Abstract
SEC35 was identified in a novel screen for temperature-sensitive mutants in the secretory pathway of the yeast Saccharomyces cerevisiae (Wuestehube et al., 1996. Genetics. 142:393–406). At the restrictive temperature, the sec35-1 strain exhibits a transport block between the ER and the Golgi apparatus and accumulates numerous vesicles. SEC35 encodes a novel cytosolic protein of 32 kD, peripherally associated with membranes. The temperature-sensitive phenotype of sec35-1 is efficiently suppressed by YPT1, which encodes the rab-like GTPase required early in the secretory pathway, or by SLY1-20, which encodes a dominant form of the ER to Golgi target -SNARE–associated protein Sly1p. Weaker suppression is evident upon overexpression of genes encoding the vesicle-SNAREs SEC22, BET1, or YKT6. The cold-sensitive lethality that results from deleting SEC35 is suppressed by YPT1 or SLY1-20. These genetic relationships suggest that Sec35p acts upstream of, or in conjunction with, Ypt1p and Sly1p as was previously found for Uso1p. Using a cell-free assay that measures distinct steps in vesicle transport from the ER to the Golgi, we find Sec35p is required for a vesicle docking stage catalyzed by Uso1p. These genetic and biochemical results suggest Sec35p acts with Uso1p to dock ER-derived vesicles to the Golgi complex.
Protein transport through the secretory pathway occurs via transport vesicles under the direction of a large set of protein components (Rothman, 1994). The process can be divided into three stages: (a) vesicle budding, (b) vesicle docking, and (c) membrane fusion, with distinct sets of proteins mediating each phase. The budding step involves recruitment of coat proteins to the membrane and culminates with the release of coated vesicles (Schekman and Orci, 1996). The docking reaction is likely to require a set of integral membrane proteins on the vesicle and target membranes, termed v-SNAREs1 and t-SNAREs (vesicle- and target membrane-soluble N-ethylmaleimide–sensitive fusion protein [NSF] attachment protein [SNAP] receptors, respectively), that are thought to confer specificity through their pair-wise interactions (Söllner et al., 1993b ). Small GTP-binding proteins of the rab family also assist in the docking process (Ferro-Novick and Novick, 1993), but their precise function is not known. The fusion step ensues after docking and results in the delivery of the vesicular cargo to the next compartment in the secretory pathway.
Vesicular transport from the ER to the Golgi apparatus in the yeast Saccharomyces cerevisiae has been extensively characterized. Transport vesicle budding involves the assembly of the COPII coat, composed of the Sec13p/Sec31p (Pryer et al., 1993; Salama et al., 1993; Barlowe et al., 1994) and Sec23p/Sec24p heterodimers (Hicke and Schekman, 1989; Hicke et al., 1992), under the direction of an integral membrane protein, Sec12p (Nakano et al., 1988; Barlowe and Schekman, 1993), a small GTPase, Sar1p (Nakano and Muramatsu, 1989), and a multidomain protein, Sec16p (Espenshade et al., 1995; Shaywitz et al., 1997). Docking is thought to require a tethering event mediated by Uso1p (Cao et al., 1998), the yeast homologue of mammalian p115 (Barroso et al., 1995; Sapperstein et al., 1995), followed by or concurrent with the interaction of a set of ER to Golgi v-SNAREs, Bet1p, Bos1p, Sec22p (Newman and Ferro-Novick, 1987; Newman et al., 1990; Ossig et al., 1991; Shim et al., 1991; Søgaard et al., 1994) and perhaps Ykt6p (Søgaard et al., 1994; McNew et al., 1997), with the cognate t-SNARE on the Golgi, Sed5p (Hardwick and Pelham, 1992). For some time it was thought that fusion may be initiated by disassembly of the v/t-SNARE complex (Söllner et al., 1993a ) by yeast SNAP, Sec17p, (Griff et al., 1992) and NSF, Sec18p (Eakle et al., 1988; Wilson et al., 1989). However, this concept has been challenged by studies with a yeast system that reconstitutes homotypic vacuolar fusion, which suggests the action of Sec18p is before vesicle docking (Mayer et al., 1996; Mayer and Wickner, 1997). In addition, a prefusion role for NSF has been supported by the recent finding that liposomes bearing SNAREs alone can fuse in the absence of NSF (Weber et al., 1998).
Several proteins involved in the regulation of yeast ER to Golgi v/t-SNARE complex assembly have been identified, including Ypt1p, Uso1p, and Sly1p. Ypt1p is a member of the rab family of small GTP-binding proteins that have been identified as important components of almost every stage in the secretory pathway (Ferro-Novick and Novick, 1993). Hydrolysis of GTP by rab-like proteins has been hypothesized to provide the regulatory switch that controls the fidelity of vesicular transport (Bourne, 1988). A second protein, Uso1p (Nakajima et al., 1991), appears to function in the same pathway as Ypt1p (Sapperstein et al., 1996), and both proteins have been demonstrated to be essential for SNARE complex assembly (Søgaard et al., 1994; Sapperstein et al., 1996; Lupashin and Waters, 1997). The third protein, Sly1p, is associated with the t-SNARE Sed5p (Søgaard et al., 1994). SLY1 is an essential gene in yeast (Dascher et al., 1991; Ossig et al., 1991), and Sly1p is required for ER to Golgi transport in vitro (Lupashin et al., 1996) and in vivo (Ossig et al., 1991). However, several lines of evidence, particularly from Sly1p homologues in other organisms, indicate that Sly1p may also function as a negative regulator of v/t-SNARE complex assembly, perhaps by preventing the association of the v- and t-SNAREs (Hosono et al., 1992; Pevsner et al., 1994; Schulze et al., 1994). A dominant allele of SLY1, termed SLY1-20, is capable of suppressing mutations in YPT1 and USO1, including complete deletions (Dascher et al., 1991; Sapperstein et al., 1996). Thus, in the presence of Sly1-20p, two components required for SNARE complex assembly are no longer essential. We have proposed a model (Sapperstein et al., 1996; Lupashin and Waters, 1997) in which Ypt1p and Uso1p function to relieve the inhibitory action of Sly1p on SNARE complex assembly. In this model Sly1-20p can be thought of as a noninhibitory form of SLY1 that renders Ypt1p and Uso1p superfluous.
