Abstract
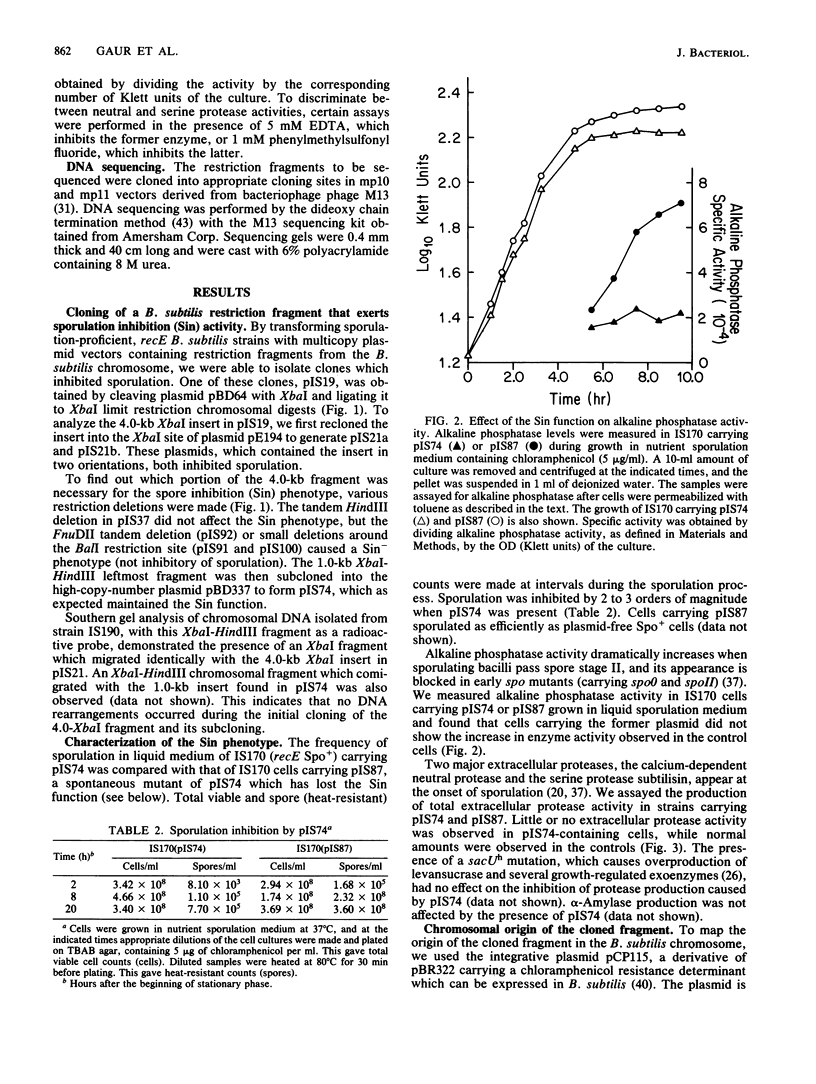
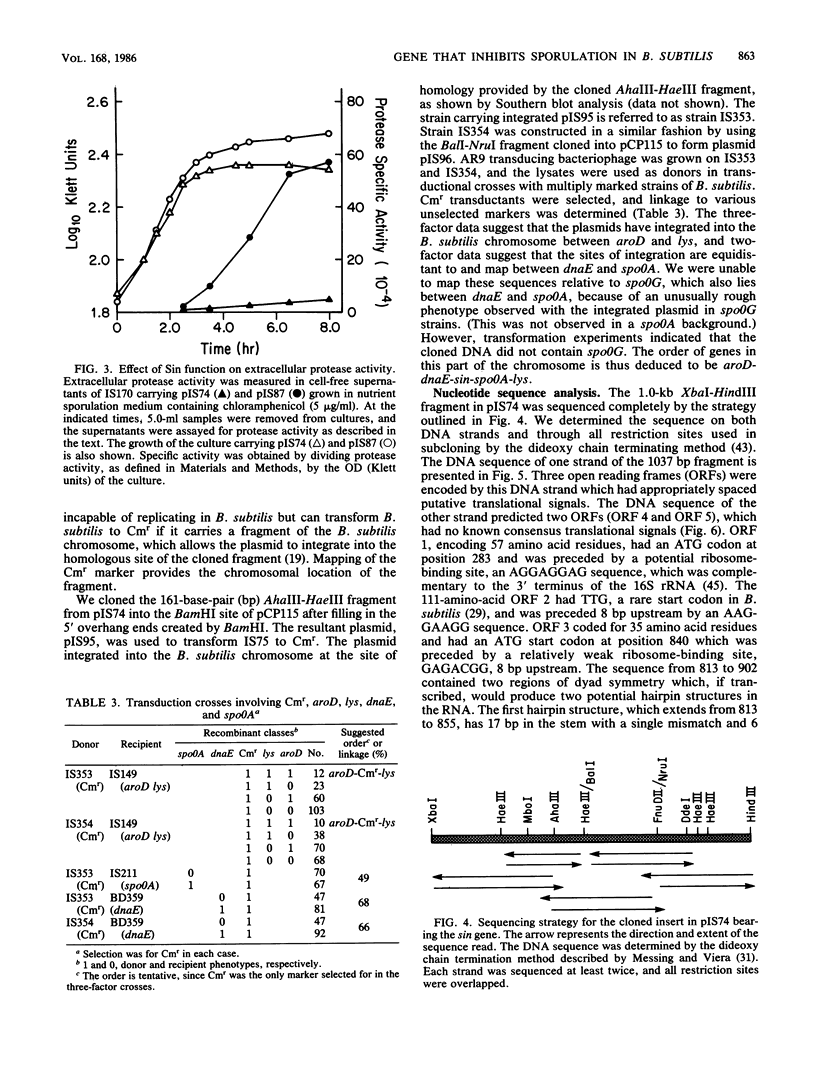
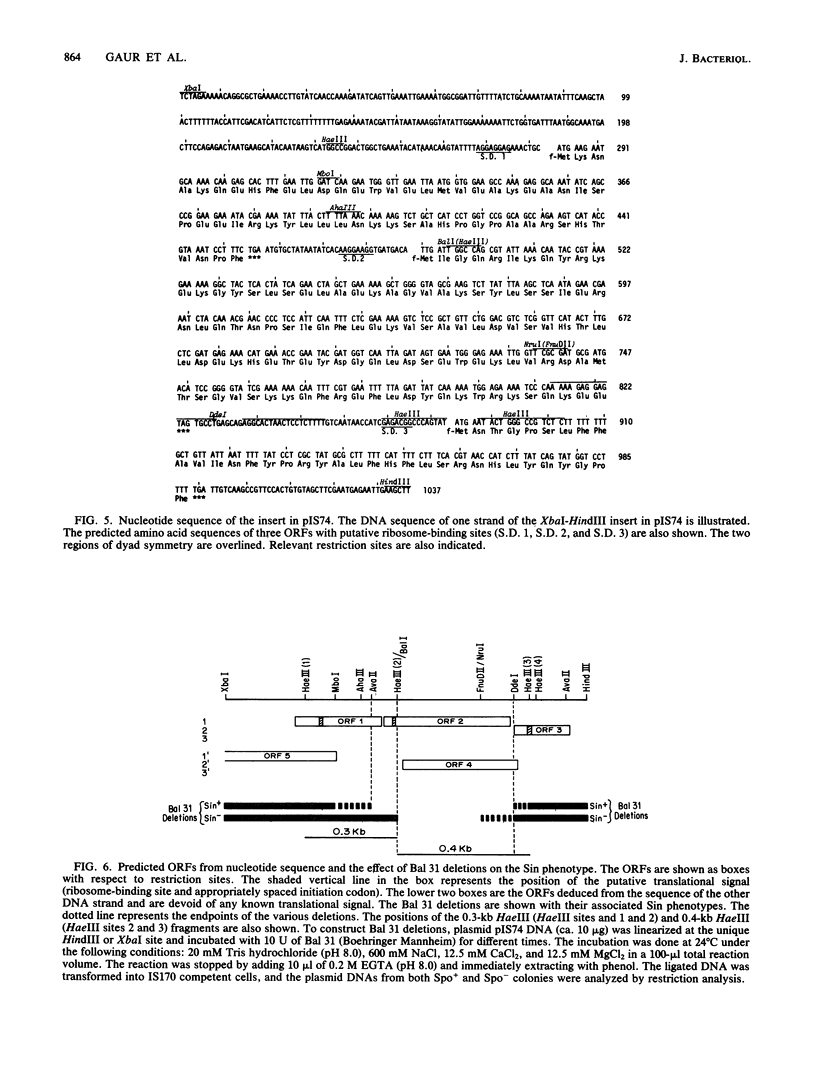
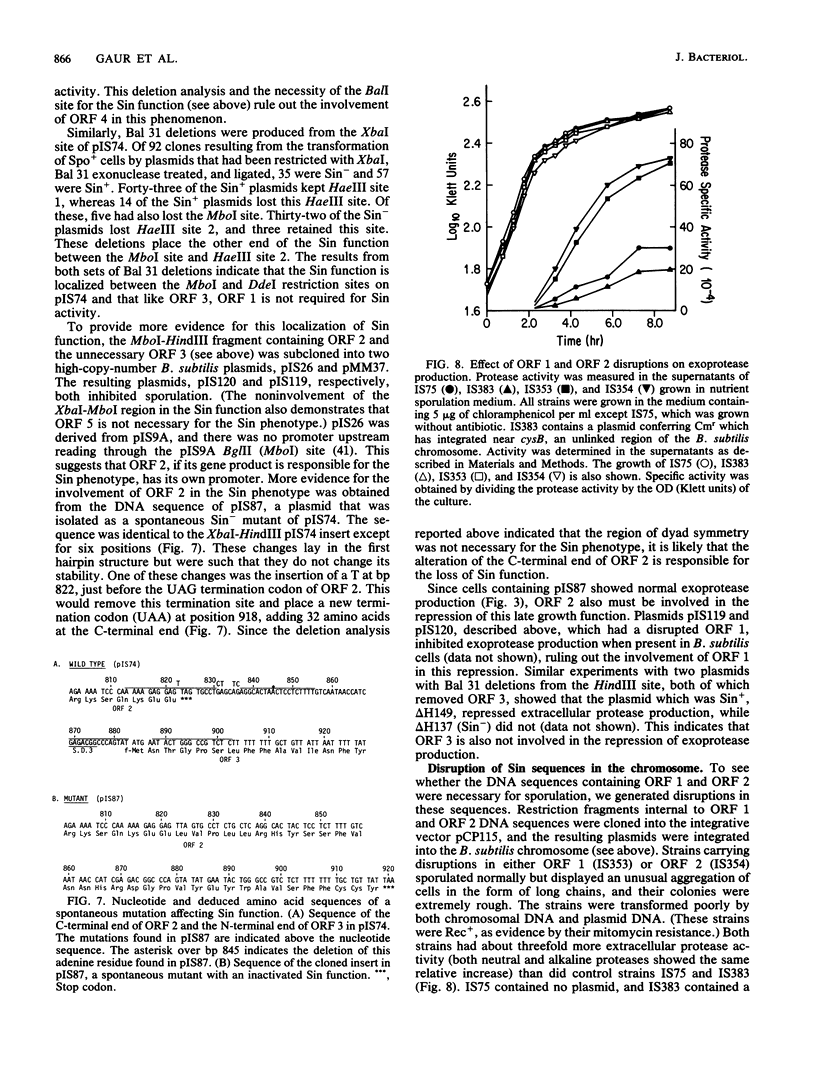
We have isolated a 1.0-kilobase fragment of the Bacillus subtilis chromosome which, when present in high-copy-number plasmids, caused a sporulation-proficient strain to become phenotypically sporulation deficient. This is referred to as the sporulation inhibition (Sin) phenotype. This DNA fragment, in multicopy, also inhibited the production of extracellular protease activity, which normally appears at the beginning of stationary growth. The origin of the fragment was mapped between the dnaE and spo0A genes on the B. subtilis chromosome, and its complete DNA sequence has been determined. By analysis of various deletions and a spontaneous mutant the Sin function was localized to an open reading frame (ORF) predicted from the DNA sequence. Inactivation of this ORF in the chromosome did not affect the ability of cells to sporulate. However, the late-growth-associated production of proteases and alpha-amylase was elevated in these cells. The predicted amino acid sequence of the protein encoded by this ORF had a DNA-binding domain, typically present in several regulatory proteins. We propose that the sin ORF encodes a regulatory protein that is involved in the transition from vegetative growth to sporulation.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anagnostopoulos C., Spizizen J. REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1961 May;81(5):741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert E., Klier A., Rapoport G. Cloning and expression in Escherichia coli of the regulatory sacU gene from Bacillus subtilis. J Bacteriol. 1985 Mar;161(3):1182–1187. doi: 10.1128/jb.161.3.1182-1187.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banner C. D., Moran C. P., Jr, Losick R. Deletion analysis of a complex promoter for a developmentally regulated gene from Bacillus subtilis. J Mol Biol. 1983 Aug 5;168(2):351–365. doi: 10.1016/s0022-2836(83)80023-0. [DOI] [PubMed] [Google Scholar]
