Abstract
The short filaments extending from the cytoplasmic face of nuclear pore complexes are thought to contain docking sites for nuclear import substrates. One component of these filaments is the large O-linked glycoprotein CAN/Nup214. Immunoprecipitation studies carried out under nondenaturing conditions, and using a variety of antibodies, reveal a novel nonglycosylated nucleoporin, Nup84, that is tightly associated with CAN/Nup214. Consistent with such an association, Nup84 is found to be exposed on the cytoplasmic face of the nuclear pore complex. cDNA sequence analyses indicate that Nup84 contains neither the GLFG nor the XFXFG repeats that are a characteristic of a number of other nuclear pore complex proteins. Secondary structure predictions, however, suggest that Nup84 contains a coiled–coil COOH-terminal domain, a conclusion supported by the observation of significant sequence similarity between this region of the molecule and various members of the tropomyosin family. Mutagenesis and expression studies indicate that the putative coiled–coil domain is required for association with the cytoplasmic face of the nuclear pore complex, whereas it is the NH2-terminal region of Nup84 that contains the site of interaction with CAN/Nup214. These findings suggest a model in which Nup84 may function in the attachment of CAN/Nup214 to the central framework of the nuclear pore complex. In this way, Nup84 could play a central role in the organization of the interface between the pore complex and the cytoplasm.
Nuclear pore complexes (NPCs)1 are large and extremely elaborate structures that mediate the bidirectional traffic of macromolecules across the nuclear envelope (Melchior and Gerace, 1995). Extensive high resolution EM observations of amphibian oocyte NPCs have recently laid the foundations of a consensus model of NPC architecture (for reviews see Goldberg and Allen, 1995; Panté and Aebi, 1994), the central feature of which is a massive symmetrical framework (120 × 80 nm) embedded in the double membranes of the nuclear envelope. This framework appears as eight radial multidomain spokes, connected at their distal ends, and on both their nuclear and cytoplasmic faces, by multisubunit rings (Akey, 1995; Akey and Radermacher, 1993; Hinshaw et al., 1992). The whole assembly embraces a large central gated channel, of as yet ill-defined structure, that may accommodate particles with diameters up to 26 nm, provided that they bear specific nuclear import/export signals. In addition to the central channel, the framework itself contains eight additional peripheral channels that are located in the vicinity of the junction between the inner and outer nuclear membranes. These peripheral channels are thought to permit the free exchange of small molecules (<10 nm in diameter) between the nucleus and the cytoplasm.
In addition to the central framework or ring–spoke complex, NPCs also possess extensive peripheral structures extending into both the cytoplasm and the nuclear interior (Ris, 1991). Projecting from the cytoplasmic ring are eight short (∼100 nm) filaments which are thought to contain docking sites for proteins en route to the nucleus (Panté and Aebi, 1996; Richardson et al., 1988). Protruding from the nucleoplasmic face of the central framework of the NPC are eight 50–100-nm-long filaments joined at their distal ends by a 30–50-nm-diameter ring. Together these form a structure resembling a basket or fishtrap, the function of which is currently unclear. It may however, by analogy with the cytoplasmic filaments, contain docking sites for translocating molecules (Bastos et al., 1996). From a morphological and probably functional perspective, the effect of these peripheral structures is to endow the NPC with an overall asymmetry about an axis parallel to the plane of the nuclear membranes.
Biophysical analyses of Xenopus oocyte NPCs have indicated that they have a total mass of about 125 MD (Reichelt et al., 1990), 30 times that of a ribosome. It has been suggested that the NPC may be composed of as many as 100 different protein subunits based partly on these findings. During the past few years a number of these proteins have been identified and characterized at the molecular level. However, even assuming quite generous stoichiometries, these can account for only a minor fraction of the vertebrate NPC mass (for reviews see Bastos et al., 1995; Rout and Wente, 1994). For the majority of the known vertebrate NPC proteins or nucleoporins, only sketchy information is available concerning their precise location within the NPC as well as the nature of their interactions with neighboring subunits. Nevertheless, since the NPC functions vectorially, this type of information is essential if we are to gain a clear understanding of the mechanisms of macromolecular translocation across the nuclear envelope.
We have previously described a protein complex that may be released directly from vertebrate NPCs and that contains two dissimilar subunits, a 250-kD protein modified with O-linked N-acetylglucosamine (O-GlcNAC) and a carbohydrate-free 75-kD protein (Panté et al., 1994). The estimated mass of this heterooligomer is ∼1–2 MD. It is now clear that the 250-kD glycoprotein is identical to a potential oncoprotein called CAN (Bastos, R., and B. Burke, unpublished results), rearrangements in the gene for which are associated certain leukemias (von Lindern et al., 1992). CAN was subsequently found to correspond to Nup214, an NPC protein (Kraemer et al., 1994). Using high resolution immunogold electron microscopy, CAN/ Nup214 has been localized within the NPC to the short filaments that extend from the cytoplasmic face of the ring– spoke complex (Panté et al., 1994). More recently, gene targeting experiments in embryonic stem cells have demonstrated that CAN/Nup214 is an essential NPC component (van Deursen et al., 1996). Early mouse embryos that are homozygous for the disrupted CAN/Nup214 allele are inviable and exhibit impaired nucleocytoplasmic transport accompanied by cell cycle arrest. At the same time these embryos exhibit morphologically identifiable NPCs that contain other members of the O-linked NPC glycoprotein family. This latter observation is consistent with the peripheral localization of CAN/Nup214 as well as earlier studies concerning the role of O-linked glycoproteins in NPC function and assembly (Finlay and Forbes, 1990). In this paper, we report the molecular characterization of the 75-kD protein associated with CAN/Nup214. The cDNA encodes a novel nucleoporin with a calculated molecular mass of 84 kD. Therefore, following the current nomenclature convention it will be referred to as Nup84. It does not belong to the O-linked glycoprotein family of nuclear pore complex proteins and it does not contain the characteristic XFXFG or GLFG repeats that are observed in other nucleoporins of both higher and lower cells. The predicted structure of this molecule combined with expression and deletion analyses suggests that it could be involved in the attachment of CAN/Nup214 to the cytoplasmic face of the NPC.
Materials and Methods
Cell Culture
BHK, BHKgrβ, NRK, and HeLa cells were maintained in a 7.5% CO2 atmosphere at 37°C. All of the cells were cultured in DME (GIBCO BRL, Gaithersburg, MD) containing 10% fetal bovine serum (HyClone, Logan, UT), 100 μg/ml of penicillin/streptomycin (GIBCO BRL) and 2 mM glutamine (GIBCO BRL).
Antibodies
The QE5 monoclonal antibody and a guinea pig polyclonal antibody that recognizes CAN/Nup214 have been described by Panté et al. (1994). For QE5 immunoadsorption experiments an affinity matrix of QE5 IgG covalently coupled to protein G-Sepharose using dimethylpimelimidate was prepared following published procedures (Harlow and Lane, 1988). An antibody against Nup84 was raised in rabbits by BAbCO (Berkeley, CA) using the synthetic peptide QINDIRNHVNF, which corresponds to the last 11 residues of the COOH terminus deduced from the cDNA sequence. The rabbit antipeptide antibody specific for human lamin A has been previously described (Burke, 1990). Human autoantibodies against the nuclear pore complex protein gp210 were gifts from Dr. Jean-Claude Courvalin (Institute Jacques Monod, Paris) and Dr. Marvin Fritzler (University of Calgary, Alberta, Canada). The polyclonal rabbit antibodies raised against recombinant importin-α and importin-β were a gift from Dr. Manfred Lohka (University of Calgary, Alberta, Canada) and Dr. Aurelian Radu (The Rockefeller University, New York), respectively. The monoclonal antibody against LGP120 was provided by Dr. Jean Gruenberg (University of Geneva, Switzerland). The anti-HA monoclonal antibody (12CA5) was obtained from BAbCO and the polyclonal rabbit antiHA antibody was provided by Medical and Biological Laboratories (Watertown, MA). The anti-cMyc monoclonal antibody (9E10) was obtained from the American Type Culture Collection (Rockville, MD). The rhodamine and FITC-conjugated secondary antibodies were purchased from Tago, Inc. (Burlingame, CA). For use in double label experiments, the secondary antibodies were cross adsorbed, as appropriate, against either mouse or rabbit IgG covalently coupled to cyanogen bromide-activated Sepharose CL 4B.
Purification of Nup84 and Protein Sequencing
Nup84 was purified from high salt/detergent extracts of rat liver nuclear envelopes using a QE5 affinity matrix prepared as described above. Briefly, nuclei were isolated from 30-g batches of rat livers according to published methods (Blobel and Potter, 1966) and, from these, nuclear envelopes (NE) were prepared after DNase and RNase digestion (Dwyer and Blobel, 1976). NEs were then extracted in a solution containing: 50 mM triethanolamine, pH 7.4, 500 mM NaCl, 0.5% Triton X-100, 1 mM DTT, and protease inhibitors. The soluble material was adsorbed to QE5 IgG cross-linked to protein G-Sepharose beads and, after extensive washes in the same solution, adsorbed proteins were eluted with SDSPAGE sample buffer. Typically, 4–10 μg of Nup84 protein (on the order of 50–100 pmol) could be recovered from about 100 g of liver tissue, sufficient to obtain reliable internal sequence information.
