aligned length, bp | 1400 (1054) | 1194 | 78–9780 | 1425 (1164) | 1175 | 135–13635 | 1293 (951) | 1104 | 154–8250 |
| Syn. distance Ks | 0.460 (0.145) | 0.446 | 0.057–1.646 | 0.468 (0.169) | 0.46 | 0.074–1.99 | 0.166 (0.061) | 0.163 | 0.01–0.61 |
| Syn. rate (×10−9) | 2.86 (0.91) | 2.79 | 0.35–10.0 | 2.91 (1.01) | 2.87 | 0.46–12.4 | 5.53 (2.07) | 5.54 | 0.34–20.3 |
| Nonsyn. distance Ka | 0.078 (0.095) | 0.051 | 0.00–0.609 | 0.090 (0.102) | 0.066 | 0.00–0.696 | 0.031 (0.040) | 0.018 | 0.00–0.25 |
| Nonsyn. rate (×10−9) | 0.49 (0.6) | 0.32 | 0.00–3.81 | 0.55 (0.63) | 0.39 | 0.00–3.81 | 1.05 (1.46) | 0.63 | 0.00–13.5 |
| 3′ UTR | |||||||||
| % Identity | 70.1 (11.4) | 68.6 | 40.0–98.4 | 71.0 (12.2) | 69.4 | 31.1–100 | 86.3 (8.9) | 87.7 | 48.8–100 |
| Aligned length, bp | 388 (380) | 264 | 40–3164 | 416 (432) | 263 | 40–3478 | 392 (391) | 235 | 43–2996 |
| Mutation distance K | 0.435 (0.212) | 0.416 | 0.016–1.230 | 0.447 (0.225) | 0.425 | 0.00–1.424 | 0.164 (0.152) | 0.136 | 0.00–1.179 |
| Mutation rate (×10−9) | 2.6 (1.3) | 2.6 | 0.1–7.7 | 2.6 (1.4) | 2.7 | 0.0–8.9 | 4.95 (5.1) | 4.5 | 0.0–39.3 |
Aligned Rat and Human Sequences: UTRs.
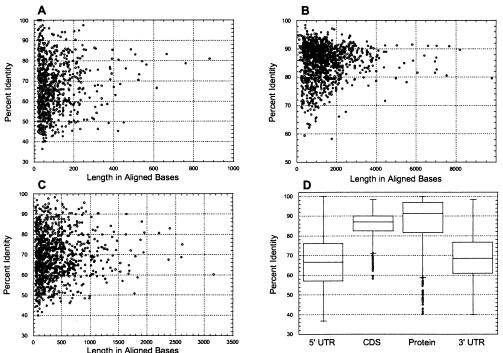
The 850 aligned 5′ UTR sequences consisted of 83,426 nucleotides. Alignment lengths ranged in size from 20 to 879 nucleotides, with a mean value of 98 (SD = 96) and a median value of 65. Fifty percent of the values were distributed within the range of 38 to 122 nucleotides and 90% of the values were between 23 and 264 nucleotides (Fig. 2A).
Figure 2.
Distributions of lengths and degrees of sequence conservation for 1,212 aligned orthologous rat and human mRNA and protein sequences. (A–C) Scatter plots of results for 5′ UTRs (A), CDSs (B), and 3′ UTRs (C). (D) Box plots of sequence conservation by region for aligned rat and human mRNAs and encoded proteins. For each category, the central box depicts the middle 50% of the data between the 25th and 75th percentile, and the enclosed horizontal line represents the median value of the distribution. Extreme values are indicated by circles that occur outside the main bodies of data.
The 1,027 aligned 3′ UTR sequences consisted of 398,199 nucleotides with alignment lengths of 40 to 3,164 nucleotides with a mean value of 388 (SD = 380) and a median of 264. Fifty percent of the lengths of 3′ UTR alignments were between 128 and 512 nucleotides and 90% of the values were between 55 and 1,127 nucleotides (Fig. 2C). On average, 3′ UTR alignments were four times longer than 5′ UTR alignments.
The mean aligned identity of human/rat 5′ UTRs was 68.4% (SD = 13.0) and the mean aligned identity of human/rat 3′ UTRs was similar at 70.1% (SD = 11.4). Median identity values for 5′ and 3′ UTRs were 66.7% and 68.6%, respectively (Table 1). For both 5′ and 3′ UTRs, the degrees of sequence conservation are broadly distributed between 37% and 100% identity (Fig. 2 A and C).