- Brehm S. P., Le Hegarat F., Hoch J. A. Deoxyribonucleic acid-binding proteins in vegetative Bacillus subtilis: alterations caused by stage O sporulation mutations. J Bacteriol. 1975 Nov;124(2):977–984. doi: 10.1128/jb.124.2.977-984.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm S. P., Le Hegarat F., Hoch J. A. Developmental modulation of deoxyribonucleic acid-binding proteins of Bacillus subtilis during sporulation stages. J Bacteriol. 1974 Dec;120(3):1443–1450. doi: 10.1128/jb.120.3.1443-1450.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V., Cannon M. C., Cannon F. C. The use of cloned nif (nitrogen fixation) DNA to investigate transcriptional regulation of nif expression in Klebsiella pneumoniae. Mol Gen Genet. 1981;184(1):102–106. doi: 10.1007/BF00271203. [DOI] [PubMed] [Google Scholar]
- Burton Z., Burgess R. R., Lin J., Moore D., Holder S., Gross C. A. The nucleotide sequence of the cloned rpoD gene for the RNA polymerase sigma subunit from E coli K12. Nucleic Acids Res. 1981 Jun 25;9(12):2889–2903. doi: 10.1093/nar/9.12.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Christison J., Martin S. M. Isolation and preliminary characterization of an extracellular protease of Cytophaga sp. Can J Microbiol. 1971 Sep;17(9):1207–1216. doi: 10.1139/m71-193. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancer B. N., Mandelstam J. Production and possible function of serine protease during sporulation of Bacillus subtilis. J Bacteriol. 1975 Feb;121(2):406–410. doi: 10.1128/jb.121.2.406-410.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelberg-Kulka H. UGA suppression by normal tRNA Trp in Escherichia coli: codon context effects. Nucleic Acids Res. 1981 Feb 25;9(4):983–991. doi: 10.1093/nar/9.4.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari E., Howard S. M., Hoch J. A. Effect of stage 0 sporulation mutations on subtilisin expression. J Bacteriol. 1986 Apr;166(1):173–179. doi: 10.1128/jb.166.1.173-179.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitt M. A., Wang L. F., Doi R. H. A strong sequence homology exists between the major RNA polymerase sigma factors of Bacillus subtilis and Escherichia coli. J Biol Chem. 1985 Jun 25;260(12):7178–7185. [PubMed] [Google Scholar]