For internal amino acid sequence analysis the procedure described in (Aebersold et al., 1987) was followed. In brief, the eluate was electrophoretically fractionated and electroblotted onto a nitrocellulose membrane and protein bands were identified by Ponceau S staining. The band corresponding to Nup84 was excised and digested in situ with trypsin. The resulting peptides were separated by micro-bore reverse-phase HPLC and sequenced in a gas-phase sequenator. All of these operations on the membrane-adsorbed protein were carried out by the Harvard University Micro-Chemistry Facility (Cambridge, MA) under the direction of Dr. W. Lane.
Cloning of cDNAs Encoding Rat and Human Nup84
One of the three peptide sequences obtained (P1) was used to design a “guessmer” of 45 bp. This is a single oligonucleotide where the substitutions at each position of ambiguity were chosen on the statistical basis of known codon utilization in the species of interest (Lathe, 1985).To clone the rat cDNA, the 45-mer was end-labeled with γ-[32P]ATP (New England Nuclear, Boston, MA) using polynucleotide kinase (New England Biolabs, Beverly, MA) and used to screen about a half million recombinants of a rat thyroid Uni-ZAP library (Stratagene, La Jolla, CA). Plaque lifts were prepared following established procedures and prehybridized at 48°C for 2–3 h in a solution containing 6× SSC and 1% Sarkosyl (ICN Biochemicals, Cleveland, OH). Hybridization was performed under the same conditions for 24 h. Filters were washed several times in the same solution at increasing temperatures up to 65°C. A single positive clone was plaque purified and pBluescript SK (−) containing the cDNA insert was excised in vivo, rescued, and sequenced.
The full-length sequence was obtained using the 5′-AmpliFINDER RACE Kit (Clontech, Palo Alto, CA). Poly (A)+ RNA for this experiment was prepared from NRK cells using the FAST TRACK mRNA isolation kit (Invitrogen, San Francisco, CA). The ZAP and the RACE clones both cloned in pBluescript SK (−) were pasted together using a convenient SphI site in the overlapping region.
To clone the human homologue of Nup84, a lambda gt10 human T cell library (generously provided by Dr. Frank McKeon, Harvard Medical School, Boston, MA) was screened with the 500-bp EcoRI-NsiI cDNA fragment corresponding to the 5′ region of the original rat clone. Hybridizations were carried out overnight at 42°C in a solution containing 5× SSC, 50% formamide, 5× Denhardt's solution, 0.5% SDS, and 100 μg/ml denatured salmon sperm DNA. Filters were washed several times in 2× SSC/0.5% SDS at the same temperature and once in 1× SSC/0.5% SDS at 52°C. Two positive clones were plaque purified. Phage DNA was prepared from 50-ml lysates and cDNA inserts were subcloned into pBS and sequenced.
Sequence Analysis and Prediction of Secondary Structure
The complete sequence of the cDNA was carried out by PCR using fluorescently tagged dideoxynucleoside terminators by the Biopolymer Facility at the Harvard Medical School. Sequence database searching was performed using BLAST (Altschul et al., 1990). The sequence alignments were accomplished employing the pair-wise alignment program PILEUP (GCG package). For the secondary structure predictions the neural network method implemented by the program PHD (Rost and Sander, 1993) was used.
Northern Blot Analysis and In Vitro Translation Assay
A multiple rat tissue Northern blot containing 2 μg of poly A+ per lane was probed with a 500-bp EcoRI-NsiI fragment from the original rat cDNA clone according to the manufacturer's procedure (Clontech). For the in vitro translation assay, the full-length cDNA of Nup84 (EcoRIXhoI) was subcloned into pSP73 (Promega, Madison, WI). 1 μg of pSP73Nup84 was in vitro transcribed using the SP6 polymerase and translated using a TNT-coupled reticulocyte lysate system (Promega) according to the manufacturer's protocol in a final volume of 25 μl per assay.
Immunofluorescence Microscopy
Cells grown on glass coverslips were fixed with formaldehyde and labeled with antibodies according to previously described procedures (Ash et al., 1977). In short, cells were fixed for 20 min at room temperature in 3% formaldehyde (prepared from paraformaldehyde dissolved at 80°C in phosphate-buffered saline). After PBS washes, the fixed cells were permeabilized for 5 min at room temperature with 0.2% Triton X-100 in PBS and labeled with appropriate primary and secondary antibodies. In addition some samples were stained with the DNA-specific Hoechst dye 33258 (Hoechst, La Jolla, CA) to reveal the cell nuclei. For differential permeabilization experiments (in which only the cytoplasmic face of the nuclear envelope is exposed to antibody) cells were instead treated for 15 min at 4°C with 0.004% digitonin in PBS (Panté et al., 1994). The digitonin was prepared from a 10% stock solution in dimethyl sulfoxide. Integrity of the nuclear membranes in digitonin permeabilized cells was always confirmed using a rabbit anti–lamin A antibody (Burke, 1990). Specimens were observed and photographed under appropriate illumination with either a Zeiss Axiophot microscope equipped with a X63 PlanApo objective lens or a Leica DMRB microscope equipped with a X63 PL APO NA1.4 objective.
Immunoprecipitations and Immunoblotting
For long-term labeling, transfected (below) or mock transfected cells grown in 35-mm tissue culture dishes were labeled overnight with 50 μCi 35S-Trans label (ICN) in 1 ml growth medium containing methionine and cysteine at 10% of their normal concentrations. Immunoprecipitation analysis was carried out as we have previously described. In short, the cells were washed once in PBS and then lysed in a buffer (TNX) containing 50 mM triethanolamine (TEA), 500 mM NaCl, 0.5% Triton X-100, 1 mM DTT, 1 mM PMSF, and 1:1,000 CLAP (10 mg/ml in DMSO of each of the following, chymostatin, leupeptin, antipain, and pepstatin).The lysate was centrifuged for 10 min at full speed in an Eppendorf cooled to 4°C. To the supernatant was added either 20 μl of a QE5 affinity matrix consisting of a 50% suspension of protein G-Sepharose to which QE5 IgG had been chemically cross-linked (above) or 5 μl of appropriate antiserum/ascites fluid and 20 μl protein A-Sepharose (50% suspension). The mixture was then rotated overnight at 4°C. The following morning the QE5 beads were washed five times in either the same buffer or in a higher stringency buffer containing 50 mM TEA, 100 mM NaCl, 0.5% Triton X-100, 0.1% SDS, 1 mM DTT, 1 mM PMSF, and 1:1,000 CLAP (Panté et al., 1994). After two final washes in 50 mM Tris, pH 7.4, the QE5 beads were suspended in SDS-polyacrylamide gel sample buffer and fractionated by electrophoresis (Laemmli, 1970). On completion of electrophoresis, gels, where necessary, were blotted onto nitrocellulose filters (BA85; Schleicher and Schuell, Keene, NH [Burnette, 1981]) using a semi-dry blotting apparatus manufactured by Hoeffer Scientific Instruments, Inc. (San Francisco, CA). Filters were blocked, labeled with primary antibodies, and then developed with peroxidase conjugated secondary antibodies exactly as previously described (Burke et al., 1982). Alternatively, radioactive gels were examined by fluorography following impregnation with Amplify (Amersham, Arlington Heights, IL) and exposure to Kodak X-Omat AR x-ray film at −70°C.
For pulse-chase experiments, BHK cells in 35-mm petri dishes were preincubated for 10 min at 37°C in DME lacking methionine and cysteine. The cells were subsequently labeled for periods up to 30 min in 0.5 ml of methionine- and cysteine-free DME containing 5% dialyzed (against PBS) fetal bovine serum and 60 μCi 35S-Trans label. For the chase (where appropriate), 1 ml of complete BHK growth medium, containing 10 times the normal concentration of methionine and cysteine, was added directly to the cells and the incubation continued for the desired time. Pulse labeled cells were then lysed on ice in 1 ml of a low salt solution containing 50 mM TEA, 0.5% Triton X-100, 1 mM DTT, 1 mM PMSF and 1:1,000 CLAP. The lysed cells were centrifuged for 2 min at 3,000 rpm in an Eppendorf centrifuge maintained at 4°C. The supernatant was removed and NaCl added to a final concentration of 500 mM from a 4 M stock. The pellet was reextracted in 1 ml of the high salt TNX buffer (above). Both the low salt supernatant and the high salt pellet extract were cleared by centrifugation in the cold for 10 min at full speed in an Eppendorf centrifuge. The samples were subsequently processed for immunoprecipitation as described above.
Expression of Rat Nup84 and CAN/Nup214 in Mammalian Cells
The cDNA coding for the rat Nup84 was cloned into the mammalian CMV driven expression vector pcDNA3 (Invitrogen) containing a 200-bp 5′-untranslated sequence from human lamin A and either an NH2-terminal HA or cMyc epitope tag. The cloning strategy took advantage of the polymerase chain reaction and utilized oligonucleotide primers incorporating suitable restriction sites at their 5′ ends (Xho1 or Cla1). The HAtagged construct of CAN/Nup214 was kindly provided by Dr. Gerard Grosveld (St. Jude Children's Research Hospital, Memphis, TN). The HA-tagged Nup153 construct has been previously described (Bastos et al., 1996). For immunofluorescence experiments, typically 1 μg of cesium chloride-purified plasmid DNA was introduced into cells growing on coverslips using the calcium phosphate method. For immunoprecipitation and immunoblot analysis the DNA was introduced using the Lipofectamine reagent exactly as described by the manufacturer (GIBCO BRL).