We believe that the ability of SLY1-20 to suppress defects in upstream docking regulators can be used to identify additional components involved in the regulation of vesicular docking. We have undertaken a genetic screen (to be presented elsewhere) to isolate novel components in this pathway which, when mutated, depend upon Sly1-20p for viability. In the course of this work, we discovered that two recently identified mutants, sec34 and sec35, can be suppressed by SLY1-20 and thus satisfy the criterion of our screen. These mutants were isolated in a novel screen to identify components involved in transport at any step between the ER and the trans-Golgi network (i.e., the Kex2p compartment) in yeast (Wuestehube et al., 1996). Both sec34 and sec35 accumulate the core-glycosylated form of secretory proteins at the nonpermissive temperature, indicating a block in ER to Golgi transport. Furthermore, electron microscopy indicated that both sec34 and sec35 accumulate numerous vesicles upon shift to the restrictive temperature (Wuestehube et al., 1996), a hallmark of genes whose protein products are involved in the docking or fusion phase of transport (Kaiser and Schekman, 1990). In this report we describe the cloning of SEC35 and analysis of its genetic interactions with other secretory genes. Strong genetic interaction between SEC35 and SLY1, YPT1, and USO1 suggests that Sec35p may function in vesicle docking. To test this possibility, we devised an in vitro transport assay that depends on the addition of purified Sec35p and Uso1p. Vesicles synthesized in the absence of functional Sec35p do not fuse with the Golgi compartment and remain as freely diffusible intermediates. Upon addition of Sec35p and Uso1p, vesicles dock to the Golgi and proceed to membrane fusion. Requirements for Sec35p at the vesicle docking step correlates our genetic experiments with the biochemically distinguishable steps of vesicle docking and membrane fusion.
Materials and Methods
Strains and Microbial Techniques
The S. cerevisiae strains used in this work are described in Table I. The sec35-1 strain RSY962 (Wuestehube et al., 1996) was crossed to RSY255 to obtain a sec35-1 haploid strain with additional auxotrophies. A temperature-sensitive ura3 leu2 segregant, GWY93, was used for subsequent analyses. The SEC35 deletion strain, GWY128, was constructed using the γ-transformation method of one-step gene replacement (for review see Rose, 1995) in which the complete open reading frame (ORF) is replaced by sequences contained in the yeast integrating vector, pRS305 (Sikorski and Hieter, 1989). The sec35Δ plasmid, pSV31 (see below), was linearized with SmaI and used to transform the diploid strain, GWY30. Leu+ transformants were purified and the deletion was confirmed by PCR using primers in LEU2 and flanking the deletion. A sec32-1 strain containing the ura3-52 mutation, GWY138, was isolated as a temperature-sensitive segregant from a cross between RSY954 (sec32-1) and RSY271 (sec18-1), which could be restored to temperature resistance by the introduction of pAN109 (2 μm BOS1). The sec32-1 mutation is allelic to BOS1, as determined by complementation analysis (Wuestehube et al., 1996). The sec34-2 strain (RSY960) was crossed to RSY255 to generate a temperature-sensitive sec34-2 strain with additional auxotrophies (GWY95). To create diploid strains heterozygous for sec35-1 and ypt1-3, GWY93 was mated to RSY976. The strain heterozygous for sec35-1 and uso1-1 was generated through mating RSY962 to GWY67. The Escherichia coli strain used in this work was XL1-Blue (Stratagene, Inc., La Jolla, CA).
Table I.
Strains Used in This Work
| Strain | Genotype | Source | ||
|---|---|---|---|---|
| RSY255 | MATα ura3-52 leu2-3,−112 | R. Schekman (University of California, Berkeley, CA) | ||
| RSY962 | MATa sec35-1 lys2-801 | R. Schekman | ||
| RSY960 | MATa sec34-2 lys2-801 | R. Schekman | ||
| RSY271 | MATα sec18-1 ura3-52 his4-612 | R. Schekman | ||
| RSY976 | MATa ypt1-3 ura3-52 | R. Schekman | ||
| RSY942 | MATa sec22-3 ura3-52 lys2-801 | R. Schekman | ||
| RSY944 | MATa bet1-1 ura3-52 lys2-801 | R. Schekman | ||
| RSY954 | MATa sec32-1 lys2-801 leu2-3, −112 | R. Schekman | ||
| RSY1074 | MATα sly1ts ura3-1 leu2-3, −112 trp1 ade2-1 | R. Schekman | ||
| RSY277 | MATα sec21-1 ura3 | R. Schekman | ||
| NY405 | MATa sec4-8 ura3-52 | P. Novick (Yale University School of Medicine, New Haven, CT) | ||
| NY420 | MATa sec19-1 ura3-52 | P. Novick | ||
| SFNY314 | MATa bet3-1 ura3-52 leu2-3, −112 | S. Ferro-Novick (Yale University) | ||
| CKY100 | MATa sec27-1 ura3 leu2 | C. Kaiser (Massachusetts Institute of Technology, Cambridge, MA) | ||
| EGY101 | MATa ret1-1 ura3 leu2 his3 trp1 suc2-Δ9 | S. Emr (University of California, San Diego, CA) | ||
| SFT1-1 | MATα URA3::sft1-1 sft1Δ::LEU2 ura3-52 leu2-3,−112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 | H. Pelham (Medical Research Council, Cambridge, UK) | ||
| GWY30 | MATa/α ura3-52/ura3-52 leu2Δ1/leu2Δ1 trp1 Δ63/trp1 Δ63 | This laboratory | ||
| GWY67 | MATα uso1-1 ura3-52 leu2-3, −112 | This laboratory | ||
| GWY93 | MATα sec35-1 ura3-52 leu2-3, −112 | This study | ||
| GWY128 | MATa/α sec35Δ::LEU2/SEC35 ura3-52/ura3-52 leu2Δ1/leu2Δ1 trp1Δ63/trp1Δ63 | This study | ||
| GWY138 | MATa sec32-1 ura3-52 his4-612 lys2-801 | This study | ||
| GWY95 | MATa sec34-2 ura3-52 leu2-3, −112 lys2-801 | This study |
Bacterial strains were grown on standard media (Miller, 1972) and transformation was performed as described (Hanahan, 1983). Transformation of yeast was performed by the method of Elble (1992) with the exception of the library transformation, which was performed by the method of Schiestl and Geitz (1989). Plasmids were isolated from yeast as described (Sapperstein et al., 1996). Yeast strains were maintained on rich media (YPD) containing 1% Bacto-yeast extract, 2% Bacto-peptone, and 2% glucose, or on synthetic media containing 0.67% yeast nitrogen base without amino acids, 2% glucose, and the appropriate supplements (Rose et al., 1990). Synthetic complete media (SC) contains all supplements necessary to support yeast growth. Diploid strains were sporulated at room temperature in liquid media containing 1% potassium acetate and 0.02% glucose. Unless otherwise noted, all low-temperature incubations of yeast strains were at room temperature, which was ∼21°C.