- Gryczan T. J., Contente S., Dubnau D. Characterization of Staphylococcus aureus plasmids introduced by transformation into Bacillus subtilis. J Bacteriol. 1978 Apr;134(1):318–329. doi: 10.1128/jb.134.1.318-329.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Dubnau D. Construction and properties of chimeric plasmids in Bacillus subtilis. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1428–1432. doi: 10.1073/pnas.75.3.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T., Contente S., Dubnau D. Molecular cloning of heterologous chromosomal DNA by recombination between a plasmid vector and a homologous resident plasmid in Bacillus subtilis. Mol Gen Genet. 1980 Feb;177(3):459–467. doi: 10.1007/BF00271485. [DOI] [PubMed] [Google Scholar]
- Haldenwang W. G., Banner C. D., Ollington J. F., Losick R., Hoch J. A., O'Connor M. B., Sonenshein A. L. Mapping a cloned gene under sporulation control by inserttion of a drug resistance marker into the Bacillus subtilis chromosome. J Bacteriol. 1980 Apr;142(1):90–98. doi: 10.1128/jb.142.1.90-98.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch J. A. Genetics of bacterial sporulation. Adv Genet. 1976;18:69–98. doi: 10.1016/s0065-2660(08)60437-x. [DOI] [PubMed] [Google Scholar]
- Jolliffe L. K., Doyle R. J., Streips U. N. Extracellular proteases modify cell wall turnover in Bacillus subtilis. J Bacteriol. 1980 Mar;141(3):1199–1208. doi: 10.1128/jb.141.3.1199-1208.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landick R., Vaughn V., Lau E. T., VanBogelen R. A., Erickson J. W., Neidhardt F. C. Nucleotide sequence of the heat shock regulatory gene of E. coli suggests its protein product may be a transcription factor. Cell. 1984 Aug;38(1):175–182. doi: 10.1016/0092-8674(84)90538-5. [DOI] [PubMed] [Google Scholar]
- Lepesant J. A., Kunst F., Lepesant-Kejzlarová J., Dedonder R. Chromosomal location of mutations affecting sucrose metabolism in Bacillus subtilis Marburg. Mol Gen Genet. 1972;118(2):135–160. doi: 10.1007/BF00267084. [DOI] [PubMed] [Google Scholar]