Results
Identification, Purification, and cDNA Cloning of Nup84
We have previously described a monoclonal antibody (QE5) that recognizes three members of the O-linked glycoprotein family of nuclear pore complex proteins: p62, Nup153, and CAN/Nup214 (Panté et al., 1994). However, QE5 immunoprecipitates prepared under nondenaturing conditions were found to contain four additional polypeptides of apparent molecular mass of 75, 58, 54, and 45 kD (Panté et al., 1994). p58, p54, and p45 had been shown by others to be associated with p62 (Finlay et al., 1991; Guan et al., 1995; Kita et al., 1993), while p75 appeared to be a novel protein and was associated with CAN/Nup214 in a large (1–2 MD) complex. In view of the location of CAN/ Nup214 in the cytoplasmic filaments of the NPC, this 75kD protein was predicted to be a nucleoporin exposed on the cytoplasmic face of the NPC (Panté et al., 1994).
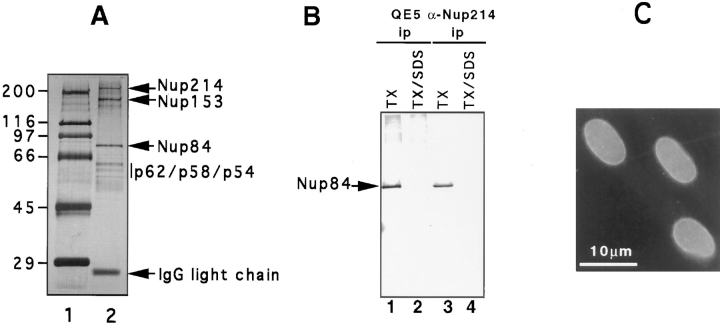
To further characterize this protein we took advantage of its association with CAN/Nup214 and hence its appearance in QE5 immunoprecipitates. An affinity matrix of QE5 IgG covalently attached to protein G-Sepharose beads was used to immunoadsorb high salt/detergent extracts of purified rat liver nuclear envelopes. The immunoadsorbed material contained six out of the seven proteins described above in roughly stoichiometric quantities (the wash conditions used here tend to strip p45 from the immunoprecipitate) and is documented in a pilot experiment shown in Fig. 1 A. Large scale QE5 immunoprecipitates prepared in this way were fractionated by electrophoresis to yield microgram quantities of gel purified protein that was subsequently used for amino acid sequence analysis. Typically, between 50 and 100 pmol of the 75-kD protein was obtained from ∼100 g of rat liver tissue.
Figure 1.
(A) Nucleoporins immunoadsorbed from rat liver nuclear envelopes using QE5-Sepharose beads (lane 2). The material shown is derived from 1 to 2 g of rat liver and is revealed by silver staining. Approximately 50– 100× this quantity was used for limited amino acid sequence analysis of Nup84. Molecular weight markers (indicated in kD) are shown in lane 1. (B) Western blot analysis of QE5 and antiNup214 immunoprecipitates of NRK cells using an antipeptide antibody against Nup84. Immunoprecipitates were washed either in a low stringency buffer containing 0.5% Triton X-100 (TX, lanes 1 and 3) that preserves the Nup214–Nup84 interaction, or in a high stringency buffer containing 0.5% Triton X-100 plus 0.1% SDS (TX/SDS, lanes 2 and 4). This latter buffer breaks the interaction between Nup214 and Nup84 (Panté et al., 1994). (C) Indirect immunofluorescence labeling of NRK cells using the anti-peptide antibody against Nup84. The nuclear envelope is clearly labeled.
We obtained three endoproteolytic fragments of the 75-kD protein. One of these peptides (highlighted sequence in Fig. 2) was long enough for us to design a “guessmer” of 45 nucleotides that was then used to screen a rat thyroid lambda Uni-ZAP cDNA library. A single clone of 2,056-bp was isolated containing a single continuous open reading frame of 1,833-bp with a TAG termination codon followed by a 3′-untranslated region containing a polyadenylation signal as well as a poly-A tail. This open reading frame contained all three peptide sequences obtained directly from the 75-kD protein (underlined sequences in Fig. 2). This clone did not, however, contain the complete cDNA. A Northern blot analysis performed using this clone as a probe hybridized with a single transcript of ∼2.6 kb. Although variable in its expression levels, it is found in all tissues as one would expect for a nucleoporin (data not shown). The size of the transcript and the lack of an ATG initiation codon indicated that we were missing around 400–500 bp from the 5′-end. To obtain the full-length cDNA we carried out a modified version of the RACE (rapid amplification of cDNA ends) technique (Edwards et al., 1991). A clone of about 600-bp containing a single putative ATG initiation codon (Kozak, 1987) in the correct reading frame was finally obtained.
Figure 2.

Complete amino acid sequence of Nup84 derived from cDNA sequence analysis. The boxed segments correspond to the COOH-terminal predicted coiled–coil regions, which are most closely related to members of the tropomyosin family. The primary structure of the underlined peptides was initially elucidated by limited amino acid sequence analysis of Nup84 tryptic fragments. The highlighted peptide (P1), also obtained by amino acid sequence analysis, was used in the design of the oligonucleotide employed in the isolation of the cDNA. The GenBank accession number of the complete cDNA sequence is U93692.
The full-length cDNA encodes a protein (Fig. 2) composed of 742 amino acid residues with a calculated molecular mass of 83,531. Since it is a nuclear pore complex protein (discussed in more detail below), it will be referred to as Nup84 following the current nomenclature convention. It has a calculated pI of 6.04, very similar to that estimated by two-dimensional gel electrophoresis (isoelectric focusing) of the QE5 immunoprecipitate. When translated in vitro, the full-length cDNA produced a single product of the expected size (data not shown).
Nup84 Is A Novel Nucleoporin That Contains Neither XFXG Nor GLFG Repeats and Has a Predicted Coiled–Coil COOH Terminus
A BLAST search with the Nup84 sequence revealed that it is a novel protein of bipartite primary structure. The COOH-terminal third of the Nup84 polypeptide, while unique, does exhibit significant sequence similarity to a number of coiled–coil molecules, most notably to the members of the tropomyosin family, for which an experimental three-dimensional model is available (Phillips et al., 1986). The identity between rat brain tropomyosin (Lees-Miller et al., 1990) and residues 500–742 of Nup84 is about 25%, with similarity scores up to 50% (Fig. 2). These values are strong indicators of a common structure and suggest that the COOH-terminal domain of Nup84 forms a coiled–coil structure (Sander and Schneider, 1991). Consistent with the sequence comparison data, a general agreement was found between the secondary structure predicted for the Nup84 COOH-terminal region and the three-dimensional structure of tropomyosin (Fig. 2). The NH2-terminal region of Nup84 displays no significant similarity to any other known protein sequences. Secondary structure predictions for this part of the polypeptide suggest a globular domain with a balanced presence of helices and B-strands. Finally, Nup84 contains neither the XFXFG nor the GLFG repeats that have been found in a number of other nucleoporins, including its partner, CAN/Nup214. While this manuscript was under review, Fornerod et al. (1997) described Nup88, a human CAN/Nup214-associated protein. It is clear that Nup88 represents the human homologue of rat Nup84 with which it shares 89% sequence identity.
Nup84 Is Associated with CAN/Nup214
To confirm that Nup84 does indeed correspond to the 75-kD protein identified in nondenaturing QE5 immunoprecipitates, we raised an antibody against a COOH-terminal peptide predicted from the Nup84 cDNA sequence. When immunoblots of QE5 immunoprecipitates from NRK cell extracts were probed with the antipeptide serum, a single band of the appropriate size was recognized (Fig. 1 B, lane 1). In contrast, no signal was observed if immunoprecipitates were subjected to stringent wash conditions that break the interaction between CAN/Nup214 and its associated protein (Fig. 1 B, lane 2). Similar analyses of immunoprecipitates carried out with an anti-CAN/Nup214 polyclonal antibody rather than with QE5 gave identical results (Fig. 1 B, lanes 3 and 4). While this Nup84 antibody was not ideal for immunocytochemistry, immunofluorescence analysis of lightly fixed NRK cells revealed labeling of the nuclear envelope, consistent with an association of Nup84 with CAN/Nup214 (Fig. 1 C).
To better examine the intracellular distribution of Nup84, an influenza virus hemagglutinin epitope tag was placed at its NH2 terminus and the protein expressed in BHK cells. Immunofluorescence microscopy using an anti-HA antibody revealed a labeling pattern consistent with association of HA-Nup84 with NPCs. At low levels of expression there is clear punctate staining of the nuclear envelope (Fig. 3 A), indistinguishable from what is seen with expressed HA-CAN/Nup214 (Fig. 3 B) as well as with HANup153 as we have recently reported (Bastos et al., 1996). An identical labeling pattern is also observed with the antinucleoporin monoclonal antibody QE5 (Bastos et al., 1996; Panté et al., 1994). At higher expression levels, the progressive accumulation of both proteins in the cytoplasm becomes apparent (Fig. 3, C and D). As will be expanded upon below, under these conditions Nup84 and CAN/Nup214 exhibit quite divergent behavior.