Although numerically insignificant (<2% of total), there are 16 cases of 5′ UTRs longer than 1,000 nucleotides in our data set. The two longest are those of human adenylyl cyclase mRNA (2,094 bases, GenBank acc. no. Z35309) and a rat ataxin mRNA (1,894 bases, GenBank acc. no. X91619). Likewise, 22 3′ UTRs (2.1% of total) are longer than 3,000 nucleotides, and the longest are those of the human ataxin mRNA (7,274 bases, GenBank acc. no. X79204) and a human cyclin D2 mRNA (5,339 bases, GenBank acc. no. D13639). The extraordinary lengths of these UTRs are not because of the insertion of repetitive elements (see below).
Aligned Rat and Human Sequences: CDSs.
The 1,212 CDSs consisted of 1,696,766 nucleotides. Alignment lengths ranged in size from 78 to 9,780 nucleotides, with a mean value of 1,400 (SD = 1,054) and a median value of 1,194. The distribution of aligned CDS sizes is narrow (Fig. 2B), with 50% of the alignments between 732 and 1,689 nucleotides in length, and 90% in the range of 446–2,567 nucleotides. The mean aligned identity of human/rat CDSs is 85.9% (SD = 6.0), and the mean aligned identity of human/rat proteins is 88.0% (SD = 11.8). The median identity values for CDSs and proteins are 87% and 91.3%, respectively (Table 1). As previously shown for human and mouse sequences (10), conservation is more narrowly distributed for nucleotide sequences compared with protein sequences they encode: 90% of CDSs are between 74% and 93% identical, whereas 90% of protein sequences are 63–100% identical (Fig. 2D). Fifty-three (4.3% of 1,212) proteins were 100% identical in sequence between humans and rats (http://www.ncbi.nlm.nih.gov/Makalowski). At the other extreme, some human/rat protein pairs shared only ≈40% identical amino acid residues.
Aligned Rat and Mouse Sequences: UTRs.
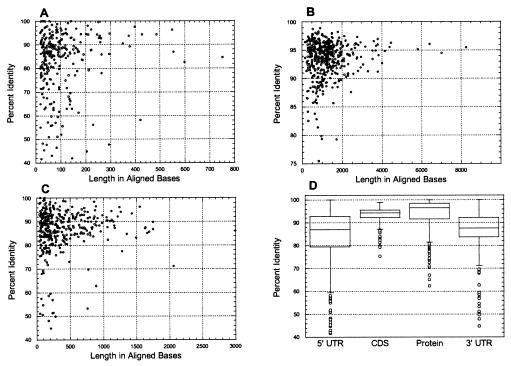
The 297 5′ UTR sequences consisted of 28,850 nucleotides. Alignment lengths ranged between 20 and 752 nucleotides, with a mean value of 97 (SD = 99) and a median value of 64 (Table 1). The distribution of 5′ UTR alignment lengths was very similar to that observed in the rat–human data set, with 50% of the values in the range of 37–120 nucleotides and 90% of the values between 22 and 264 nucleotides (Fig. 3A). The 371 3′ UTR sequences consisted of 145,310 nucleotides with alignment lengths of 43 to 2,996 nucleotides. The mean length was 391 (SD = 391) nucleotides and the median value was 235 (Table 1). Again, the distribution of 3′ UTR alignment lengths was very similar to that of the rat–human data, with 50% of the values between 128 and 525 nucleotides and 90% of the values between 58 and 1,156 nucleotides (Fig. 3C). On average, 3′ UTR alignments were four times longer than 5′ UTR alignments.
Figure 3.
Distributions of lengths and degrees of sequence conservation for 470 aligned orthologous mouse and rat mRNA and protein sequences. (A–C) Scatter plots of results for 5′ UTRs (A), CDSs (B), and 3′ UTRs (C). (D) Box plots as described in the legend to Fig. 2.
The mean aligned identity of mouse/rat 5′ UTRs was 84.5% (SD = 12.9) and the mean aligned identity of mouse/rat 3′ UTRs was higher at 87.3% (SD = 8.9). The median identity values for 5′ and 3′ UTRs were 87.1% and 87.7%, respectively (Table 1). For both 5′ and 3′ UTRs, the degrees of sequence conservation were broadly distributed between 41.7% and 100% identity (Fig. 3 A and C).