- Lewandoski M., Dubnau E., Smith I. Transcriptional regulation of the spo0F gene of Bacillus subtilis. J Bacteriol. 1986 Nov;168(2):870–877. doi: 10.1128/jb.168.2.870-877.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki S., Kobayashi Y. New mutation affecting the synthesis of some membrane proteins and sporulation in Bacillus subtilis. J Bacteriol. 1984 Jul;159(1):228–232. doi: 10.1128/jb.159.1.228-232.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin J. R., Murray C. L., Rabinowitz J. C. Unique features in the ribosome binding site sequence of the gram-positive Staphylococcus aureus beta-lactamase gene. J Biol Chem. 1981 Nov 10;256(21):11283–11291. [PubMed] [Google Scholar]
- Meeks-Wagner D., Wood J. S., Garvik B., Hartwell L. H. Isolation of two genes that affect mitotic chromosome transmission in S. cerevisiae. Cell. 1986 Jan 17;44(1):53–63. doi: 10.1016/0092-8674(86)90484-8. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Mitchell A. P., Herskowitz I. Activation of meiosis and sporulation by repression of the RME1 product in yeast. 1986 Feb 27-Mar 5Nature. 319(6056):738–742. doi: 10.1038/319738a0. [DOI] [PubMed] [Google Scholar]
- Mizuno T., Chou M. Y., Inouye M. A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA). Proc Natl Acad Sci U S A. 1984 Apr;81(7):1966–1970. doi: 10.1073/pnas.81.7.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagami Y., Tanaka T. Molecular cloning and nucleotide sequence of a DNA fragment from Bacillus natto that enhances production of extracellular proteases and levansucrase in Bacillus subtilis. J Bacteriol. 1986 Apr;166(1):20–28. doi: 10.1128/jb.166.1.20-28.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Piggot P. J., Coote J. G. Genetic aspects of bacterial endospore formation. Bacteriol Rev. 1976 Dec;40(4):908–962. doi: 10.1128/br.40.4.908-962.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot P. J., Hoch J. A. Revised genetic linkage map of Bacillus subtilis. Microbiol Rev. 1985 Jun;49(2):158–179. doi: 10.1128/mr.49.2.158-179.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price C. W., Gitt M. A., Doi R. H. Isolation and physical mapping of the gene encoding the major sigma factor of Bacillus subtilis RNA polymerase. Proc Natl Acad Sci U S A. 1983 Jul;80(13):4074–4078. doi: 10.1073/pnas.80.13.4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna N., Dubnau E., Smith I. The complete DNA sequence and regulatory regions of the Bacillus licheniformis spoOH gene. Nucleic Acids Res. 1984 Feb 24;12(4):1779–1790. doi: 10.1093/nar/12.4.