Figure 3.
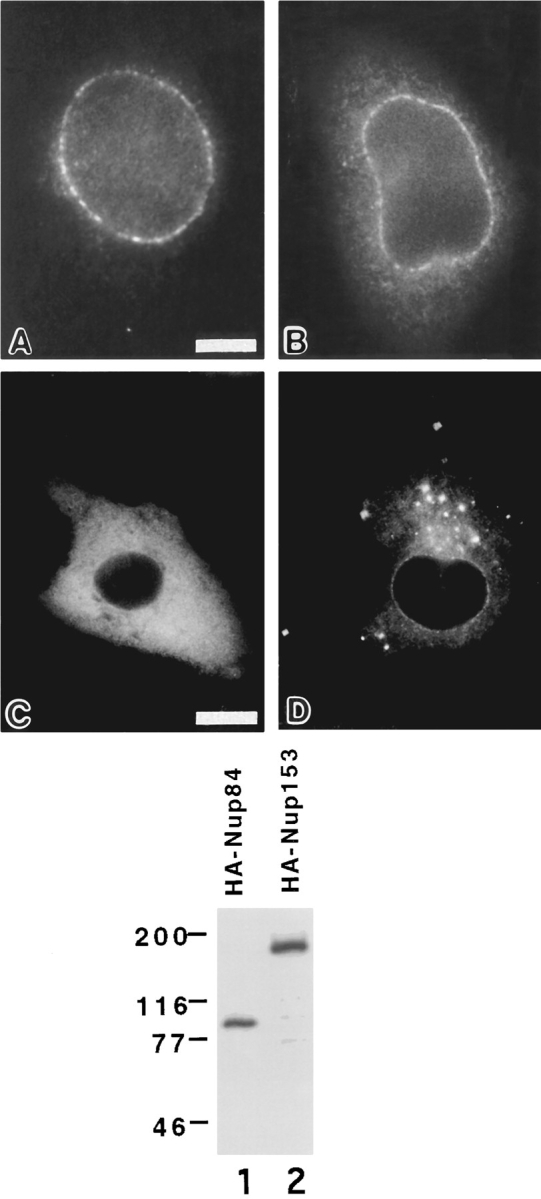
Indirect immunofluorescence analysis of BHK cells expressing HA-Nup84 (A, C) or HA-Nup214 (B, D) at either low (A, B) or high (C, D) levels. The monoclonal antibody used is 12CA5, which recognizes the HA epitope. At low expression levels both proteins localize to the nuclear envelope, whereas at higher levels they spill over into the cytoplasm. At these higher levels their localizations appear quite distinct. While the cytoplasmic distribution of HA-Nup84 is largely uniform, HA-Nup214 tends to concentrate in unusual cytoplasmic foci. The immunoblot is of low stringency QE5 immunoprecipitates of BHK cells expressing either HA-Nup84 (lane 1) or HA-Nup153 (lane 2). The blot is probed with the anti-HA antibody (12CA5). Since HA-Nup84 is not recognized by QE5, its appearance in the immunoprecipitate must be by virtue of its association with CAN/ Nup214. HA-Nup153 (lane 2) provided a positive control for the QE5 immunoprecipitation (the monoclonal antibody QE5 recognizes Nup153 and p62 in addition to CAN/Nup214). Bars: (A, B) 5 μm and (C, D) 10 μm.
A further experiment was performed to determine if the expressed HA-Nup84 associated with endogenous CAN/ Nup214. For this purpose detergent (Triton X-100) extracts were prepared from BHK cells expressing HANup84. These extracts were then immunoprecipitated with QE5 and analyzed by Western blot employing the 12CA5 anti-HA monoclonal antibody. As shown in Fig. 3 (lane 1), HA-Nup84 was clearly detectable in the immunoprecipitate. Since Nup84 does not contain an epitope recognized by QE5, its presence in the immunoprecipitate has to be due to the interaction with CAN/Nup214. The results described here (Figs. 1 and 3), taken together with previously published biochemical data (Panté et al., 1994), confirm that Nup84 and CAN/Nup214 are physically associated.
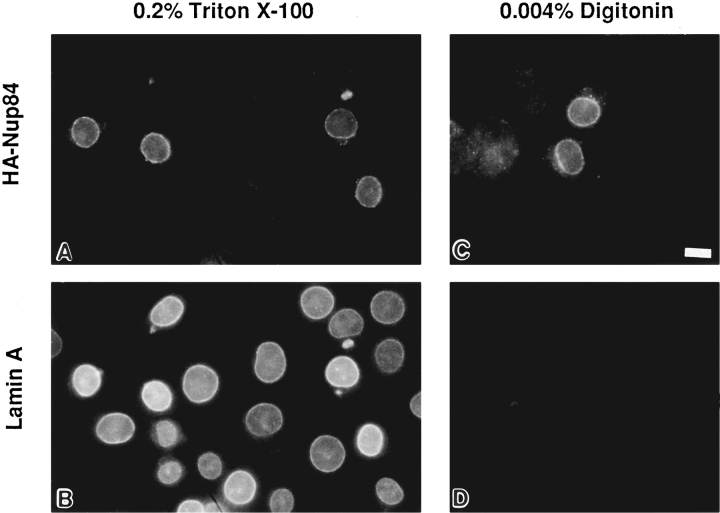
Since CAN/Nup214 is a component of the cytoplasmic filaments of the NPC we would expect to find that its associated protein, Nup84, also faces the cytoplasm. To determine whether this is the case, we performed immunofluorescence microscopy on BHK cells, expressing HA-Nup84, which were permeabilized with 0.004% digitonin on ice. Under these conditions, the plasma membrane is permeabilized but the NE remains intact (Adam et al., 1990). Therefore only those proteins on the cytoplasmic face of the NPC can be accessible to the antibodies. As illustrated in Fig. 4, the digitonin permeabilized cells show clear nuclear envelope labeling when the anti-HA antibody was used (Fig. 4 C), but no labeling at all when an anti–lamin A antibody was used (Fig. 4 D). This finding strongly suggests that Nup84 is indeed exposed on the cytoplasmic face of the NPC.
Figure 4.
Differential permeabilization of BHK cells transiently expressing HA-Nup84. Cells are double labeled with the monoclonal anti-HA (12CA5, A and C) and a rabbit anti-peptide antibody against lamin A (B and D). While 0.2% Triton X-100 (A, B) permeabilizes both the plasma membrane and the nuclear membranes and allows labeling by both antibodies, 0.004% digitonin selectively permeabilizes only the plasma membrane. In this case, HA-Nup84 is detected at the nuclear periphery but lamin A, which is localized on the nucleoplasmic face of the nuclear envelope, remains unlabeled. These results indicate that HA-Nup84 is exposed on the cytoplasmic face of the nuclear envelope. Bar, 10 μm.
Nup84 and CAN/Nup214 Associate prior to Their Stable Integration into NPCs
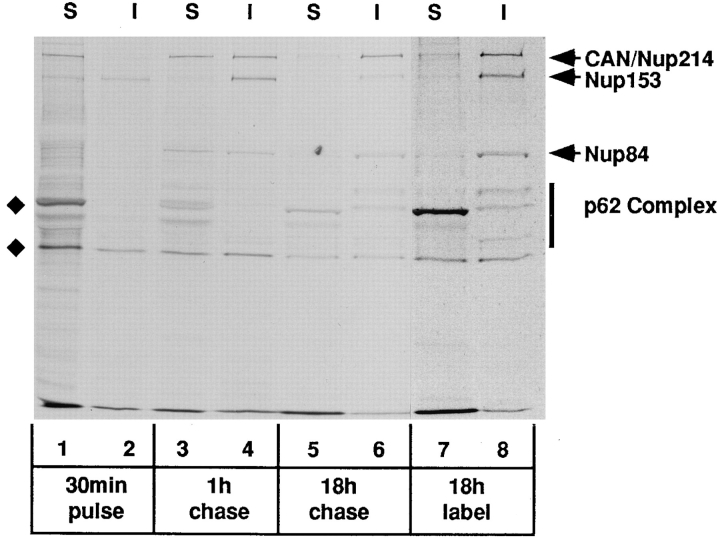
Previous studies by Gerace and co-workers revealed that many NPC glycoproteins (including CAN/Nup214) are resistant to solubilization by low salt buffers containing Triton X-100 (Snow et al., 1987). Furthermore, purified nuclear envelopes extracted in this way, while lacking both inner and outer nuclear membranes, exhibit morphologically identifiable NPCs anchored to a residual nuclear lamina (Gerace et al., 1984). We have taken advantage of these observations to determine whether the stable assembly of CAN/Nup214 into NPCs occurs before or after its association with Nup84. To first define the solubility properties of total cellular CAN/Nup214, BHK cells that had been labeled for 18 h (essentially to equilibrium) with 35STrans label were lysed in a Triton-containing low salt (0 mM NaCl) buffer. After centrifugation, the pellet fraction was reextracted at high salt (500 mM NaCl). Both high and low salt extracts were then analyzed by immunoprecipitation using the QE5 monoclonal antibody under conditions that preserve the interaction of CAN/Nup214 with Nup84. As shown in Fig. 5 (lanes 7 and 8) and consistent with the published studies (Snow et al., 1987), 80–90% of the CAN/ Nup214 is resistant to solubilization at low salt. The opposite result, however, was obtained when only newly synthesized CAN/Nup214 was observed. After a 30-min pulse label, greater than 90% of the newly synthesized CAN/ Nup214 was found to be soluble at low salt (Fig. 5, lanes 1 and 2). During a subsequent chase (up to 18 h) in medium containing excess unlabeled methionine and cysteine, CAN/Nup214 acquired resistance to solubilization with a half-time of ∼1 h (Fig. 5, lanes 3–6). Since all available data indicate that CAN/Nup214 is localized exclusively to NPCs and at the same time cannot be extracted at low salt, this solubility shift is most likely to reflect the stable integration of newly synthesized CAN/Nup214 into pore complexes. A telling feature of this experiment is that newly synthesized Nup84 associates with CAN/Nup214 very rapidly; in fact it becomes just detectable within the time frame of the 30-min pulse. Furthermore, the complex formed by these two molecules can be found in the low salt extract (Fig. 5, lanes 1 and 3), which would imply that CAN/ Nup214 and Nup84 associate before their stable assembly into NPCs. Nup153, like CAN/Nup214, exhibits a similar decrease in solubility soon after synthesis, but with a somewhat shorter half-time. In contrast, a longer half-time (>1 h) is observed for p62.