Plasmids
The plasmids used in this work are shown in Table II; plasmid construction was as follows: SEC35 (ORF YGR120c) was subcloned from a genomic library plasmid as an NsiI/BamHI fragment and ligated into the LEU2 centromere (CEN) vector, pRS415, that had been previously digested with PstI and BamHI, creating pSV15. ORF YGR122w was isolated from a genomic library plasmid by digestion with PstI and XhoI and then insertion into PstI- and XhoI-digested pRS415 to create pSV23. SEC35 was excised from pSV15 as a KpnI/SacI fragment and ligated into three similarly digested vectors: (a) a URA3 CEN vector, pRS416, to generate pSV16; (b) a URA3 integrating vector, pRS306, to create pSV18; and (c) a URA3 2-μm vector, pRS424, to generate pSV17. To construct the plasmid encoding the glutathione-S-transferase (GST)–Sec35 fusion protein, pSV29, the SEC35 ORF was amplified from pSV16 by PCR, placing a BamHI site adjacent to the sequence encoding the first amino acid of SEC35 and an EcoRI site 3′ to the stop codon (5′ primer: 5′ cgc-gga-tcc-atg-gtc-aac-agt-cat-ag 3′; 3′ primer: 5′ ccg-gaa-ttc-gtt-ttc-tcc-caa-cta-tg 3′). The PCR product was digested with BamHI and EcoRI and then ligated into pGEX-2T (Pharmacia Biotech, Inc., Piscataway, NJ) that had also been digested with BamHI and EcoRI. The plasmid encoding the His6-Sec35p fusion protein, pSV20, was generated by first subcloning the above PCR product into pRS416. SEC35 was then removed from this plasmid as a BamHI/HindIII fragment and then ligated into pQE-30 (QIAGEN, Inc., Santa Clarita, CA) that had been similarly digested. To generate the plasmid used for sequencing the sec35-1 allele, pSV42, the above primers were used to amplify the ORF using genomic DNA isolated from the sec35-1 strain as a template. The PCR product was digested with BamHI and EcoRI and ligated into a similarly digested pRS416. The plasmid used to create the SEC35 deletion, pSV31, was constructed in two steps. First, a 921-bp region immediately 3′ to SEC35 was amplified by PCR, including an XbaI site adjacent to the stop codon of SEC35 (5′ primer: 5′ gct-cta-gag-ttg-gga-gaa-aac-ttt-c 3′; 3′ primer: 5′ cgc-tgt-agt-att-ctg-agg 3′). This product was cleaved with XbaI and BamHI (at a site 826 bp from the 3′ end of SEC35), and ligated into a similarly digested LEU2 integrating vector, pRS305. Second, a 1,300-bp fragment that encodes sequence 5′ to SEC35 was isolated from pSV16 using a HindIII site just upstream of the first amino acid in SEC35 and a second HindIII site in the polylinker of pSV16. This fragment, when ligated into the HindIII site of the plasmid generated in the first step, resulted in a construct in which sequences flanking SEC35 are present in the polylinker of pRS305 with the region 3′ of the ORF in front of the region 5′ of the ORF. To create pSV11, SLY1-20 was excised from pYCp50-SLY1-20 as a SpeI/ClaI fragment (generated from a partial digest with SpeI because there is a SpeI site internal to the gene) and ligated into a similarly digested vector, pRS416. To isolate BOS1 from the genomic fragment that contains several additional genes, the gene was liberated from pAN109 as a NruI/KpnI fragment and ligated into pRS416, which had been digested with SmaI and KpnI, generating pSK101.
Table II.
Plasmids Used in This Work
| Plasmid | Description | Source | ||
|---|---|---|---|---|
| pJG103 | 2 μm SEC22 URA3 | S. Ferro-Novick | ||
| pSFN2d | 2 μm BET1 URA3 | S. Ferro-Novick | ||
| pAN109 | 2 μm BOS1 URA3 | S. Ferro-Novick | ||
| pNB167 | 2 μm YPT1 URA3 | S. Ferro-Novick | ||
| pGR3 | 2 μm BET3 URA3 | S. Ferro-Novick | ||
| pSED5 | 2 μm SED5 URA3 | H. Pelham | ||
| pSFT1 | 2 μm SFT1 URA3 | H. Pelham | ||
| pNB657 | 2 μm SEC19 URA3 | P. Novick | ||
| pNB142 | 2 μm SEC4 URA3 | P. Novick | ||
| pYEpSNC1 | 2 μm SNC1 LEU2 | J. Gerst (Weismann Institute of Science, Rehovot, Israel) | ||
| pYEpSNC2 | 2 μm SNC2 LEU2 | J. Gerst | ||
| pSLY1-LEU2 | 2 μm SLY1 LEU2 | H.D. Schmitt (Max Planck Institute, Gottingen, Germany | ||
| pSK60 | 2 μm YKT6 URA3 | This laboratory | ||
| pSK47 | 2 μm USO1 URA3 | This laboratory | ||
| pPI2 | 2 μm VTI1 URA3 | This laboratory | ||
| pSK101 | 2 μm BOS1 URA3 | This study | ||
| pSV17 | 2 μm SEC35 URA3 | This study | ||
| pNB166 | CEN YPT1 URA3 | S. Ferro-Novick | ||
| pYCp50-SLY1-20 | CEN SLY1-20 URA3 | H.D. Schmitt | ||
| pSV11 | CEN SLY1-20 URA3 | This study | ||
| pSV15 | CEN SEC35 LEU2 | This study | ||
| pSV16 | CEN SEC35 URA3 | This study | ||
| pSV23 | CEN YGR122w LEU2 | This study | ||
| pSV42 | CEN sec35-1 URA3 | This study | ||
| pSV18 | YIp SEC35 URA3 | This study | ||
| pSV31 | YIp sec35Δ LEU2 | This study | ||
| pSV29 | GST-Sec35p | This study | ||
| pSV20 | His6-Sec35p | This study | ||
| pRS424 | YEp URA3 | P. Hieter (Johns Hopkins School of Medicine, Baltimore, MD) | ||
| pRS415 | YCp LEU2 | P. Hieter | ||
| pRS416 | YCp URA3 | P. Hieter | ||
| pRS305 | YIp LEU2 | P. Hieter | ||
| pRS306 | YIp URA3 | P. Hieter | ||
| pGEX-2T | GST-fusion vector | Pharmacia Biotech, Inc., Piscataway, NJ | ||
| pQE-30 | His6-fusion vector | QIAGEN, Inc., Santa Clarita, CA |
Cloning of SEC35
To clone SEC35, GWY93 was transformed with two YCp50-based URA3 libraries containing inserts of 10–15 kb and 15–20 kb (A1 and B1, respectively) (Rose et al., 1987). Temperature-resistant transformants were selected directly by incubation at 37.5°C for 2 d on SC-URA media. After confirming the temperature resistance of each colony by retesting on YPD media at 37.5°C, plasmids were isolated from each temperature-resistant transformant and used to retransform the sec35-1 strain. Plasmids that conferred temperature resistance were subjected to restriction digest analysis to eliminate duplicate isolates and to identify overlapping fragments. Five plasmids from each library were end-sequenced by the dideoxy chain termination method (Sanger et al., 1977) using primers that flank the insert site in YCp50 (YEp24-F and YEp24-R; Sapperstein et al., 1996), and the resulting sequences were used to search the Saccharomyces Genome Database with the BLAST algorithm (Altschul et al., 1990). A 9-kb region was common to all the library clones and contained two uncharacterized ORFs, YGR120c and YGR122w, and the gene encoding an ammonium transporter, MEP1 (Marini et al., 1994). Both novel ORFs were subcloned individually into pRS415, generating pSV16 and pSV23 (refer to plasmid construction).
