Abstract
The availability of the complete sequence of Escherichia coli strain MG1655 provides the first opportunity to assess the overall impact of horizontal genetic transfer on the evolution of bacterial genomes. We found that 755 of 4,288 ORFs (547.8 kb) have been introduced into the E. coli genome in at least 234 lateral transfer events since this species diverged from the Salmonella lineage 100 million years (Myr) ago. The average age of introduced genes was 14.4 Myr, yielding a rate of transfer 16 kb/Myr/lineage since divergence. Although most of the acquired genes subsequently were deleted, the sequences that have persisted (≈18% of the current chromosome) have conferred properties permitting E. coli to explore otherwise unreachable ecological niches.
The lack of complex morphological characteristics and a robust fossil record have impeded efforts to understand the processes mediating differentiation and speciation in prokaryotes. Although vast numbers of mutations are introduced into bacterial populations in each generation, it is difficult to account for the ability of bacteria to respond to new selection pressures and to exploit new environments by the accumulation of point mutations alone; for example, no phenotype distinguishing the closely related taxa Escherichia coli and Salmonella enterica can be attributed to point mutational processes. Hence, the rapid adaptation of bacteria to novel environments often is ascribed to genes acquired through horizontal, interspecific gene transfer (1). Transferred sequences have a large impact on bacterial evolution; for example, the incorporation of a DNA fragment conferring virulence characteristics can transform a benign strain of E. coli into a pathogen in a single step (2–4), and the conversion from antibiotic sensitivity to resistance typically is caused by the acquisition of sequences rather than by the point mutational evolution of existing genes (5).
Horizontal transfer, even at very low levels, produces a mosaic chromosome comprised of genes of differing ancestries and durations in the genome. In some cases, it is possible to establish the evolutionary history of a gene by examining its distribution among several related bacteria; if a gene is confined to one taxon or species, it is more likely to have been acquired through gene transfer than to have been lost independently from multiple lineages. This phylogenetic approach has yielded evidence that several genes, including those constituting the lac operon, arose in E. coli through horizontal transfer (6, 7).
The DNA sequence of the gene itself also can provide clues to its origin and ancestry within a genome. Bacterial species display wide variation in overall GC content, but the genes within a particular species’ genome are fairly similar in base composition and patterns of codon usage (8–11). Consequently, sequences that are new to a bacterial genome, i.e., those introduced through horizontal transfer, often bear unusual sequence characteristics and thus can be distinguished from ancestral DNA. Evidence that base composition and codon usage patterns are reliable indicators of horizontal transfer has come from comparative analyses of the Escherichia coli and Salmonella enterica chromosomes (7, 12–14). The vast majority of genes confined to only one of these two enteric species, such as those responsible for coenzyme B12 biosynthesis (cbi/cob), citrate utilization (tct), and host cell recognition and invasion (inv/spa) of Salmonella, and lactose (lac) and phosphonate (phn) utilization in E. coli, have anomalous nucleotide compositions and do not use the synonymous codons typically used by these species.
Although each of these cases exemplifies the potential role of horizontal genetic transfer in shaping the character of bacterial species, quantification of the total contribution of horizontal transfer on bacterial evolution has been difficult. To establish the overall contribution of horizontal transfer in the evolution of bacterial chromosomes, it is necessary to have (i) means of identifying all of the horizontally transferred sequences from a complete genomic sequence and (ii) methods of estimating the age of genes since their introduction into a genome. By analyzing the complete nucleotide sequence of the E. coli genome, the present study provides the first accurate appraisal of the rate of horizontal transfer on an evolutionary timescale, its effects on the organization of a bacterial genome, and its role in bacterial diversification and speciation.
MATERIALS AND METHODS
From the complete sequence of E. coli strain MG1655 (15), protein coding regions initially were identified as atypical if their GC contents at first and third codon positions were two or more SEs higher or lower than the respective means for all genes in the genome. We also plotted the χ2 of codon usage for each gene—which assesses the degree of bias in the use of synonymous codons vs. the expectation based on nucleotide composition of codon positions—against its Codon Adaptation Index, which measures the degree of bias toward the subset of codons used by highly expressed genes in E. coli (16, 17). From this plot, it is possible to recognize genes whose atypical base composition results from the prevalence of codons preferentially used by E. coli and to identify genes transferred into E. coli from organisms of similar base composition but of very different codon usage patterns. (Because these genes use codons not used by E. coli and do not use codons preferred by E. coli, they show a strong bias in codon usage but a low Codon Adaptation Index.) Next, we determined whether ORFs were situated at a specific chromosomal position containing other horizontally transferred sequences, which is indicative of acquisition in a single transfer event. This would recognize, for example, otherwise typical genes within a translationally coupled operon of horizontally transferred sequences.