Figure 5.
Pulse–chase analysis of NPC proteins in BHK cells labeled with 35S-Trans label. The cells were either labeled continuously for 18 h (lanes 7 and 8) or pulse labeled for 30 min (lanes 1–6) followed by a “chase” of 1 h (lanes 3 and 4) or 18 h (lanes 5 and 6) in medium containing excess unlabeled methionine/cysteine. At the appropriate time, the cells were lysed in a low salt buffer containing Triton X-100. The distributions of CAN/Nup214 and Nup84 between soluble and insoluble fractions were then determined by immunoprecipitation analysis using the QE5 monoclonal antibody (which recognizes CAN/Nup214, Nup153, and p62). Total or bulk CAN/Nup214 (and associated Nup84) observed in the samples derived from the continuously labeled cells (lanes 7 and 8) is almost completely (80–90%) resistant to solubilization. In contrast, following the 30-min labeling period, 90% of newly synthesized CAN/Nup214 is found in the soluble fraction (lanes 1 and 2) and, furthermore, coprecipitates with Nup84. Both of these proteins are subsequently “chased” into the insoluble fraction with a half-time of about 1 h (lanes 3 and 4). These findings are consistent with the notion that the two proteins associate before stable integration into the NPC. While derived from the same experiment and run on the same gel, exposure times for lanes 1–6 versus lanes 7 and 8 have been adjusted (11 d and 8 h, respectively) to yield comparable intensities. The two bands indicated by the diamonds, the upper of which migrates just ahead of the p62-associated protein, p58, are both observed with nonimmune Sepharose beads.
Nup84 May Function in the Attachment of CAN/ Nup214 to the NPC
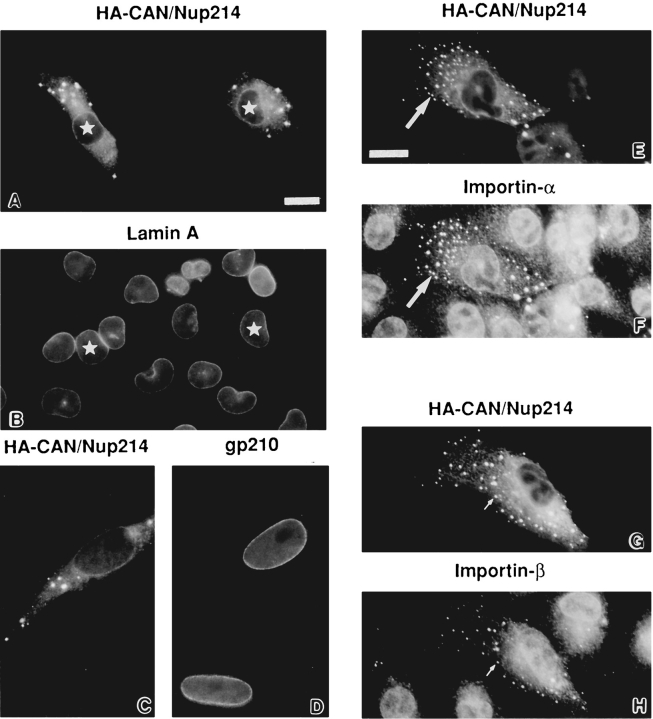
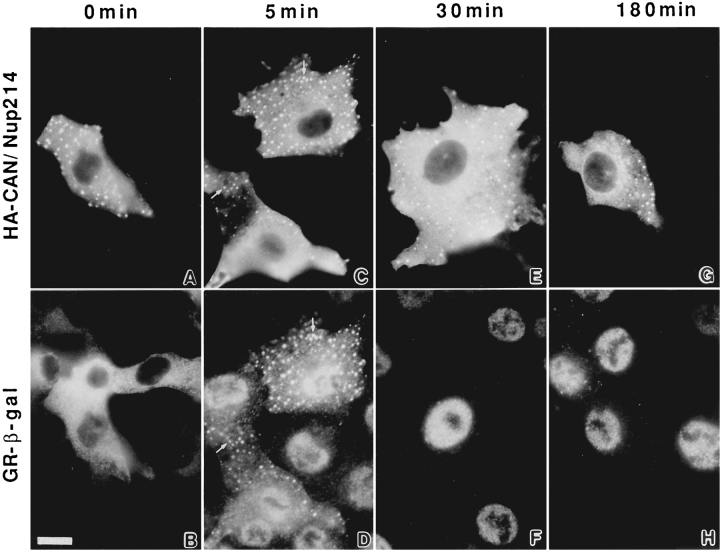
Clues to the possible function of Nup84 were provided by a series of transfection experiments, some in combination with CAN/Nup214. When HA-tagged CAN/Nup214 is introduced alone into BHK cells it associates with the nuclear envelope (Fig. 3). Like Nup84, as its expression level rises and all available nuclear envelope binding sites are saturated, CAN/Nup214 spills over into the cytoplasm (Fornerod et al., 1995). At this point, however, all similarity in the behavior of the two proteins vanishes. In contrast to Nup84, which appears to distribute evenly throughout the cytoplasm (Fig. 3 C), CAN/Nup214 localizes in part to brightly labeled micron-scale foci visible against a general cytoplasmic background (Fig. 3 D). To date, we have unfortunately been unable to identify an ultrastructural correlate for these structures. However, they do not appear to contain other nuclear envelope constituents such as lamins (Fig. 6, A and B) or the NPC proteins Nup153 (not shown) and gp210 (Fig. 6, C and D). Furthermore, these structures are not labeled by antibodies against either the endoplasmic reticulum or a lysosomal marker, the transmembrane gycoprotein LGP120 (not shown). Proteins that were, however, associated with these foci were the two subunits of the NLS receptor, importins or karyopherins-α and β (Fig. 6, E–H). Consistent with this finding, we also noticed the transient association of a nuclear import substrate with these structures (Fig. 7). This observation was made in BHKgrβ cells (Bastos et al., 1996), which constitutively express a glucocorticoid receptor–β-galactosidase fusion protein (grβ). This fusion protein localizes exclusively to the cytoplasm until the addition of dexamethasone to the culture medium, whereupon it translocates rapidly and quantitatively into the nucleus (Picard and Yamamoto, 1987). When this translocation process was examined in cells expressing high levels of HA-CAN/Nup214, the uniformly cytoplasmic grβ was found to redistribute to the foci as well as to the nucleus within a few minutes of dexamethasone treatment. Over a period of 1–2 h, grβ associated with the foci was lost.
Figure 6.
Double indirect immunofluorescence microscopy of BHK cells (A, B, E–H) and HeLa cells (C and D) transiently expressing HACAN/Nup214. Cells were labeled with a monoclonal antibody against the HA epitope (12CA5, A, C, F, and H) as well as rabbit antibodies against lamin A (B), importin-α (F), and importin-β (H) and a human autoantibody recognizing the NPC membrane protein gp210 (C). The same nuclei in A and B are indicated with stars. CAN/Nup214 cytoplasmic foci containing importins-α and -β are indicated with arrows in E–H. While the importins clearly associates with the CAN/Nup214 cytoplasmic foci, these structures are negative for lamin A and gp210. Bars (A–D and E–H) 10 μm.
Figure 7.
Double indirect immunofluorescence microscopy of BHKgrβ cells transiently expressing HA-CAN/Nup214. 24 h posttransfection and before fixation, the cells were treated with dexamethasone for periods up to 3 h as indicated at the top of the figure. The primary antibodies were 12CA5 (anti-HA, upper row) and a rabbit anti–β-galactosidase (corresponding fields, lower row). It is evident that there is a transient association of glucocorticoid receptor–β-galactosidase with the CAN/Nup214 foci shortly after the addition of dexamethasone (indicated with arrows in C and D). This is consistent with the observation that importins-α and -β are also associated with these structures (Fig. 6). Bar, 10 μm.