Four individual 5′ UTRs (1.4% of total) consist of more than 1,000 nucleotides, and the longest one was that of mouse brain potassium channel protein (1,456 bases, GenBank acc. no. Y00305). Three 3′ UTRs (0.8% of total) were longer than 2,000 nucleotides, and the longest was that of the mouse insulin-like growth factor binding protein 5 (4,358 bases, GenBank acc. no. L12447). None of these long UTRs contain repetitive elements.
Aligned Rat and Mouse Sequences: CDSs.
The 470 CDSs consisted of 591,861 nucleotides. Alignment lengths ranged in size from 159 to 8,250 nucleotides, with a mean value of 1,292 (SD = 923) and a median value of 1,114 (Table 1). For coding sequences, 50% of aligned lengths are between 708 and 1,548 nucleotides and 90% are between 324 and 2,889 nucleotides (Fig. 3B). The mean aligned identity of mouse/rat CDSs is 93.5% (SD = 3.2) and the mean aligned identity of mouse/rat proteins is 94.0% (SD = 6.4). The median values for CDSs and proteins are 94% and 96.4%, respectively (Table 1). Fifty percent of CDSs are within an identity range of 92–96%, and 90% of protein sequences are within an identity range of 88–97%. Among 470 analyzed proteins, 23 (5%) share an identical amino acid sequence.
Aligned Mouse and Human Sequences.
Data on 1,196 mouse and human ortholog pairs was reported previously (10). Subsequent findings indicated that some sequences in this data set actually represent paralogs. Therefore these sequences were removed to create a revised data set of 1,138 mouse–human ortholog pairs. Summary statistics have been recalculated and the revised values are included in Table 1. Also included in Table 1 are new calculations of evolutionary distances (see below).
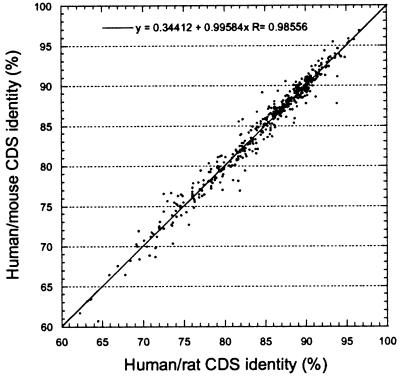
Ortholog Authentication.
Because the divergence times between humans and rats and between humans and mice should be the same, the overlapping set of 470 human, rat, and mouse sequence triplets provides an opportunity to validate the conclusion of orthology for all human–rodent sequence pairs. The correlation between human/rat and human/mouse coding sequence identities was plotted (Fig. 4) and the distances of all points from the regression line were calculated. Three hundred and forty-four points (77.5%) lie <1 SD from the regression line and 425 (92.8%) points are <2 SD. Only six points (1.3%) lie >3 SD from the line. From the normal distribution one can expect two points to occur >3 SD from the line, and examples in excess of this might represent paralogous sequence pairs. An extrapolation from this analysis indicates that no more than 10 (0.5%) sequence pairs have been misidentified as orthologs in the entire human/rodent data set.
Figure 4.
Correlation of coding sequence identities between orthologous human/mouse and human/rat sequence pairs.
Analysis of Evolutionary Distances.
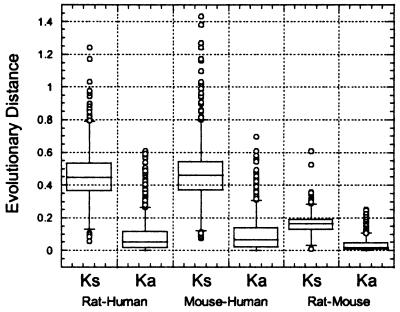
For rat and human genes (Fig. 5), the nonsynonymous nucleotide substitution distance, Ka, ranges from 0 to 0.609, with a length-weighted mean Ka of 0.078 (SD = 0.095). Synonymous substitution distances, Ks, range between 0.057 and 1.646, with a length-weighted mean value of 0.460 (SD = 0.145). As shown in Fig. 6, the average values of mutation distances in untranslated regions are similar to Ks, with K = 0.486 for 5′ UTRs (SD = 0.260) and K = 0.413 for 3′ UTRs (SD = 0.212).
Figure 5.