The list of horizontally transferred genes recovered by these procedures then was examined to identify known native genes that exhibit atypical base compositions for other reasons, such as the amino acid content of the encoded protein. For example, the prevalence of lysine residues in certain ribosomal proteins contributes to the uncharacteristically low GC contents of the coding sequences. Finally, a blast search (18) was performed on all ORFs to detect similarities with genes of closely related species. The list of the horizontally transferred genes in E. coli MG1655 detected by these procedures is available at ftp://ftp.pitt.edu/dept/biology/lawrence/.
RESULTS AND DISCUSSION
The Amount of Horizontally Transferred DNA in E. coli.
The complete 4,639,221-bp sequence of the chromosome of E. coli strain MG1655 has been resolved (15), and our analysis indicated that 755 (17.6%) of the 4288 ORFs in the genome originated through horizontal gene transfer. Although similar to previous estimates based on limited portions of the E. coli chromosome (7, 13, 14), this value represents the minimum amount of DNA acquired through transfer because it does not include sequences obtained from organisms with nucleotide compositions and codon usage patterns closely resembling those of E. coli. These 755 horizontally transferred genes constitute 547.8 kb of DNA and were introduced into the E. coli genome in at least 234 events. Although the phenotypes provided by the majority of these genes are unknown, some of the loci with characterized functions are noted in Fig. 1.
Figure 1.
Distribution of horizontally transferred DNA in the E. coli MG1655 chromosome. Within each centisome, each bar denotes a continuous segment of transferred DNA containing one or more ORFs; and the length of each bar represents its size rounded to the nearest 500 bp. Features of transferred regions, such as duration in the chromosome and the identification of repeated and mobile elements, follow the notation presented in the key. The age of each continuous segment of DNA was inferred from the ages of genes successfully analyzed by back-amelioration; segments lacking genes of known age are shown in black, and no segment comprised genes with significantly different ages. Positions of the replication origin (oriC) and terminus (terC), as well as the identity of the specific tRNA loci found to be adjacent to a horizontally transferred region, are noted on the left of the open bar representing the MG1655 chromosome. The nomenclature for phage and IS elements, and for genes of known function contained within a particular transferred segment, are shown within the corresponding bar. The identities of insertion sequences are noted except as follows: adjacent IS911/(fragment)/IS3 are located within minute 5; adjacent IS3/IS600 are located within minute 8; and adjacent IS2/IS30 are located within minute 31.
Distribution of Horizontally Transferred DNA Within the E. coli Chromosome.
Among strains of E. coli, the segment surrounding the replication terminus displays the most variation in chromosome size and organization, presumably because of elevated levels of recombination in that region (19). The quadrant of the E. coli MG1655 chromosome spanning the replication terminus (minutes 23–47) has experienced a substantially higher number of horizontal transfer events (36% of the 755 events) than has the remainder of the genome, and it also contains the largest amount of horizontally transferred DNA, due principally to the presence of three prophages.
Horizontally transferred genes in the E. coli chromosome often are situated adjacent to a tRNA locus (Fig. 1), which implicates bacteriophages as vehicles for their introduction because several lysogenic coliphages insert preferentially at tRNA loci (20–22). Although tRNA genes are known to serve sporadically as chromosomal insertion sites in bacterial taxa as diverse as Corynebacterium (23), Dichelobacter (24), Haemophilus (25), Helicobacter (26), Mycobacterium (27), Pseudomonas (28), Rhizobium (29), Salmonella (30), and Yersinia (31), the extent of their use as integration sites in E. coli is unexpected. For the majority of transfer events occurring at tRNA loci in the E. coli MG1655 chromosome, there are no remnants of phage-like sequences; however, ORFs corresponding to phage Sf6 and P4 integrases occur immediately downstream of the tRNAargW and tRNAleuX loci, respectively. The leuX locus also has served as the integration site for a 190-kb pathogenicity island detected in a uropathogenic strain of E. coli (32, 33).