For integrative mapping, pSV18 was digested at a unique site within SEC35 (SnaBI) and used to transform GWY93. The resulting temperature-resistant strain was mated to a wild-type strain of the opposite mating type, and the diploid was sporulated and subjected to tetrad analysis.
Sequencing of the sec35-1 Allele
The UV-generated sec35-1 allele was sequenced by the Princeton University sequencing facility (Princeton, NJ) from plasmid pSV42 (see above). The ORF was sequenced using primers homologous to the polylinker of pRS416 (T3: 5′ aat-taa-ccc-tca-cta-aag-gg 3′; and KS: 5′ tcg-agg-tcg-acg-ggt-atc 3′).
Suppression Analysis
To test for suppression of the sec35-1 strain, GWY93 was transformed with the following plasmids (see Table II): pSV16, pSV11, pSK60, pSK47, pJG103, pSFN2d, pSK101, pNB166, pNB167, pGR3, pSED5, pSFT1, pNB657, pNB142, pPI2, pSLY1-LEU2, pYEpSNC1, and pYEpSNC2. Purified transformants were grown to stationary phase at 21°C in synthetic media. Suppression was tested by spotting the saturated culture and five 10-fold dilutions onto YPD plates followed by incubation for 4 d at 37.5°C. Suppression of the secretory mutants shown in Table I by pSV17 (2 μm of SEC35 URA3) was tested by streaking purified transformants for single colonies on YPD plates followed by incubation at 37.5°C. Suppression of the bet3-1 strain was assessed at 30°C.
To test the suppression of the SEC35 deletion strain, the sec35Δ/SEC35 heterozygous deletion strain (GWY128) was transformed with pSV16, pSV11, and pNB167. Purified transformants were sporulated and dissected on YPD plates. Suppression of the deletion was assessed after 5 (pSV16) or 7 (pSV11 and pNB167) d of growth at 21°C.
Metabolic Labeling and Immunoprecipitation of Carboxypeptidase Y
Strains were grown at room temperature in SC media lacking methionine (SC-MEI) or SC-URA-MET media to an OD595 of 0.3–0.6. For each strain, one OD unit of cells was pelleted, resuspended in 0.5 ml of the same media in which it was grown, and then preshifted to 39°C for 45 min. This incubation and all those subsequent were conducted at 39°C in a shaking water bath. Cells were radiolabeled for 10 min with 50 μCi Tran35S-label (ICN Radiochemicals, Irvine, CA), followed by a 20-min chase, initiated by the addition of a one twenty-fifth volume of chase mix (250 mM cysteine, 250 mM methionine). The reactions were stopped by addition of an equal volume of 20 mM sodium azide and a one-fiftieth volume of 10 mg/ml cycloheximide, and then placed on ice for 10 min. Cell lysis and immunoprecipitation of carboxypeptidase Y (CPY) was performed as previously described (Sapperstein et al., 1996). Antibodies against CPY were provided by S. Emr (University of California, San Diego, CA).
Purification of Recombinant Proteins from E. coli
To purify GST–Sec35p, the fusion protein was expressed in XL1-Blue from pSV29. After growth of a 500-ml bacterial culture at 37°C to an OD595 of 1.0, expression of the fusion protein was induced by the addition of isopropylthiogalactoside to 0.5 mM. After a 4-h induction period, cells were harvested (7,000 g for 10 min at 4°C in a rotor [model SA600; Sorvall Inc., Newtown, CT]), resuspended in 25 ml PBS, and then lysed by sonication. To prevent nonspecific protein–protein interaction, Triton X-100 (Sigma Chemical Co., St. Louis, MO) was added to 1%, and the lysate was rocked at 4°C for 30 min. After removal of debris and insoluble protein by centrifugation (12,000 g for 10 min at 4°C in a rotor [model SA600; Sorvall Inc.]), the lysate was incubated with 2 ml of glutathione–Sepharose beads (Pharmacia Biotech, Inc., Piscataway, NJ) with gentle end-over-end mixing for 2 h at 4°C. The beads were washed twice with 25 ml PBS/1% Triton X-100 and loaded into a 2-ml column. The fusion protein was eluted with 5 ml of 20 mM reduced glutathione in 50 mM Tris-Cl, pH 8.0.
To purify His6-Sec35p, a 500-ml culture of XL1-Blue transformed with pSV20 was grown, induced, and then processed as described above. The clarified bacterial sonicate containing 1% Triton X-100 was incubated with 2 ml Ni2+-NTA-agarose (QIAGEN, Inc., Santa Clarita, CA) for 2 h at 4°C with end-over-end mixing. The beads were washed twice with 25 ml PBS/1% Triton X-100 and then twice with 25 ml wash buffer (50 mM sodium phosphate, pH 6.0, 300 mM NaCl, 10% glycerol; QIAGEN, Inc., Santa Clarita, CA). The beads were transferred to a 2-ml column and His6-Sec35p was eluted with 250 mM imidazole in wash buffer. For use in the in vitro assay (see below), His6-Sec35p was further purified by size-exclusion chromatography to ensure that it was monomeric: 200 μl of a sample containing 5 mg of protein was chromatographed through a 24-ml Superose 12 column (model HR10/30; Pharmacia Biotech, Inc.) at 0.3 ml/ min in 25 mM Hepes/KOH, pH 7.0, 150 mM KOAc, 1 mM DTT.