Analysis of evolutionary distances for orthologous sequence pairs.
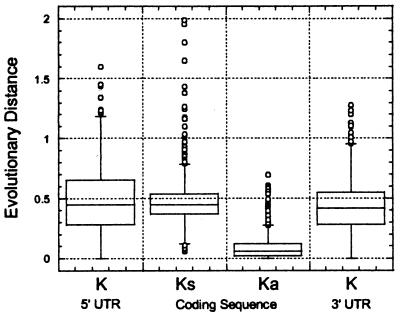
Figure 6.
Analysis of evolutionary distances in untranslated and coding regions of human–rodent mRNA sequences.
Similar values characterize the mouse/human data set (Fig. 5). Ka ranges from 0 to 0.696, with a length-weighted mean Ka of 0.090 (SD = 0.102). Ks ranges from 0.074 to 1.99, with a length-weighted mean of 0.460 (SD = 0.176). As shown in Fig. 6, the average values of mutation distances in UTRs are similar to Ks, with K = 0.493 for 5′ UTRs (SD = 0.273) and K = 0.447 for 3′ UTRs (SD = 0.225).
Rats and mice diverged as species about 10–15 million years ago, whereas the human/rodent divergence time is usually taken to be the time of the great mammalian radiation of 80 million years ago (4). Thus lower Ka and Ks values (Table 1, Fig. 5) in rodent species reflect a shorter period of time for substitutions to have occurred. Ka values for the rat/mouse samples are narrowly distributed between 0 and 0.250, with a length-weighted mean value of 0.035 (SD = 0.040). Ks ranges from 0.010 to 0.610, with a length-weighted mean value of 0.167 (SD = 0.061). The average K in 3′ UTRs equals 0.164 (SD = 0.152) and is almost identical with that at synonymous sites, but the K for 5′ UTRs is significantly higher, with a value of 0.212 (SD = 0.224).
Correlations of Mutation Rates Among Coding and Noncoding Regions of mRNAs.
1,880 unique human/rodent mRNA pairs were analyzed for possible correlations in intrasequence changes. The correlation coefficient was strongest (r = 0.46) between 3′ UTR and CDS sequences and weakest (r = 0.29) between 5′ and 3′ untranslated sequences (r = 0.29). r = 0.32 for 5′ UTR and CDS sequences. Correlation between synonymous and nonsynonymous changes was also assessed and appears to be relatively high (r = 0.56). Correlation coefficient graphs for all of these cases are available at http://www.ncbi.nlm.nih.gov/Makalowski.
Splice Junctions and Interspersed Repeats.
The presence of intron sites and repetitive elements in the untranslated portions of mRNAs have important implications for gene mapping, cloning, and sequence analysis (14). The occurrence of splice junctions, and short and long interspersed repetitive elements (SINEs and LINEs, respectively) in our human–rodent data set was determined as described previously (10).
We found evidence for a single splice junction in only 8 of 4,571 human and rodent 3′ UTRs surveyed. In 7 of the 8 cases, the splice junctions occur within the 35 bases distal to the stop codon. In the remaining instance (mRNA for rat hepatic leukemia factor, acc. no. S79820), the splice junction was found 165 bases distal to the stop codon. In 5′ UTRs, splice junctions occur more frequently, being present in 46 of 4,447 mRNAs examined. In 12 cases there was more than one splice junction in a single 5′ UTR and as many as four in the 5′ UTR of the adenosine A1 receptor (acc. no. L22214). Although splice junctions in 5′ UTRs are more broadly distributed than in 3′ UTRs, 15 of them occur within first 50 nucleotides upstream of the initiation codon, with the closest splice site only 6 bases upstream from the coding region in the mRNA for human cAMP-dependent protein kinase (acc. no. M33336). The most distant splice junction occurs in the 5′ UTR of ataxin mRNA 5′ and is 769 bases upstream from the initiation codon.
A number of different studies have shown that repetitive elements are present in about 10% of mammalian mRNA (10, 15, 16). These elements may be found in all mRNA regions, with the highest probability of occurrence in the 3′ UTR and the lowest in coding sequences. Among rodent sequences in our data set, repeats were found in 197 of 2,283 (8.6%) of 3′ UTRs. These repeats consisted of 239 fragments of SINEs, 33 fragments of LINEs, 35 long terminal repeats (LTRs), and 13 fragments of transposons. In total, repetitive sequences accounted for 13% of the total bases in rodent 3′ UTRs. Among human sequences in our data set, repeats were found in 186 of 1,879 (9.9%) of 3′ UTRs. These repeats consisted of 160 fragments of SINEs, 45 fragments of LINEs, 9 LTRs, and 21 transposons and account for 17.8% of the total bases in human 3′ UTRs.