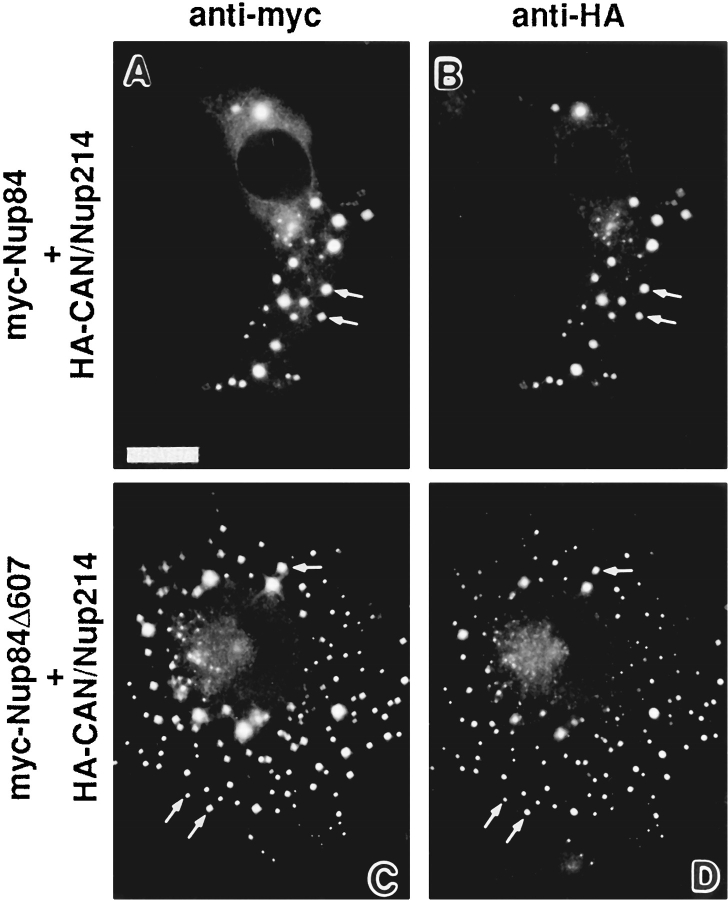
When BHK cells were cotransfected with Myc-Nup84 and HA-CAN/Nup214, each protein was found to strongly influence the other's distribution (Fig. 8, A and B). In the case of Myc-Nup84 it localized both to the nuclear envelope and to the CAN/Nup214 foci with little left free in the cytoplasm. Similarly, cytosolic HA-CAN/Nup214 was found to concentrate largely within the foci. These data indicate that high concentrations of HA-CAN/Nup214 divert cytoplasmic Myc-Nup84 into the foci, while excess Myc-Nup84 promotes a further accumulation of HA-CAN/Nup214 within these structures. This colocalization and codependent redistribution clearly implies that these two proteins interact in vivo.
Figure 8.
Double indirect immunofluorescence microscopy of BHK cells cotransfected with expression constructs containing either HA-Nup214 and Myc-Nup84 (A and B) or HA-Nup214 and Myc-Nup84Δ607 (C and D). The antibodies used were a monoclonal anti-Myc (9E10, A and C) and a rabbit anti–HA (B and D). It is clear from this figure and from the results presented in Figs. 3 and 9 that there is a codependent distribution of HANup214 and both Myc-Nup84 and Myc-Nup84Δ607. Both Nup84 constructs concentrate in HA-Nup214 foci in the cytoplasm (indicated by the arrows) while there is diminution in the amount of HA-Nup214 localized to the nuclear envelope (compare with Figs. 3 and 5). These results provide strong, albeit circumstantial, evidence that both Myc-Nup84 and Myc-Nup84Δ607 possess the capacity to associate with Nup214. Bar, 10 μm.
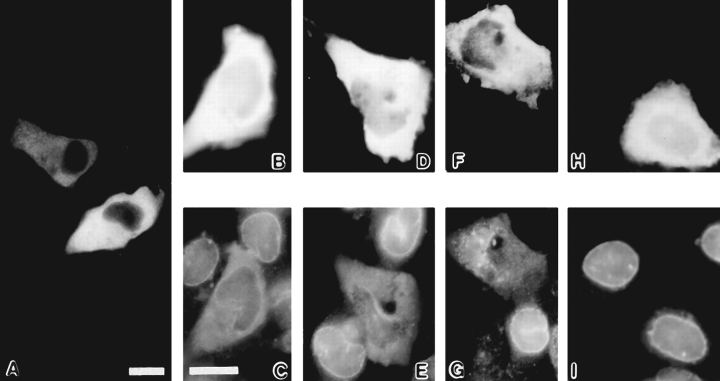
To investigate these interactions further we made a Myc-Nup84 mutant missing 140 residues from the COOH terminus (Myc-Nup84Δ607). This mutant therefore lacked about two thirds of the presumed coiled–coil tail. When expressed in BHK cells, Myc-Nup84Δ607 localized only to the cytoplasm with no detectable association with the nuclear envelope at any expression level (Fig. 9 A). However, cells expressing high levels of this mutant also exhibited an accumulation of some endogenous (possibly only newly synthesized) CAN/Nup214 in the cytoplasm (Fig. 9, B–G). In contrast, overexpression of full-length MycNup84 had no such effect (Fig. 9, H and I). In cells transfected with both Myc-Nup84Δ607 and HA-CAN/Nup214, a codependent redistribution of both proteins was again observed (Fig. 8, C and D). Both Myc-Nup84Δ607 and HA-CAN/Nup214 localized exclusively to the foci with neither exhibiting any clear nuclear envelope association (Fig. 8, C and D). These results would suggest that while Myc-Nup84Δ607 has lost the ability to associate with the nuclear envelope it retains the capacity to interact with CAN/Nup214. As will be expanded upon below, these findings are consistent with a model in which Nup84 cooperates in the assembly of CAN/Nup214 into the NPC.
Figure 9.
Expression of HA-Nup84Δ607 in BHK cells. HA-Nup84Δ607, detected with 12CA5, localizes exclusively to the cytoplasm regardless of expression level (A). No association with the nuclear envelope is evident. In B–G, the effects of HA-Nup84Δ607 expression (B, D, and F) on endogenous CAN/Nup214 localization (C, E, and G), detected with guinea pig anti–CAN/Nup214 are shown. It is apparent that HA-Nup84Δ607 overexpression causes mislocalization of some of the endogenous CAN/Nup214 to the cytoplasm. For comparison, the effects of full-length HA-Nup84 (H) are also shown. In this case there is no obvious effect on endogenous CAN/Nup214 localization (I). Bars (A and B–I) 10 μm.
Discussion
Nup84 represents a new NPC protein identified on the basis of its interaction with CAN/Nup214. Although devoid of characteristic peptide repeats such as XFXFG or GLFG, Nup84 can be added to the growing list of nucleoporins from a variety of sources that contain putative coiled–coil domains. Indeed, it has recently been proposed (Hurwitz and Blobel, 1995) that this feature may represent a major structural motif through which nucleoporins interact. Examples of such a role for coiled–coils are provided by several nucleoporins from both yeast and higher cells. The vertebrate nucleoporin p62, which forms a complex with three additional NPC proteins, p58, p54, and p45 (Finlay et al., 1991; Guan et al., 1995; Kita et al., 1993; Panté et al., 1994), has an α-helical coiled–coil COOH-terminal domain of about the same size as that of Nup84 (Buss et al., 1994). This region of p62 has been shown to contain the site of interaction with p54 (Buss and Stewart, 1995) that itself has a predicted α-helical coiled–coil domain (Hu et al., 1996). Predicted coiled–coils have been found in the peptide repeat-containing yeast nucleoporins Nsp1, Nup49, Nup57, Nup159, as well as in two other NPC proteins NIC96 (Grandi et al., 1993) and Nup82 (Grandi et al., 1995a ). Hurt and co-workers have shown, both biochemically and genetically, that Nsp1, Nup49, Nup57, and NIC96 are all associated and that, as in the case of p62/ p54, this association may be mediated by their helical domains (Grandi et al., 1995b ).
The finding that Nup84 and CAN/Nup214 are associated raises the issue of where this association initially takes place, either in the cytosol soon after synthesis or within the pore complex itself. Pulse–chase experiments involving differential salt extraction procedures indicate that newly synthesized CAN/Nup214 and Nup84 associate fairly swiftly (within 30 min of synthesis), at a time when they are still soluble at low salt. Since NPC-associated CAN/Nup214 is resistant to solubilization under these conditions, this suggests that association of CAN/Nup214 with Nup84 occurs in the cytoplasm, or at least before their stable assembly into NPCs. This view is supported by the results of the cotransfection studies involving MycNup84 and HA-CAN/Nup214, which clearly indicate that these proteins must be capable of interacting while still cytoplasmic. By analogy with other complex self-assembly systems such as bacteriophage T4, the association of Nup84 and CAN/Nup214 to form a soluble heterooligomer might actually be a prerequisite for the incorporation of either molecule into the NPC. The same could also be true for the p62/58/54/45 subcomplex since our preliminary observations suggest that these proteins at least associate relatively rapidly (Fig. 5 and unpublished results). In contrast, the rate at which p62 becomes nuclear envelope associated (half-time of 6–8 h in nonsynchronized BRL cells) is exceedingly slow (Davis and Blobel, 1986, 1987). In this way, de novo formation of NPCs during interphase may involve the prefabrication of NPC modules which are subsequently assembled to yield mature NPCs. Certainly this type of modular NPC assembly, at least with respect to p62, appears to be the rule during nuclear reformation at the end of mitosis (Finlay et al., 1991).