Antibody Production and Purification
To generate antigen for antibody production, Sec35p was excised from the purified GST–Sec35p fusion protein by cleavage with thrombin (Boehringer Mannheim, Indianapolis, IN) during dialysis against 50 mM Tris-Cl, pH 8.0, for 12 h at 4°C. The GST moiety and any uncleaved fusion protein was removed by passage of the material over a 2-ml glutathione– Sepharose column and collecting the Sec35p-containing column flow-through. Recombinant Sec35p was used for the preparation of polyclonal antibodies in rabbits by standard protocols (Harlow and Lane, 1988) at the Princeton University (Princeton, NJ) animal facility. A Sec35p affinity column, generated by coupling 20 mg of His6-Sec35p (purified as described above) to 3.5 ml cyanogen bromide-activated Sepharose according to the manufacturer's specifications (Pharmacia Biotech, Inc.), was used to affinity purify antibodies to Sec35p (Harlow and Lane, 1988).
Preparation of Protein Extracts and Immunoblotting
Total yeast protein extracts were prepared as described (Ohashi et al., 1982). Samples were separated by SDS-12% PAGE, transferred to nitrocellulose, and then probed with the appropriate primary antibodies according to standard protocols (Harlow and Lane, 1988). Affinity-purified antibodies against Sec35p were made as described above, antibodies against Sed5p were generated and affinity purified as described previously (Lupashin and Waters, 1997), and monoclonal antibodies against phosphoglycerate kinase (PGK) were from Molecular Probes, Inc. (Eugene, OR). Horseradish peroxidase–conjugated secondary antibodies (Bio-Rad Laboratories, Hercules, CA) were used at a dilution of 1:3,000. Incubations with both primary and secondary antibodies were for 1 h at room temperature. Immunoblots were developed with the chemiluminescent detection kit (Amersham Corp., Arlington Heights, IL).
Subcellular Fractionation and Extractions
A 250-ml culture of RSY255 was grown in YPD media to an OD595 of 0.5– 1.0, washed in distilled water, and then resuspended at 30 OD/ml in ice-cold buffer 88 (20 mM Hepes/KOH, pH 7.0, 150 mM KOAc, 5 mM Mg[OAc]2) containing 1 mM DTT, 1 mM PMSF, 5 mM 1,10-phenanthroline, 2 μM pepstatin A, 2 μg/ml aprotinin, and 0.5 μg/ml leupeptin. One-half the sample volume of acid-washed glass beads (0.45-mm diameter; Thomas Scientific, Swedesboro, NJ) was added to the mixture, which was then vortexed six times for 20 s with 1-min incubations on ice between each burst. To remove unlysed cells, the crude yeast lysate was centrifuged at 1,000 g for 3 min at 4°C in a rotor (model SA600; Sorvall, Inc.). The supernatant was diluted to 5 mg/ml in buffer 88 and used for several manipulations. First, to examine the total membrane-associated pool of Sec35p, the S1 supernatant was layered over a 14% sucrose cushion (in buffer 88), centrifuged at 175,000 g for 60 min at 4°C in a rotor (model TLA100.2; Beckman Instruments, Palo Alto, CA), and separated into the S175 and P175 fractions. Second, the S1 supernatant was centrifuged at 10,000 g for 15 min at 4°C in a rotor (model SA600; Sorvall, Inc.) to generate the supernatant (S10) and pellet (P10) fractions. The S10 was layered over a 14% sucrose cushion (in buffer 88) and then centrifuged at 175,000 g to generate the supernatant (S175) and pellet (P175) fractions. Finally, to perform protein extractions, 0.5 ml of the S1 supernatant was mixed with an equal volume of either buffer 88 or one of the three extraction solutions (2% Triton X-100 in buffer 88, 2M NaCl in buffer 88, and 200 mM Na2CO3 in H2O) and then incubated on ice for 30 min. The extracts were then layered over a 14% sucrose cushion adjusted to the final concentration of the extraction solution (1% Triton X-100 in buffer 88, 1 M NaCl in buffer 88 or 100 mM Na2CO3 in H2O, respectively), centrifuged at 175,000 g for 60 min, and then separated into the S175 and P175 fractions. For all 175,000 g centrifugation steps, 0.8 ml of sample was layered over a 0.2 ml sucrose cushion and, after centrifugation, the supernatant fractions were removed in a total of 0.9 ml, thus distributing the cushion between the supernatant and pellet fractions.
In Vitro ER to Golgi Transport Assay
Yeast semi-intact cells from either wild-type (RSY255) or the sec35-1 strain (RSY962) were prepared from log phase cultures of strains grown at 23°C and stored frozen at −70°C (Baker et al., 1988). Before assays, a tube of cells was quickly thawed and washed three times in buffer 88 to remove cytosol, with each wash followed by centrifugation at 12,000 rpm in a refrigerated centrifuge (model 5417; Eppendorf Scientific, Inc., Hamburg, Germany). [35S]-pre-pro-α-factor was posttranslationally translocated into ER membranes of semi-intact cells at 10°C as previously described (Baker et al., 1988). After washing semi-intact cells with buffer 88, vesicle docking and transport assays were performed at the indicated temperatures (Barlowe, 1997; Cao et al., 1998). Briefly, the addition of purified COPII proteins buds ER-derived vesicles from semi-intact cells that are freely diffusible and remain in the supernatant fraction after centrifugation at 14,000 rpm. Protease-protected glycosylated [35S]pro-α-factor ([35S]gp-α-factor) contained in these vesicles was quantified after solubilization and precipitation with concanavalin A–Sepharose. For transport assays, COPII proteins in addition to purified Uso1p, LMA1 and His6-Sec35p were added as indicated in the figure legends, and then the amount of Golgi-modified [35S]gp-α-factor was measured by immunoprecipitation with anti-α1,6-mannose–specific antibodies. The data presented is the average of duplicate determinations and the error bars represent the range.
Results
Cloning of SEC35
SEC35 was cloned by complementation of the temperature-sensitive phenotype of the sec35-1 strain. The sec35-1 strain was transformed with a CEN-based library, and temperature-resistant transformants were selected directly by incubation at the restrictive temperature. Greater than 50 genome equivalents were screened and 32 temperature- resistant colonies were obtained. To test plasmid linkage of the temperature-resistant phenotype, plasmids were isolated from each of the temperature-resistant colonies and used to retransform the sec35-1 strain. The ends of the genomic inserts of 10 plasmids that restored temperature resistance were sequenced, and the resulting nucleotide sequences were used to search the Saccharomyces Genome Database (http://genome-www.stanford.edu/Saccharomyces/). Each of the plasmids contained a fragment of chromosome VII, and a 9-kb region bearing two uncharacterized ORFs and a gene encoding an ammonium transporter, MEP1 (Marini et al., 1994), was common to all of them. Each of the two novel ORFs was subcloned into a CEN-plasmid and tested for its ability to complement the temperature-sensitive phenotype of the sec35-1 strain. Only one of the two ORFs, designated YGR120c in the Saccharomyces Genome Database, restored growth at 37.5°C to wild-type levels.