Inspection of Fig. 1 demonstrates that 25 of the 37 (68%) IS sequences in this strain are associated with other horizontally transferred DNA, which constitutes only 11% of total chromosome length. This distribution could mean (i) that IS elements are introduced along with horizontally transferred segments, (ii) that the present positions of IS elements in horizontally transferred regions reflect transposition events that were not detrimental to the host, or (iii) that IS elements directly promote in the integration of foreign DNA. Each class of IS elements is associated with acquired DNA to a characteristic degree, thus, for example, six of seven IS2 elements, but none of three IS186 elements, are associated with other acquired DNA—which is not expected if their distribution resulted from simple transposition into horizontally transferred regions. Based on the frequent location of IS elements at the junction between native and transferred DNA, it is most likely that IS elements mediate the insertion of foreign plasmids by replicate-cointegrate formation and did not subsequently insert later into horizontally transferred regions. Therefore, these data support a model whereby mobile genetic elements facilitate the integration of horizontally transferred episomes into the bacterial chromosome, which results in the introduction of novel DNA.
Amelioration of Horizontally Transferred Genes.
Variation in base composition and codon usage patterns among bacterial species (which serves as the basis for identifying horizontally transferred genes) is due largely to biases in the mutation rates at each of the four bases, such that the frequency of G/C to A/T mutations is not same as that of the reverse mutations. These mutational biases, termed “directional mutation pressure” by Sueoka (34–36), vary between species and are apparent in the base composition at each position of codons; the differences in base composition among bacterial species are most pronounced at third codon positions, where the majority of sites are synonymous and closely approximate neutral substitutions.
At the time of introduction, horizontally transferred genes have the base composition and codon usage pattern of the donor genome. But because transferred genes are subject to those mutational processes affecting the recipient genome, the acquired sequences will incur substitutions and eventually come to reflect the DNA composition of the new genome. This process of “amelioration”—whereby a sequence adjusts to the base composition and codon usage of the resident genome—is a function of the relative rate of G/C to A/T mutations. Based on substitution rates estimated for E. coli and the mutational bias of this species, it is possible to predict the amount of time required after transfer for a transferred gene to fully resemble native DNA. And because the first, second, and third position of codons are subject to different selective constraints, the nucleotide composition at each position of horizontally transferred genes ameliorates at a characteristic rate (16).
Variation in the rate of amelioration at each codon position furnishes a property unique to horizontally transferred genes and allows us to estimate the amount of time the gene has been residing in a genome. Whereas recently transferred genes show the patterns of nucleotide composition typical of the donor genome, and fully ameliorated genes show the nucleotide compositions of the recipient E. coli genome, the nucleotide compositions of genes in the process of amelioration do not resemble those of either the donor or recipient genomes. Moreover, because the nucleotide compositions of bacterial genomes result from characteristic patterns of mutations (16, 37), genes in the process of amelioration do not resemble any genome. Because of these mutational processes, the GC content of each codon position of horizontally transferred genes can be “back-ameliorated” until the base compositions at all codon positions fit those typically observed in a bacterial species (16). This permits one to estimate of the amount of time that a horizontally transferred gene has been ameliorating (i.e., residing) in the genome and hence provides the age of acquired sequences.
Rate of Horizontal Transfer in E. coli.
To determine the age of acquired genes in the E. coli MG1655 genome, the 755 genes identified as having been transferred horizontally were sorted into 108 groups based on their GC contents at each codon position. [Genes at the same chromosomal location were sorted into different pools if their nucleotide compositions were sufficiently different to indicate independent origin. For example, the lacI and lacZ genes (≈56% GC) are not pooled with the lacYA genes (≈43% GC) because this gene cluster evidently was assembled from independent sources.] Each of these pools was subjected to reverse amelioration analysis with the use of the substitution rates at synonymous and nonsynonymous sites determined for E. coli (16).
Divergence at synonymous and nonsynonymous sites have been calculated as 94% and 3.9%, respectively (38), which, on the assumption of a divergence time between E. coli and S. enterica of 100 million years (Myr) (39, 40), correspond to rates of 0.47% and 0.0195% per Myr per lineage. Estimates of amelioration rates and durations in the chromosome assume that transferred genes are evolving at the same average rate as are the rest of the genes in the E. coli genome. Highly expressed genes providing essential functions show reduced rates of evolution at both nonsynonymous (because of selection on protein function) and synonymous (because of selection on codon usage) sites. However, horizontally transferred genes would not specify essential functions and are not expected to be highly expressed; therefore, evolution at synonymous sites is not likely to be constrained by selection on codon usage bias. Although it is possible that horizontally transferred genes are under relaxed selection for function—which would increase substitution rates at nonsynonymous sites—allowance for reasonably accelerated rates of evolution resulting from relaxed selection does not significantly alter the results. Therefore, these parameters, as derived by Sharp (38), provide a suitable estimate of the rate of evolution of horizontally transferred DNA.