The association with CAN/Nup214, a component of the NPC cytoplasmic filaments, suggests that Nup84 should be exposed on the cytoplasmic face of the nuclear envelope, a prediction borne out by differential permeabilization studies of transfected BHK cells. Appropriate targeting of Nup84 is contingent upon the integrity of it its COOH-terminal coiled–coil domain since a mutant in which two thirds of this region is deleted (Nup84Δ607) is found to remain cytosolic. This is in marked contrast to the behavior of full-length Nup84, and also to CAN/Nup214, which both localize exclusively to the nuclear periphery when expressed at low levels. Interaction with CAN/Nup214 is apparently not sufficient to correctly localize Nup84. This view is based on our observations that Nup84Δ607, like full-length Nup84, codistributes with CAN/Nup214 when both proteins are expressed at high levels in BHK cells. These results may at first seem paradoxical since one might expect that interaction of Nup84Δ607 with CAN/ Nup214 should be able to confer NPC association on the former. The fact that it does not suggests a scheme in which Nup84 functions in the integration of CAN/Nup214 into the NPC. This could potentially involve Nup84 acting as a linker between CAN/Nup214 and the central region of the NPC. In such a model Nup84 would be expected to interact with CAN/Nup214 via its globular NH2-terminal domain, while its COOH-terminal α-helical domain would mediate attachment to the central framework of the NPC. A variation on this theme would be to suggest that the association of Nup84 with CAN/Nup214 results in the formation of a new binding domain that is required for the interaction of the CAN/Nup214-Nup84 module with the NPC central framework. Such a role for Nup84 in CAN/ Nup214 NPC-association is lent some credence by the finding that overexpression of Nup84Δ607 results in a partial accumulation of endogenous (most likely newly synthesized) CAN/Nup214 to the cytoplasm. The relative dispositions of these two proteins, as well as their interactions within the NPC, will be more precisely defined as antiNup84 antibodies suitable for high resolution EM-immunolocalization become available.
Overexpression of Nup84 (as well as Nup84Δ607) yielded no detectable transport related phenotype, at least in terms of nuclear protein import and poly (A)+ RNA export (Enarson, M., R. Bastos, and B. Burke, data not shown). However, its partner protein, CAN/Nup214, when expressed at high levels was found to cause the transient diversion of an inducible nuclear import substrate (grβ) into unusual cytoplasmic foci. These foci were also found to contain both importins-α and -β but were devoid of other NPC and nuclear envelope proteins (as well as ER and lysosomal proteins). At the same time no obvious effect could be observed on RNA trafficking (Bastos et al., 1996). These findings would be consistent with CAN/ Nup214 or another associated nucleoporin containing docking sites for incoming nuclear proteins complexed with the NLS receptor. Support for such a role is provided by the earlier observations of Richardson et al. (1988) and more recently of Panté and Aebi (1996) that a nuclear import substrate, nucleoplasmin-coated colloidal gold particles, associates with the cytoplasmic filaments of the NPC at an early step in translocation. Furthermore, biochemical data indicate that the NLS receptor complex associates in vitro with the peptide repeat domains of certain nucleoporins (Morioianu et al., 1995; Radu et al., 1995), CAN/ Nup214 being one of these, an interaction that is regulated by essential soluble import factors, including the small GTP-binding protein Ran (Melchior et al., 1995; Nehrbass and Blobel, 1996; Paschal and Gerace, 1995; Rexach and Blobel, 1995). A role for CAN/Nup214 in nuclear protein import is also indicated by the recent knockout data of Grosveld and colleagues (van Deursen et al., 1996). In addition, nuclear accumulation of poly (A)+ RNA in CAN/ Nup214(−/−) embryos suggested a potential function in RNA export. However, as pointed out by van Deursen et al. (1996), this could be secondary to the import defect.
In summary, we have identified a new nucleoporin, Nup84, which is proposed to function in the assembly of CAN/Nup214 on the cytoplasmic face of the NPC. Since CAN/Nup214 or an additional associated protein may contain docking sites for incoming nuclear proteins, the role of Nup84 may be to organize these sites at the cytoplasmic periphery of the NPC. This proposed role in CAN/ Nup214 attachment clearly implies that Nup84 may interact with additional NPC components at the base of the cytoplasmic filaments, most likely in the cytoplasmic ring of the central framework. Since this is a largely uncharted region, the identification of new Nup84-associated proteins should provide important new information concerning the organization of the cytoplasmic face of the NPC.
Acknowledgments
We thank Dr. Gerard Grosveld for kindly providing us with an HAtagged CAN/Nup214 construct. We thank also Dr. Jean Claude Courvalin, Dr. Marvin Fritzler, and Dr. Aurelian Radu for their generous gift of antibodies. We would like to especially acknowledge both Frank McKeon and Manfred Lohka for advice, reagents, and valuable discussions, and Isabel McMorrow for her help during the early part of this work. Thanks are also due to Cristina Pujades, Lorenza Lanini, Roydon Price, and Amy Hudson for reagents and helpful debate among other things.
This work was supported by grants to Brian Burke from the Alberta Heritage Foundation for Medical Research, the Medical Research Council of Canada, and the National Institutes of Health.
Footnotes
Please address all correspondence to Brian Burke, Faculty of Medicine, University of Calgary, 3330 Hospital Drive NW, Calgary, Alberta T2N 4N1, Canada. Tel.: (403) 220-7287. Fax: (403) 270-0979. e-mail: bburke@acs.ucalgary.ca
R. Bastos' present address is Institute Jacques Monod, Département Biologie Cellulaire, Université Paris 7, Tour 43, 2 Place Jussieu, 75251 Paris Cedex 05, France.
1. Abbreviation used in this paper: NPC, nuclear pore complex.
References
- Adam SA, Sterne-Marre RE, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebersold RH, Leavitt J, Saavedra RA, Hood LE, Kent SBH. Internal amino acid sequence analysis of proteins separated by one- or twodimensional gel electrophoresis after in situ protease digestion on nitrocellulose. Proc Natl Acad Sci USA. 1987;84:6970–6974. doi: 10.1073/pnas.84.20.6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey CW. Structural plasticity of the nuclear pore complex. J Mol Biol. 1995;248:273–293. doi: 10.1016/s0022-2836(95)80050-6. [DOI] [PubMed] [Google Scholar]
- Akey CW, Radermacher M. Architecture of the Xenopus nuclear pore complex revealed by three dimensional cryo-electron microscopy. J Cell Biol. 1993;122:1–20. doi: 10.1083/jcb.122.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ash JF, Louvard D, Singer SJ. Antibody-induced linkages of plasma membrane proteins to intracellular actomyosin-containing filaments in cultured fibroblasts. Proc Natl Acad Sci USA. 1977;74:5584–5588. doi: 10.1073/pnas.74.12.5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos R, Lin A, Enarson M, Burke B. Targeting and function in mRNA export of nuclear pore complex protein Nup153. J Cell Biol. 1996;134:1141–1156. doi: 10.1083/jcb.134.5.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos R, Panté N, Burke B. Nuclear pore complex proteins. Int Rev Cytol. 1995;162B:257–302. doi: 10.1016/s0074-7696(08)62619-4. [DOI] [PubMed] [Google Scholar]
- Blobel G, Potter VR. Nuclei from rat liver: isolation method that combines purity with high yield. Science (Wash DC) 1966;154:1662–1664. doi: 10.1126/science.154.3757.1662. [DOI] [PubMed] [Google Scholar]
- Burke B. On the cell-free association of lamins A and C with metaphase chromosomes. Exp Cell Res. 1990;186:169–176. doi: 10.1016/0014-4827(90)90223-w. [DOI] [PubMed] [Google Scholar]
- Burke B, Griffiths G, Reggio H, Louvard D, Warren G. A monoclonal antibody against a 135k Golgi membrane protein. EMBO J. 1982;2:1621–1628. doi: 10.1002/j.1460-2075.1982.tb01364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette WN. ‘Western blotting': electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981;112:195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Buss F, Kent H, Stewart M, Bailler SB, Hanover JA. Role of different domains in the self-association of rat nucleoporin p62. J Cell Sci. 1994;107:631–638. doi: 10.1242/jcs.107.2.631. [DOI] [PubMed] [Google Scholar]
- Buss F, Stewart M. Macromolecular interactions in the nucleoporin p62 complex of rat nuclear pores: binding of nucleoporin p54 to the rod domain of p62. J Cell Biol. 1995;128:251–261. doi: 10.1083/jcb.128.3.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis LI, Blobel G. Identification and characterization of a nuclear pore complex protein. Cell. 1986;45:699–709. doi: 10.1016/0092-8674(86)90784-1. [DOI] [PubMed] [Google Scholar]