To determine whether YGR120c encoded authentic Sec35p or a low copy number suppressor, integrative mapping was performed. A plasmid bearing YGR120c and URA3 was cleaved at a unique site within the coding sequence of YGR120c to direct integration into its genomic locus in the sec35-1 strain. The resulting Ura+ temperature-resistant strain was mated to a wild-type strain and the diploid was sporulated and dissected. Of the 24 tetrads examined, all segregants were temperature resistant. Since the temperature-sensitive phenotype did not segregate away from the integrated plasmid that was marked by URA3, the cloned gene is very tightly linked to the temperature-sensitive sec35-1 mutation. Therefore, integrative mapping confirms that YGR120c is SEC35.
Sequence of Sec35p and Sec35-1p
The predicted protein sequence encoded by SEC35 is shown in Fig. 1 a. Sec35p is composed of 275 residues and has a calculated mass of 31.8 kD and a pI of 4.56. Sec35p lacks any hydrophobic stretch of sufficient length to act as a transmembrane domain, nor does it contain consensus sequences for lipid modification; thus, it is predicted to be a cytosolic or peripheral membrane protein. Sec35p shows no significant propensity to form coiled coils as predicted by the PairCoil algorithm (Berger et al., 1995), although the COILS program (Lupas et al., 1991) does identify two regions of ∼20 amino acids with greater than 70% probability of forming coiled coils. Database searches reveal no homologues of Sec35p.
Figure 1.
Predicted amino acid sequence of Sec35p and Sec35-1p. (a) Protein sequence of Sec35p. SEC35 was sequenced as part of the Yeast Genome Sequencing project and corresponds to ORF YGR120c, GenBank/EMBL/DDBJ accession number Z72905. (b) Sequence of wild-type SEC35 from bp 613 to 628 with the transversion mutation at bp 624 (t to a) and corresponding codon change (tyrosine to ochre) present in the sec35-1 allele indicated with arrows.
Sequencing of the sec35-1 allele revealed a single base pair change at base pair 624 (t to a) resulting in the conversion of a tyrosine codon to an ochre stop codon (Fig. 1 b). The sec35-1 allele thus encodes a truncated protein with a predicted mol wt of 24 kD. Because the strain containing this truncation displays no growth defect at temperatures from 21° to 30°C, the carboxy-terminal 68 amino acids of the protein are not crucial to the function of the protein in this temperature range.
Deletion of SEC35 Results in a Severe Growth Defect
To determine whether SEC35 is an essential gene, the entire ORF was replaced in a diploid strain by the sequences in a LEU2 integrating plasmid (refer to Materials and Methods). After sporulation of the heterozygous deletion strain, tetrads were dissected on YPD plates and incubated at 30°C. Upon incubation for 7 d, tetrads were observed to contain two distinct sizes of colonies, with each tetrad containing two large and two (or, less frequently, one) very small segregants (Fig. 2 a). The small segregants were all Leu+, indicating the presence of the disruption (marked by LEU2), whereas the large segregants were all Leu−. Thus, SEC35 is not an essential gene although the small colony size of the sec35Δ haploid indicates a severe growth defect in the absence of SEC35.
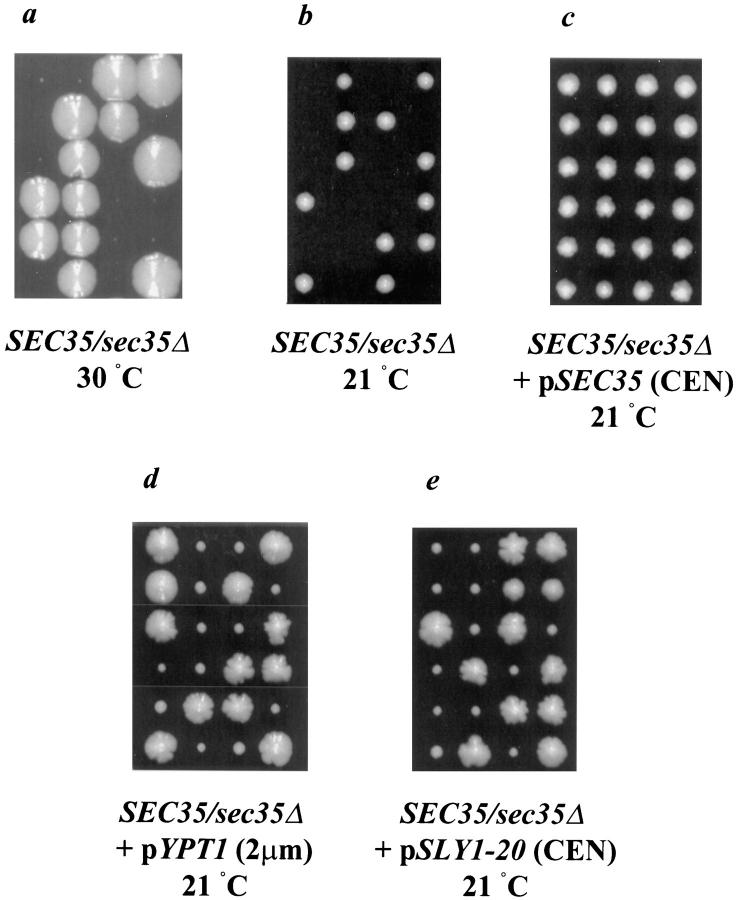
Figure 2.
A haploid strain containing a deletion of SEC35 exhibits a severe growth defect at 30°C and is inviable at 21°C; the cold-sensitive lethality can be rescued by the expression of SEC35, low levels of SLY1-20, or high levels of YPT1. The sec35Δ/SEC35 heterozygous deletion strain alone (a and b), or transformed with pSEC35 (CEN) (c), pYPT1 (2μm) (d), or pSLY1-20 (CEN) (e) was sporulated and dissected. The resulting tetrads were incubated at 30°C for 7 d (a) or at 21°C for 5 d (b and c) or 7 d (d and e) on YPD plates. In c–e, only tetrads containing four viable spores are shown; tetrads with two or three viable spores were assumed to result from tetrads in which one or both of the sec35Δ haploid spores failed to receive the suppressing plasmid.