In contrast, the frequency of repetitive elements in 5′ UTRs is much lower. Repeats were found in only 53 of 1,826 human 5′ UTRs (2.9%) and in 73 of 2,187 rodent 5′ UTRs (3.2%). In rodent sequences, 66 SINEs, 9 LINEs, and 8 LTR fragments account for 12.7% of the total bases in 5′ UTRs. In human 5′ UTRs there were 38 SINE fragments, 13 LINEs, 5 LTRs, and 4 transposons that constituted 28.7% of the total bases.
DISCUSSION
Comparative analysis of biological characteristics has a long and fruitful history, and it is becoming increasingly possible to carry out such studies in a comprehensive manner at the molecular level. A complete description of the comparative genomics of two organisms includes alignments of all ancestrally related (homologous) sequences, and this is already being accomplished for a number of microbial species (17, 18). But we are far from the goal of being able to describe mammalian genomes at this level of detail. Nevertheless, comparative maps of the human and mouse genomes are available and currently contain nearly 1,800 loci in 201 conserved linkage groups (19–21). Comparative studies of genomic sequence have been performed on a limited number of available large contigs (reviewed in ref. 5). The present work reports an analysis of 2,820 coding and noncoding, transcribed, orthologous sequence pairs from mice, rats, and humans (Fig. 1). These 2,820 sequence pairs correspond to 1,880 unique human–rodent gene products that represent approximately 2–4% of transcribed mammalian protein-encoding genes. Despite this small percentage, we believe that this collection is representative of the genome as a whole, for reasons presented earlier (10).
Previous conclusions about the rates of evolution of mammalian genes have been based on rather small samples of sequence data (4, 22–24). For example, Li (4) reported a range of nonsynonymous mutation rates of 0.00 to 3.06 substitutions per site per 109 years, with an average value of 0.74 (SD = 0.67), based on an analysis of 47 human and rodent ortholog pairs. On the basis of the present analysis of 1,880 human-rodent ortholog pairs (see below), mammalian genes appear to be evolving significantly more slowly than previously thought, with a mean value of 0.52 (SD = 0.59) and a median value of only 0.32 substitution per site per 109 years (Tables 1 and 2). Average rates of synonymous nucleotide substitutions were also found to be lower than previous estimates: 2.92 × 10−9 (this study) compared with 3.51 × 10−9 (4).
Table 2.
Average properties of orthologous human and rodent mRNAs
| Property | 5′ UTR | CDS | 3′ UTR |
|---|---|---|---|
| No alignments examined | 1,416 | 1,880 | 1,590 |
| Average length* | 115 | 1,450 | 411 |
| 75th percentile | 143 | 1,773 | 543 |
| 95th percentile | 309 | 3,390 | 1,228 |
| 99th percentile | 532 | 6,543 | 2,069 |
| Average % identity | 67 | 85 | 69 |
| Average mutation distance | K = 0.455 | Ks = 0.467 | K = 0.410 |
| Ka = 0.084 | |||
| Frequency of splice junction, % | 1.03 | ND | 0.17 |
| Frequency of repetitive element, % | 3.14 | ND | 9.20 |
In the 470 cases in which the same human mRNA sequence matched to both mouse and rat orthologs (Fig. 1), only one sequence pair was chosen, on the basis of the most complete (longest) rodent sequence available. ND = not determined.
Excludes poly(A) and gaps in alignment.
An interesting question, vis-à-vis the neutral theory of molecular evolution, is whether there is any evidence that substitution rates are correlated among coding and noncoding regions of mRNAs. Our survey of human and rodent sequences shows significant positive correlation between substitution rates in coding and untranslated parts of messages and a tendency for substitution rates in untranslated regions to be lower for more conserved proteins and higher for less conserved ones. Our results also demonstrate a statistically significant correlation between substitution rates at synonymous and nonsynonymous sites (r = 0.57 and 0.54 for human/rodent and rat/mouse data, respectively). This phenomenon in particular has been observed in previous studies on much smaller data sets: r = 0.51 for 26 mammalian gene pairs (22), r = 0.45 for 363 mouse/rat orthologs (25), and r = 0.57 for 72 human/calf orthologs (24). This correlation between substitution rates at synonymous and nonsynonymous sites is in disagreement with the neutral theory of molecular evolution (26). No satisfactory explanation has been found for this phenomenon.