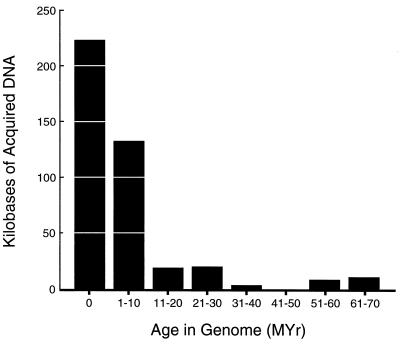
Based on the degree of amelioration, the oldest horizontally transferred genes in the E. coli genome were acquired nearly 100 Myr ago, just after the estimated time of divergence of E. coli and S. enterica; however, most of the transferred DNA has a relatively recent origin in the E. coli chromosome (Fig. 2). Given this distribution, the average age of horizontally transferred genes is 6.7 Myr, which, to a first approximation, yields a rate of accumulation of 64.2 kb/Myr. However, much of the very recently acquired DNA includes IS elements, remnants of prophages, and other sequences that are unlikely to contain genes that are directly advantageous to the host. Also, the age distribution of introduced sequences in E. coli MG1655 also suggests that very few acquired genes are maintained for more than 10 Myr (Fig. 2). Exclusion of such recently acquired sequences, i.e., sequences present for <1 Myr, yields an the average age of horizontally transferred genes of 14.4 Myr. Given the total amount of acquired DNA in the E. coli MG1655 genome, these data predict that horizontally transferred genes have been accumulating at a rate of 16 kb/Myr. Therefore, E. coli chromosome has gained ≈1,600 kb of novel genes through horizontal transfer since diverging from Salmonella enterica ≈100 Myr ago.
Figure 2.
Age distribution of horizontally transferred DNA in the E. coli MG1655 chromosome. Bars represent the amount of protein-coding, horizontally transferred DNA of the specified age present in the 571 genes analyzed by back amelioration. The remaining 184 genes could not be successfully analyzed because of aberrant nucleotide compositions, which could result from horizontal transfer among more than one genome; most of the genes comprising this group are encoded by prophages and mobile genetic elements.
Although chromosome size in natural isolates of E. coli can vary by as much as 1,000 kb (41–44), the E. coli genome has not been steadily increasing in size because chromosome lengths are conserved in E. coli and S. enterica. This implies that, on an evolutionary timescale, increases in genome size due to the acquisition of horizontally transferred sequences are offset by equivalent losses of DNA through deletion. Unlike ancestral genes, most acquired sequences do not confer a long term selective advantage to the host and are, consequently, likely to be lost by deletion. These processes result in an extremely dynamic genome in which substantial amounts of DNA are introduced into (and deleted from) a chromosome, which may effectively change the ecological and pathogenic character of a bacterial species.
Three age classes of horizontally transferred genes are noted in Fig. 1. The age assigned to a horizontally transferred segment reflects the average of its constituent genes; and in almost all cases, the ages of adjacent genes within a region were congruent. As suggested in Fig. 2, most of the horizontally transferred segments are of relatively recent origin (light blue bars in Fig. 1); and these genes are expected to have a variable distribution among E. coli isolates. In contrast, the subset of genes showing evidence of long term amelioration (dark blue bars) are likely to be fixed in the E. coli population, and such genes may well have contributed to the differentiation of E. coli from Salmonella.
Impact of Horizontally Transferred DNA.
The 1,600-kb of horizontally transferred DNA sampled by E. coli since its divergence from S. enterica has included a large number of operons that could provide novel functions immediately on introduction (45). Although point mutations introduce ≈22 kb of variant DNA per Myr (16), the types of information introduced by these processes are very different. Stepwise mutational changes only rarely confer novel functions, whereas traits encoded by acquired DNA will occasionally confer the ability to explore new environments (1). As a result, none of the phenotypic characteristics that distinguish E. coli and S. enterica are attributable to the divergence of homologous genes by mutation; instead, all of the species-specific traits derive from functions encoded by horizontally transferred genes (e.g., lactose utilization, citrate utilization, indole production, propanediol utilization) or from the loss of ancestral DNA (e.g., alkaline phosphatase). Likewise, horizontal transfer has played a significant role in the emergence of pathogenic enteric bacteria (46), and several bacterial pathogens have acquired clusters of virulence genes that display atypical base compositions and reside at tRNA loci (47, 48). These pathogenicity islands are not present in related nonpathogenic species and often encode determinants responsible for establishing specific interactions with a host. Therefore, our results support the hypothesis that the diversification of enteric bacteria into discrete species occurs by a very different process than that proposed for eukaryotes: Bacterial speciation is likely to be driven by a high rate of horizontal transfer, which introduces novel genes, confers beneficial phenotypic capabilities, and permits the rapid exploitation of competitive environments.