- Davis LI, Blobel G. Nuclear pore complex contains a family of glycoproteins that includes p62: glycosylation through a previously unidentified cellular pathway. Proc Natl Acad Sci USA. 1987;84:7552–7556. doi: 10.1073/pnas.84.21.7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer N, Blobel G. A modified procedure for the isolation of a pore complex lamina fraction from rat liver nuclei. J Cell Biol. 1976;70:581–591. doi: 10.1083/jcb.70.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JB, Delort J, Mallet J. Oligodeoxyribonucleotide ligation to single-stranded cDNAs: a new tool for cloning 5′ ends of mRNAs and for constructing cDNA libraries by in vitro amplification. Nucleic Acids Res. 1991;19:5227–5232. doi: 10.1093/nar/19.19.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay D, Meier E, Bradley P, Horecka J, Forbes DJ. A complex of nuclear pore proteins required for pore function. J Cell Biol. 1991;114:169–183. doi: 10.1083/jcb.114.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay DR, Forbes DJ. Reconstitution of biochemically altered nuclear pores: transport can be eliminated and restored. Cell. 1990;60:17–29. doi: 10.1016/0092-8674(90)90712-n. [DOI] [PubMed] [Google Scholar]
- Fornerod M, Boer J, van Baal S, Jaegle M, von Lindern M, Murti KG, Davis D, Bonten J, Buijs A, Grosveld G. Relocation of the carboxyterminal part of CAN from the nuclear envelope to the nucleus as a result of leukemia-specific chromosome rearrangements. Oncogene. 1995;10:1739–1748. [PubMed] [Google Scholar]
- Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti KG, Fransen J, Grosveld G. The human homologue of CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerace L, Comeau C, Benson M. Organization and modulation of nuclear lamina structure. J Cell Sci Suppl. 1984;1:137–160. doi: 10.1242/jcs.1984.supplement_1.10. [DOI] [PubMed] [Google Scholar]
- Goldberg MW, Allen TD. Structural and functional organization of the nuclear envelope. Curr Opin Cell Biol. 1995;7:301–309. doi: 10.1016/0955-0674(95)80083-2. [DOI] [PubMed] [Google Scholar]
- Grandi P, Doye V, Hurt EC. Purification of NSP1 reveals complex formation with GLFG nucleoporins and a novel nuclear pore protein NIC96. EMBO J. 1993;12:3061–3071. doi: 10.1002/j.1460-2075.1993.tb05975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Emig S, Weise C, Hucho F, Pohl T, Hurt EC. A novel nuclear pore protein Nup82p which specifically binds to a fraction of Nsp1p. J Cell Biol. 1995a;130:1263–1273. doi: 10.1083/jcb.130.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Schlaich N, Tekotte H, Hurt EC. Functional interaction of Nic96p with a core nucleoporin complex consisting of Nsp1p, Nup49p and a novel protein Nup57p. EMBO J. 1995b;14:76–87. doi: 10.1002/j.1460-2075.1995.tb06977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan T, Müller S, Klier G, Panté N, Blevitt JM, Haner M, Paschal B, Aebi U, Gerace L. Structural analysis of the p62 complex, an assembly of O-linked glycoproteins that localizes near the central gated channel of the nuclear pore complex. Mol Biol Cell. 1995;6:1591–1603. doi: 10.1091/mbc.6.11.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow, E., and D. Lane. 1988. Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 726 pp.
- Hinshaw JE, Carragher BO, Milligan RA. Architecture and design of the nuclear pore complex. Cell. 1992;69:1133–1141. doi: 10.1016/0092-8674(92)90635-p. [DOI] [PubMed] [Google Scholar]
- Hu T, Guan T, Gerace L. The molecular and functional characterization of the p62 complex, an assembly of nuclear pore complex glycoproteins. J Cell Biol. 1996;134:589–601. doi: 10.1083/jcb.134.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz ME, Blobel G. NUP82 is an essential nucleoporin required for poly (A)+RNA export. J Cell Biol. 1995;130:1275–1281. doi: 10.1083/jcb.130.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita K, Omata S, Horigome T. Purification and characterization of a nuclear pore glycoprotein complex containing p62. J Biochem Tokyo. 1993;113:377–382. doi: 10.1093/oxfordjournals.jbchem.a124054. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer D, Wozniak RW, Blobel G, Radu A. The human CAN protein, a putative oncogene product associated with myeloid leukemogenesis, is a nuclear pore complex protein that faces the cytoplasm. Proc Natl Acad Sci USA. 1994;91:1519–1523. doi: 10.1073/pnas.91.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature (Lond) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lathe R. Synthetic oligonucleotide probes deduced from amino acid sequence data. Theoretical and practical considerations. J Mol Biol. 1985;183:1–12. doi: 10.1016/0022-2836(85)90276-1. [DOI] [PubMed] [Google Scholar]
- Lees-Miller JP, Goodwin LO, Helfman DM. Three novel brain tropomyosin isoforms are expressed from the rat alpha-tropomyosin gene through the use of alternative promoters and alternative RNA processing. Mol Cell Biol. 1990;10:1729–1742. doi: 10.1128/mcb.10.4.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior F, Gerace L. Mechanisms of nuclear protein import. Curr Opin Cell Biol. 1995;7:310–318. doi: 10.1016/0955-0674(95)80084-0. [DOI] [PubMed] [Google Scholar]
- Melchior F, Guan T, Yokoyama N, Nishimoto T, Gerace L. GTP hydrolysis by Ran occurs at the nuclear pore complex in an early step of protein import. J Cell Biol. 1995;131:571–581. doi: 10.1083/jcb.131.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morioianu J, Hijikata M, Blobel G, Radu A. Mammalian karyopherin a1b and a2b heterodimers: a1 or a2 subunit binds nuclear localization signal and b subunit interacts with peptide repeat-containing nucleoporins. Proc Natl Acad Sci USA. 1995;92:6532–6536. doi: 10.1073/pnas.92.14.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrbass U, Blobel G. Role of the nuclear transport factor p10 in nuclear import. Science (Wash DC) 1996;272:120–122. doi: 10.1126/science.272.5258.120. [DOI] [PubMed] [Google Scholar]
- Panté N, Aebi U. Towards understanding the three-dimensional structure of the nuclear pore complex at the molecular level. Curr Opin Struct Biol. 1994;4:187–196. [Google Scholar]
- Panté N, Aebi U. Sequential binding of import ligands to distinct nucleopore regions during their nuclear import. Science (Wash DC) 1996;273:1729–1732. doi: 10.1126/science.273.5282.1729. [DOI] [PubMed] [Google Scholar]
- Panté N, Bastos R, McMorrow I, Burke B, Aebi U. Interactions and three-dimensional localization of a group of nuclear pore complex proteins. J Cell Biol. 1994;126:603–617. doi: 10.1083/jcb.126.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschal BM, Gerace L. Identification of NTF2, a cytosolic factor for nuclear import that interacts with nuclear pore complex protein p62. J Cell Biol. 1995;129:925–937. doi: 10.1083/jcb.129.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips GN, Jr, Fillers JP, Cohen C. Tropomyosin crystal structure and muscle regulation. J Mol Biol. 1986;192:111–131. doi: 10.1016/0022-2836(86)90468-7. [DOI] [PubMed] [Google Scholar]
- Picard D, Yamamoto KR. Two signals mediate hormone-dependent nuclear localization of the glucocorticoid receptor. EMBO J. 1987;6:3333–3340. doi: 10.1002/j.1460-2075.1987.tb02654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu A, Moore MS, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- Reichelt R, Holzenburg A, Buhle EL, Jarnik M, Engel A, Aebi U. Correlation between structure and mass distribution of the nuclear pore complex and of distinct pore components. J Cell Biol. 1990;110:883–894. doi: 10.1083/jcb.110.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- Richardson WD, Mills AD, Dilworth SM, Laskey RA, Dingwall C. Nuclear protein migration involves two steps: rapid binding at the nuclear envelope followed by slower translocation through the nuclear pores. Cell. 1988;52:655–664. doi: 10.1016/0092-8674(88)90403-5. [DOI] [PubMed] [Google Scholar]
- Ris H. The 3-D structure of the nuclear pore complex as seen by high voltage electron microscopy and high resolution low voltage scanning electron microscopy. EMSA Bull. 1991;21:54–56. [Google Scholar]
- Rost B, Sander C. Improved prediction of protein secondary structure by use of sequence profiles and neural networks. Proc Natl Acad Sci USA. 1993;90:7558–7562. doi: 10.1073/pnas.90.16.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Wente SR. Pores for thought: nuclear pore complex proteins. Trends Cell Biol. 1994;4:357–365. doi: 10.1016/0962-8924(94)90085-x. [DOI] [PubMed] [Google Scholar]
- Sander C, Schneider R. Database of homology-derived protein structures and the structural meaning of sequence alignment. Proteins. 1991;9:56–68. doi: 10.1002/prot.340090107. [DOI] [PubMed] [Google Scholar]
- Snow CM, Senior A, Gerace L. Monoclonal antibodies identify a group of nuclear pore complex glycoproteins. J Cell Biol. 1987;104:1143–1156. doi: 10.1083/jcb.104.5.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deursen J, Boer J, Kasper L, Grosveld G. G2arrest and impaired nucleocytoplasmic transport in mouse embryos lacking the protooncogene CAN/Nup214. EMBO J. 1996;15:5574–5583. [PMC free article] [PubMed] [Google Scholar]
- von Lindern M, van Baal S, Wiegant J, Raap A, Hagemijer A, Grosveld G. can, a putative oncogene associated with myeloid leukemogenesis, may be activated by fusion of its 3′ half to different genes: characterization of the setgene. Mol Cell Biol. 1992;12:3346–3355. doi: 10.1128/mcb.12.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]