In contrast, when dissected tetrads were incubated at 21°C a 2+:2− segregation for viability was observed (Fig. 2 b). All viable spores were Leu− and thus SEC35 is an essential gene at this temperature. Examination of the terminal phenotype of the sec35Δ haploid segregants revealed microcolonies of ∼25 cells and thus spores deleted for SEC35 can germinate and undergo four or five cell divisions before terminating vegetative growth. Transformation of the sec35Δ/SEC35 heterozygous diploid strain with a plasmid encoding SEC35 before sporulation and dissection complemented the cold-sensitive lethality of the sec35Δ haploid, resulting in 4+:0− segregation for viability (Fig. 2 c). Of the tetrads with four viable spores, the deletion (marked by LEU2) segregated 2+:2−, and the sec35Δ haploid colonies were invariably Ura+ due to their reliance on the plasmid for viability.
Multicopy Suppression Analysis
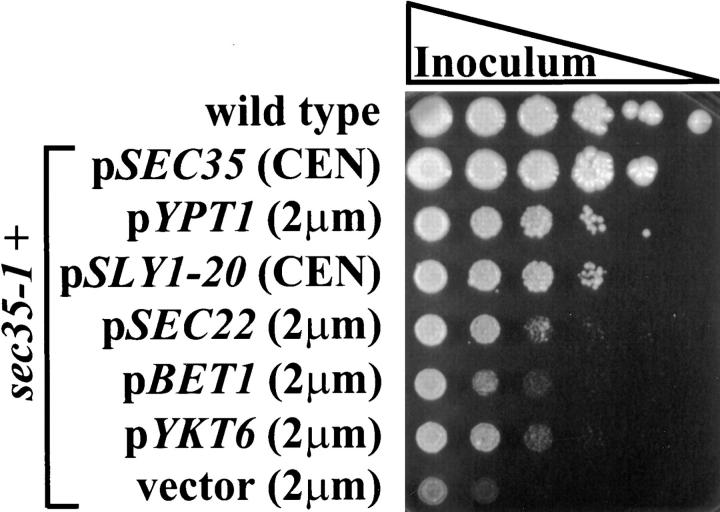
To gain insight into the function of Sec35p we undertook a systematic analysis of its genetic interactions with other genes involved in secretion. Since the sec35-1 mutant accumulates vesicles and is likely to be involved in vesicular docking or fusion, we first examined the interaction of SEC35 with other genes known to be involved in this step. To do this, we tested the ability of several known genes to act as multicopy suppressors of the sec35-1 temperature-sensitive growth defect. As shown in Fig. 3, the temperature sensitivity of sec35-1 could be suppressed by overexpression of YPT1 from a 2-μm plasmid, or by low-copy expression of the dominant mutation of SLY1, termed SLY1-20 (Ossig et al., 1991). YPT1 expressed from a low-copy (CEN) plasmid could also suppress the temperature-sensitive growth defect, although to a slightly lower level than the high-copy plasmid (data not shown). Thus, SEC35 displays a strong genetic interaction with two genes, YPT1 and SLY1, whose protein products are implicated in the regulation of vesicle docking (Rexach and Schekman, 1991; Segev, 1991; Lian et al., 1994; Søgaard et al., 1994; Lupashin et al., 1996; Lupashin and Waters, 1997). In contrast, high-copy expression of a third gene with a potential role in the regulation of vesicle docking, USO1 (Sapperstein et al., 1996), was incapable of suppressing sec35-1. Overexpression of the wild-type allele of SLY1 was also unable to suppress the growth defect of the sec35-1 strain, emphasizing the unique function of the SLY1-20 allele. To determine the specificity of the suppression of sec35-1 by YPT1, we also tested SEC4, a yeast YPT1 homologue involved in the docking stage of Golgi to plasma membrane transport (Goud et al., 1988). SEC4, however, was unable to suppress the growth defect when expressed from a 2-μm plasmid (data not shown), indicating that the suppression of sec35-1 by YPT1 is specific.
Figure 3.
Suppression of the temperature-sensitive growth phenotype of the sec35-1 strain. Wild-type and sec35-1 strains containing the indicated plasmids were grown to stationary phase in synthetic media and used for a series of 10-fold dilutions. The stationary phase culture and five serial dilutions of each strain were spotted onto YPD plates and grown for 4 d at 37.5°C.
The sec35-1 strain could also be suppressed, although to a much lesser extent, by overexpression of three v-SNAREs involved in docking vesicles to the cis-Golgi, Sec22p, Bet1p, and Ykt6p (Newman and Ferro-Novick, 1987; Ossig et al., 1991; Søgaard et al., 1994; McNew et al., 1997) (Fig. 3). A fourth v-SNARE, Bos1p, (Shim et al., 1991; Newman et al., 1992), however, was incapable of suppressing the sec35-1 defect (data not shown). Interestingly, when the genes encoding the two v-SNAREs required for Golgi to plasma membrane transport, SNC1 and SNC2 (Gerst et al., 1992; Protopopov et al., 1993), were tested for high-copy suppression of the sec35-1 temperature sensitivity, SNC2 was found to suppress the defect as well as, if not better than, each of the ER to Golgi v-SNAREs. In contrast, SNC1 had no effect (data not shown).
A number of other genes were also tested for the ability to suppress sec35-1 when expressed from 2-μm plasmids, including those encoding the ER to Golgi t-SNARE SED5 (Hardwick and Pelham, 1992), the putative intra-Golgi retrograde SNARE SFT1 (Banfield et al., 1995), the multifunctional v-SNARE VTI1 (Fischer von Mollard et al., 1997; Lupashin et al., 1997), a novel ER to Golgi transport factor BET3 (Rossi et al., 1995), and the GDP dissociation inhibitor SEC19, also known as GDI1 (Garrett et al., 1994). None of these genes improved the growth of sec35-1 at the restrictive temperature.
Because SLY1-20 and high-copy YPT1 strongly suppressed the temperature-sensitive allele of SEC35, we next tested whether they were capable of suppressing the cold-sensitive lethality of the genomic deletion. The heterozygous deletion strain was transformed with plasmids encoding either high levels of YPT1 or CEN levels of SLY1-20, sporulated, and then subjected to tetrad analysis. After incubation at room temperature for 7 d, the majority of the tetrads contained three or four viable spores (Fig. 2, d and e), rather than the 2+:2− segregation evident without the plasmid (Fig. 2 c). The sec35Δ haploid colonies (marked by LEU2) were without exception Ura+, and thus were viable by virtue of the suppressing plasmid. Suppression of the deletion is not complete, however, as indicated by the small size of the haploid colonies bearing the deletion compared with the wild-type segregants in each tetrad. The ability of SLY1-20 and high-copy YPT1 to suppress the deletion of SEC35 implies that their protein products function either downstream from, or in a parallel pathway with, Sec35p.