Acknowledgments
This work was supported by grants from the National Institutes of Health to H.O. and by grants from the Alfred P. Sloan Foundation and the David and Lucille Packard Foundation to J.G.L.
ABBREVIATION
- Myr
million years
References
- 1.Lawrence J G. Trends Microbiol. 1997;5:355–359. doi: 10.1016/S0966-842X(97)01110-4. [DOI] [PubMed] [Google Scholar]
- 2.Hacker J, Blum-Oehler G, Muhldorfer I, Tschape H. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 3.McDaniel T K, Kaper J B. Mol Microbiol. 1997;23:399–407. doi: 10.1046/j.1365-2958.1997.2311591.x. [DOI] [PubMed] [Google Scholar]
- 4.Groisman E A, Ochman H. Trends Microbiol. 1994;2:289–294. doi: 10.1016/0966-842x(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 5.Falkow S, Tompkins L S, Silver R P, Guerry P, LeBlanc D J. Ann NY Acad Sci. 1971;182:153–171. doi: 10.1111/j.1749-6632.1971.tb30654.x. [DOI] [PubMed] [Google Scholar]
- 6.Ochman H, Groisman E A. In: Molecular Ecology and Evolution: Approaches and Applications. Schierwater B, Streit B, Wagner G P, DeSalle R, editors. New York: Birkhauser; 1994. [Google Scholar]
- 7.Ochman H, Lawrence J G. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. 2nd Ed. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, D.C.: Am. Soc. Microbiol.; 1996. pp. 2627–2637. [Google Scholar]
- 8.Sueoka N, Marmur J, Doty P. Nature (London) 1959;183:1429–1431. doi: 10.1038/1831429a0. [DOI] [PubMed] [Google Scholar]
- 9.Sueoka N. J Mol Biol. 1961;3:31–40. [Google Scholar]
- 10.Sueoka N. Proc Natl Acad Sci USA. 1962;48:582–592. doi: 10.1073/pnas.48.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rolfe R, Meselson M. Proc Natl Acad Sci USA. 1959;45:1039–1042. doi: 10.1073/pnas.45.7.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guerdoux-Jamet P, Hénaut A, Nitschké P, Risler J L, Danchin A. DNA Res. 1997;4:257–265. doi: 10.1093/dnares/4.4.257. [DOI] [PubMed] [Google Scholar]
- 13.Whittam T S, Ake S. In: Mechanisms of Molecular Evolution. Takahata N, Clark A G, editors. Tokyo: Japan Scientific Society Press; 1992. pp. 223–246. [Google Scholar]
- 14.Medigue C, Rouxel T, Vigier P, Henaut A, Danchin A. J Mol Biol. 1991;222:851–856. doi: 10.1016/0022-2836(91)90575-q. [DOI] [PubMed] [Google Scholar]
- 15.Blattner F R, Plunkett G R, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 16.Lawrence J G, Ochman H. J Mol Evol. 1997;44:383–397. doi: 10.1007/pl00006158. [DOI] [PubMed] [Google Scholar]
- 17.Sharp P M, Li W-H. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Perkins J D, Heath J D, Sharma B R, Weinstock G M. J Mol Biol. 1993;232:419–445. doi: 10.1006/jmbi.1993.1401. [DOI] [PubMed] [Google Scholar]
- 20.Cheetham B F, Katz M E. Mol Microbiol. 1995;18:201–208. doi: 10.1111/j.1365-2958.1995.mmi_18020201.x. [DOI] [PubMed] [Google Scholar]
- 21.Pierson L S d, Kahn M L. J Mol Biol. 1987;196:487–496. doi: 10.1016/0022-2836(87)90026-x. [DOI] [PubMed] [Google Scholar]
- 22.Inouye S, Sunshine M G, Six E W, Inouye M. Science. 1991;252:969–971. doi: 10.1126/science.1709758. [DOI] [PubMed] [Google Scholar]
- 23.Ratti G, Covacci A, Rappuoli R. Mol Microbiol. 1997;25:1179–1181. doi: 10.1046/j.1365-2958.1997.5191887.x. [DOI] [PubMed] [Google Scholar]
- 24.Cheetham B F, Tattersall D B, Bloomfield G A, Rood J I, Katz M E. Gene. 1995;162:53–58. doi: 10.1016/0378-1119(95)00315-w. [DOI] [PubMed] [Google Scholar]