Regarding mouse and rat genes, Wolfe and Sharp (25) have analyzed a collection of 363 mouse and rat ortholog pairs (coding sequences only) and observed evolutionary distances of Ka = 0.032 (SD = 0.049) and Ks = 0.224 (SD = 0.084) at nonsynonymous and synonymous sites, respectively. In the present study of 470 mouse–rat ortholog pairs (including the 5′ and 3′ UTRs), we found a very similar evolutionary distance for nonsynonymous sites (Ka = 0.035) but a significantly lower distance (Ks = 0.167) for synonymous sites (Table 1). This latter inconsistency is because of the fact that Wolfe and Sharp (25) applied a method that is now known to underestimate the number of nonsynonymous sites and significantly overestimate the synonymous ones (27, 28). The value of Ks is similar to K (0.164) in 3′ UTRs, although 5′ UTRs appear to be evolving more rapidly (K = 0.212).
The molecular clock hypothesis postulates that the substitution rate is constant in all evolutionary lineages (29). The concept has been controversial with a wide range of views. Ochman and Wilson (30) suggested the existence of universal clock of synonymous substitution, but Goodman (31, 32) denied the existence of the molecular clock altogether. Our set of 470 orthologous sequences present in three species enabled us to test the existence of local molecular clock hypothesis in mice and rats, using human sequences as an outgroup. DNA–DNA hybridization studies suggested a constant substitution rate in mouse and rat lineages (33, 34). This finding was confirmed by analysis of nucleotide sequences using human as an outgroup (35, 36). When hamster was used as an outgroup in nucleotide sequence comparison (36), the molecular clock was constant at synonymous sites but significantly higher in mouse lineage at nonsynonymous sites. Because O’hUigin and Li (36) used only 42 genes in their analysis, we decided to reexamine the substitution rates in murine lineages, using our 10-fold larger data set.
The mean Ks between human and mouse is 0.4662 (±0.0064) and between human and rat is 0.4720 (±0.0066). The Ka between human and mouse is 0.0947 (±0.0047) and between human and rat it is 0.0972 (±0.0049). In both cases the differences in substitution rates between mouse and rat lineages are less than the standard error and thus statistically insignificant. Similarly, O’hUigin and Li (36) did not observe statistically significant differences in mouse and rat substitution rates when human was used as an outgroup, although they did observe a difference when hamster sequences were used as an outgroup. Thus it may be that human sequences are too distant from rodents to detect subtle differences in the variation of substitution distances within the murine lineage.
Because there is no significant difference between various measures of sequence properties from the 1,212 rat/human and 1,138 mouse/human comparisons (Table 1), we have combined the individual studies to provide a generalized picture of the 1,880 unique human–rodent sequence pairs (Table 2, Fig. 6). The average length of mRNAs in human and rodents [5′ UTR + CDS + 3′ UTR, excluding poly(A)] is just under 2 kb. 3′ UTRs are four times longer than 5′ UTRs on average. The mean degree of sequence identity in untranslated regions is 67–69%, whereas coding sequences are, as expected, much more highly conserved, with a mean identity of 85%. Coding sequences evolve about 1/5 as fast as noncoding sequences. Although the observed frequencies of occurrence of splice junctions in untranslated regions are low (0.17–1.03%), splice junctions are about 1/5 as likely to occur in a 3′ UTR compared with a 5′ UTR. Repetitive elements are present in 3–9% of untranslated regions and are three times more frequent in 3′ UTRs than in 5′ UTRs.
All of these sequence features have important implications for gene mapping, sequence interpretation, and functional genomics applications. For example, the fact that 3′ UTRs are more divergent than coding sequences and have a very low incidence of splice junctions validates their use for the development of gene-specific sequence tagged sites (STSs) for transcript mapping (37). These same features also make them attractive for designing or populating large-scale gene expression arrays (38, 39).