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer P., Millet J., Aubert J. P. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci U S A. 1965 Sep;54(3):704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons R. W., Kleckner N. Translational control of IS10 transposition. Cell. 1983 Sep;34(2):683–691. doi: 10.1016/0092-8674(83)90401-4. [DOI] [PubMed] [Google Scholar]
- Smith I., Paress P. Genetic and biochemical characterization of kirromycin resistance mutations in Bacillus subtilis. J Bacteriol. 1978 Sep;135(3):1107–1117. doi: 10.1128/jb.135.3.1107-1117.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stragier P., Bouvier J., Bonamy C., Szulmajster J. A developmental gene product of Bacillus subtilis homologous to the sigma factor of Escherichia coli. Nature. 1984 Nov 22;312(5992):376–378. doi: 10.1038/312376a0. [DOI] [PubMed] [Google Scholar]
- Takahashi I. Catabolite repression-resistant mutants of Bacillus subtilis. Can J Microbiol. 1979 Nov;25(11):1283–1287. doi: 10.1139/m79-202. [DOI] [PubMed] [Google Scholar]
- Trach K. A., Chapman J. W., Piggot P. J., Hoch J. A. Deduced product of the stage 0 sporulation gene spo0F shares homology with the Spo0A, OmpR, and SfrA proteins. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7260–7264. doi: 10.1073/pnas.82.21.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir J., Dubnau E., Ramakrishna N., Smith I. Bacillus subtilis spo0H gene. J Bacteriol. 1984 Feb;157(2):405–412. doi: 10.1128/jb.157.2.405-412.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Ferrari E., Chen E., Henner D. J. Identification of the pleiotropic sacQ gene of Bacillus subtilis. J Bacteriol. 1986 Apr;166(1):113–119. doi: 10.1128/jb.166.1.113-119.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa H., Kazami J., Yamashita S., Chibazakura T., Sone H., Kawamura F., Oda M., Isaka M., Kobayashi Y., Saito H. Revised assignment for the Bacillus subtilis spo0F gene and its homology with spo0A and with two Escherichia coli genes. Nucleic Acids Res. 1986 Jan 24;14(2):1063–1072. doi: 10.1093/nar/14.2.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]