- 25.Hauser M A, Scocca J J. J Bacteriol. 1992;174:6674–6677. doi: 10.1128/jb.174.20.6674-6677.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Censini S, Lange C, Xiang Z, Crabtree J E, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. Proc Natl Acad Sci USA. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee M H, Pascopella L, Jacobs W R, Jr, Hatfull G F. Proc Natl Acad Sci USA. 1991;88:3111–3115. doi: 10.1073/pnas.88.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayashi T, Matsumoto H, Ohnishi M, Terawaki Y. Mol Microbiol. 1993;7:657–667. doi: 10.1111/j.1365-2958.1993.tb01157.x. [DOI] [PubMed] [Google Scholar]
- 29.Papp I, Dorgai L, Papp P, Jonas E, Olasz F, Orosz L. Mol Gen Genet. 1993;240:258–264. doi: 10.1007/BF00277064. [DOI] [PubMed] [Google Scholar]
- 30.Blanc-Potard A B, Groisman E A. EMBO J. 1997;16:5376–5385. doi: 10.1093/emboj/16.17.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carniel E, Guilvout I, Prentice M. J Bacteriol. 1996;178:6743–6751. doi: 10.1128/jb.178.23.6743-6751.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blum G, Ott M, Lischewski A, Ritter A, Imrich H, Tschape H, Hacker J. Infect Immunol. 1994;62:606–614. doi: 10.1128/iai.62.2.606-614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ritter A, Gally D L, Olsen P B, Dobrindt U, Friedrich A, Klemm P, Hacker J. Mol Microbiol. 1997;25:871–882. doi: 10.1111/j.1365-2958.1997.mmi517.x. [DOI] [PubMed] [Google Scholar]
- 34.Sueoka N. Proc Natl Acad Sci USA. 1988;85:2653–2657. doi: 10.1073/pnas.85.8.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sueoka N. J Mol Evol. 1992;34:95–114. doi: 10.1007/BF00182387. [DOI] [PubMed] [Google Scholar]
- 36.Sueoka N. J Mol Evol. 1993;37:137–153. doi: 10.1007/BF02407349. [DOI] [PubMed] [Google Scholar]
- 37.Muto A, Osawa S. Proc Natl Acad Sci USA. 1987;84:166–169. doi: 10.1073/pnas.84.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharp P M. J Mol Evol. 1991;33:23–33. doi: 10.1007/BF02100192. [DOI] [PubMed] [Google Scholar]
- 39.Ochman H, Wilson A C. J Mol Evol. 1988;26:74–86. doi: 10.1007/BF02111283. [DOI] [PubMed] [Google Scholar]
- 40.Doolittle R F, Feng D F, Tsang S, Cho G, Little E. Science. 1996;271:470–477. doi: 10.1126/science.271.5248.470. [DOI] [PubMed] [Google Scholar]
- 41.Ochman H, Bergthorsson U. Curr Opin Genet Dev. 1995;5:734–738. doi: 10.1016/0959-437x(95)80005-p. [DOI] [PubMed] [Google Scholar]
- 42.Bergthorsson U, Ochman H. J Bacteriol. 1995;177:5784–5789. doi: 10.1128/jb.177.20.5784-5789.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harsono K D, Caspar C W, Luchansky J B. Appl Environ Microbiol. 1993;59:3141–3144. doi: 10.1128/aem.59.9.3141-3144.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bergthorsson U, Ochman H. Mol Biol Evol. 1998;15:6–16. doi: 10.1093/oxfordjournals.molbev.a025847. [DOI] [PubMed] [Google Scholar]
- 45.Lawrence J G, Roth J R. Genetics. 1996;143:1843–1860. doi: 10.1093/genetics/143.4.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barinaga M. Science. 1996;272:1261–1263. doi: 10.1126/science.272.5266.1261. [DOI] [PubMed] [Google Scholar]
- 47.Groisman E A, Ochman H. Cell. 1996;87:791–794. doi: 10.1016/s0092-8674(00)81985-6. [DOI] [PubMed] [Google Scholar]
- 48.Mecsas J, Strauss E J. Emerg Inf Dis. 1996;2:271–288. doi: 10.3201/eid0204.960403. [DOI] [PMC free article] [PubMed] [Google Scholar]