Furthermore, this large set of authenticated human–rodent ortholog pairs should be valuable for cross-referencing human–mouse, human–rat, and rat–mouse gene maps (19–21). Indeed, 1,326 (70%) of the 1,880 human orthologs (Fig. 1) are already present on an upcoming new release of the RH Consortium human gene map (unpublished observation and ref. 14). Matched rodent–human ortholog pairs also may be useful for optimizing hybridization stringency for sequence detection and gene discrimination across a broad range of sequence conservation (40). Finally, the fact that 99% of the mRNA alignments in our sample are shorter than 10 kb indicates that cDNA libraries with insert sizes in this range may be adequate for the conversion of ESTs into full-length cDNA sequences.
The distributions of sequence conservation in transcribed sequences provide a scale of comparison for interpreting the significance of sequence similarities in noncoding genomic sequences such as introns, promoters, and intergenic regions (5). They should be helpful in classifying similarities (i.e., answering the question of whether two homologous sequences are orthologs or paralogs) among human, mouse, and rat ESTs. This large set of validated ortholog pairs may be also useful as a standard for cross-referencing more distantly related vertebrate and invertebrate genomes (41).
Acknowledgments
We thank Jinghui Zhang for modifications of software tools, Peter Kuehl for determining the map locations of human sequences, Hugues Sicotte for helpful suggestions, and David Lipman for a critical reading of the manuscript.
ABBREVIATIONS
- EST
expressed sequence tag
- UTR
untranslated or noncoding region of mRNA
- CDS
protein-encoding sequence
- acc. no.
GenBank accession number
- SINE
short interspersed repetitive element
- LINE
long interspersed repetitive element
- LTR
long terminal repeat
Footnotes
A mismatch penalty of −3 and the PAM120 scoring matrix were used for DNA and protein alignments, respectively. Other parameters included: match 10, gap opening penalty 50, gap extension penalty 5, and longest penalized gap 10.
References
- 1.Bassett, D. E., Boguski, M. S. & Hieter, P. (1996) Nature (London) 589–590. [DOI] [PubMed]
- 2.Botstein D, Cherry J M. Proc Natl Acad Sci USA. 1997;94:5506–5507. doi: 10.1073/pnas.94.11.5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mushegian A R, Bassett D E, Jr, Boguski M S, Bork P, Koonin E V. Proc Natl Acad Sci USA. 1997;94:5831–5836. doi: 10.1073/pnas.94.11.5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li W-H. Molecular Evolution. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- 5.Hardison R C, Oeltjen J, Miller W. Genome Res. 1997;7:959–966. doi: 10.1101/gr.7.10.959. [DOI] [PubMed] [Google Scholar]
- 6.Camper S A, Meisler M H. Mamm Genome. 1997;8:461–463. doi: 10.1007/s003359900476. [DOI] [PubMed] [Google Scholar]
- 7.James M R, Lindpaintner K. Trends Genet. 1997;13:171–173. doi: 10.1016/s0168-9525(97)01130-x. [DOI] [PubMed] [Google Scholar]
- 8.Fitch W M. Syst Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- 9.Duret L, Mouchiroud D, Gouy M. Nucleic Acids Res. 1994;22:2360–2365. doi: 10.1093/nar/22.12.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makalowski W, Zhang J, Boguski M S. Genome Res. 1996;6:846–857. doi: 10.1101/gr.6.9.846. [DOI] [PubMed] [Google Scholar]
- 11.Benson D A, Boguski M S, Lipman D J, Ostell J. Nucleic Acids Res. 1997;25:1–6. doi: 10.1093/nar/25.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ina Y. J Mol Evol. 1995;40:190–226. doi: 10.1007/BF00167113. [DOI] [PubMed] [Google Scholar]
- 13.Kimura M. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 14.Schuler G D, Boguski M S, Stewart E A, Stein L D, Gyapay G, Rice K, White R E, Rodriguez-Tome P, Aggarwal A, Bajorek E, et al. Science. 1996;274:540–546. [PubMed] [Google Scholar]
- 15.Crampton J M, Davies K E, Knapp T F. Nucleic Acids Res. 1981;9:3821–3834. doi: 10.1093/nar/9.15.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yulug I G, Yulug A, Fisher E M. Genomics. 1995;27:544–548. doi: 10.1006/geno.1995.1090. [DOI] [PubMed] [Google Scholar]
- 17.Koonin E V. Genome Res. 1997;7:418–421. doi: 10.1101/gr.7.5.418. [DOI] [PubMed] [Google Scholar]
- 18.Tatusov R L, Koonin E V, Lipman D J. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 19.Andersson L, Archibald A, Ashburner M, Audun S, Barendse W, Bitgood J, Bottema C, Broad T, Brown S, Burt D, et al. Mamm Genome. 1996;7:717–734. doi: 10.1007/s003359900222. [DOI] [PubMed] [Google Scholar]
- 20.Eppig J T. Curr Opin Genet Dev. 1996;6:723–730. doi: 10.1016/s0959-437x(96)80027-x. [DOI] [PubMed] [Google Scholar]
- 21.DeBry R W, Seldin M F. Genomics. 1996;33:337–351. doi: 10.1006/geno.1996.0209. [DOI] [PubMed] [Google Scholar]
- 22.Graur D. J Mol Evol. 1985;22:53–62. doi: 10.1007/BF02105805. [DOI] [PubMed] [Google Scholar]
- 23.Ohta T, Ina Y. J Mol Evol. 1995;41:717–720. doi: 10.1007/BF00173150. [DOI] [PubMed] [Google Scholar]
- 24.Mouchiroud D, Gautier C, Bernardi G. J Mol Evol. 1995;40:107–113. doi: 10.1007/BF00166602. [DOI] [PubMed] [Google Scholar]
- 25.Wolfe K H, Sharp P M. J Mol Evol. 1993;37:441–456. doi: 10.1007/BF00178874. [DOI] [PubMed] [Google Scholar]
- 26.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, U.K.: Cambridge Univ. Press; 1983. [Google Scholar]
- 27.Li W-H, Wu C I, Luo C C. Mol Biol Evol. 1985;2:150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- 28.Li W-H. J Mol Evol. 1993;36:96–99. doi: 10.1007/BF02407308. [DOI] [PubMed] [Google Scholar]
- 29.Zuckerkandl E, Pauling L. In: Evolving Genes and Proteins. Bryson V, Vogel H, editors. New York: Academic; 1965. pp. 97–166. [DOI] [PubMed] [Google Scholar]
- 30.Ochman H, Wilson A C. J Mol Evol. 1987;26:74–86. doi: 10.1007/BF02111283. [DOI] [PubMed] [Google Scholar]
- 31.Goodman M. In: Molecular Evolution. Ayala F, editor. Sunderland, MA: Sinauer; 1976. [Google Scholar]
- 32.Goodman M. Prog Biophys Mol Biol. 1981;38:105–164. doi: 10.1016/0079-6107(81)90012-2. [DOI] [PubMed] [Google Scholar]
- 33.Brownell E, Krystal M, Arnheim N. Mol Biol Evol. 1983;1:29–37. doi: 10.1093/oxfordjournals.molbev.a040304. [DOI] [PubMed] [Google Scholar]
- 34.Catzeflis F M, Sheldon F H, Ahlquist J E, Sibley C G. Mol Biol Evol. 1987;4:242–253. doi: 10.1093/oxfordjournals.molbev.a040444. [DOI] [PubMed] [Google Scholar]
- 35.Li W H, Tanimura M, Sharp P M. J Mol Evol. 1987;25:330–342. doi: 10.1007/BF02603118. [DOI] [PubMed] [Google Scholar]
- 36.O’hUigin C, Li W H. J Mol Evol. 1992;35:377–384. doi: 10.1007/BF00171816. [DOI] [PubMed] [Google Scholar]
- 37.Boguski M S, Schuler G D. Nat Genet. 1995;10:369–371. doi: 10.1038/ng0895-369. [DOI] [PubMed] [Google Scholar]
- 38.Fodor S P, Rava R P, Huang X C, Pease A C, Holmes C P, Adams C L. Nature (London) 1993;364:555–556. doi: 10.1038/364555a0. [DOI] [PubMed] [Google Scholar]
- 39.Schena M, Shalon D, Davis R W, Brown P O. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 40.Hacia J C, Makalowski W, Edgemon K, Erdos M R, Robbins C M, Fodor S P A, Brody L C, Collins F S. Nat Genet. 1998;18:155–158. doi: 10.1038/ng0298-155. [DOI] [PubMed] [Google Scholar]
- 41.Sidow A. Curr Opin Genet Dev. 1996;6:715–722. doi: 10.1016/s0959-437x(96)80026-8. [DOI] [PubMed] [Google Scholar]