If Sec35p and Ypt1p were functioning in parallel pathways, one might expect the genetic interactions to be reciprocal in nature, and thus a temperature-sensitive allele of YPT1 might be suppressed by high level expression of SEC35. To test this hypothesis, the ypt1-3 strain was transformed with a 2-μm plasmid encoding SEC35 and then suppression was tested at the restrictive temperature. Although the levels of Sec35p expressed from the plasmid were ∼20× the genomic level (see Fig. 6), a high level of Sec35p was incapable of compensating for the temperature sensitivity of ypt1-3 (data not shown). This result implies that Ypt1p may function downstream of Sec35p, but the possibility of a parallel pathway cannot be completely ruled out. The ability of 2-μm–expressed SEC35 to suppress temperature-sensitive mutations in many other genes required for ER to Golgi transport was also tested. High levels of Sec35p were expressed in each of the following strains but were unable to suppress the growth defect at the restrictive temperatures: sec22-3, bos1-1, bet1-1 (ER to Golgi v-SNAREs), sec18-1 (yeast NSF), uso1-1 (ER to Golgi docking factor), sly1ts (t-SNARE–associated protein), bet3-1 (novel ER to Golgi factor), sft1-1 (putative retrograde t-SNARE), sec19-1 (GDP-dissociation inhibitor), sec4-8 (Golgi to plasma membrane rab-like GTPase), sec21-1 (γ-COP), sec27-1 (β′-COP), ret1-1 (α-COP), and sec34-2 (novel ER to Golgi factor).
Figure 6.
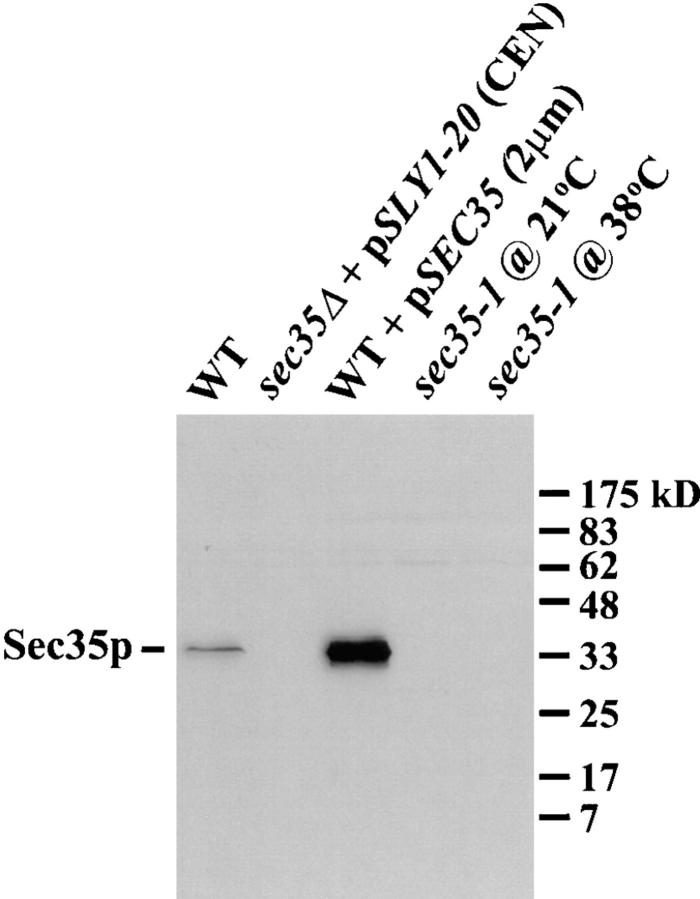
Characterization of the affinity-purified anti-Sec35p antibody. Extracts were made by glass bead lysis from logarithmically growing strains: wild-type, sec35Δ transformed with pSLY1-20 (CEN), wild-type transformed with pSEC35 (2μm), and sec35-1. All strains were incubated at 21°C before lysis except for the sec35-1 strain, which was maintained at 21°C or shifted to 38°C for 1 h before lysis. Extracts (0.2 OD595 per lane) were separated by SDS-12%PAGE and immunoblotted with affinity-purified anti-Sec35p antibodies. The migration of mol wt markers is shown on the right.
Suppression of the ER to Golgi Transport Block of the sec35-1 Strain by SLY1-20 and High-Copy YPT1
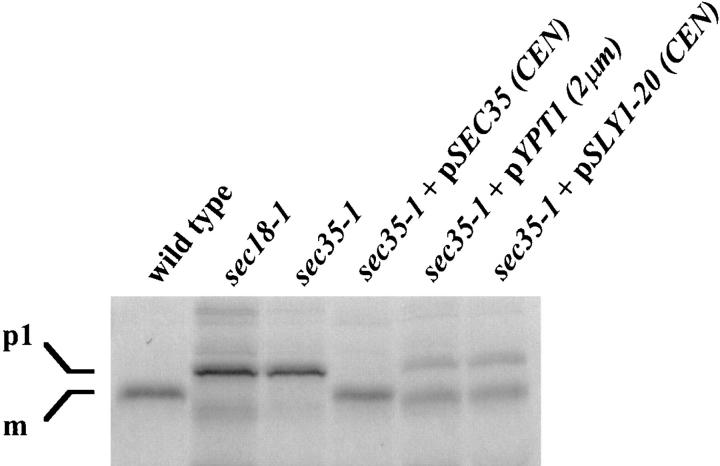
To confirm that the suppression of the temperature-sensitive growth defect of the sec35-1 strain by SLY1-20 and YPT1 was due to a direct effect on secretion, we tested whether expression of either gene could suppress the ER to Golgi transport defect. Thus, we examined the ability of CPY to transit from the ER to the vacuole in the mutant strain expressing low levels of SLY1-20 or high levels of YPT1 (Fig. 4). In wild-type cells, CPY is translocated into the ER where it is glycosylated, producing a 67-kD form termed p1 (Stevens et al., 1982). The oligosaccharides are extended in the Golgi generating the p2 form. Finally, upon transport to the vacuole, CPY is proteolytically processed to the 61-kD mature form. To examine progress of CPY through the secretory pathway, we performed a pulse-chase analysis of CPY maturation. Strains were shifted to the restrictive temperature for 45 min, labeled with Tran35S-label for 10 min, and then chased for 20 min with an excess of unlabeled cysteine and methionine. CPY was then immunoprecipitated, separated by SDS-PAGE, and then visualized by autoradiography. Wild-type and sec18-1 strains served as controls for the mature and p1 forms of CPY, respectively. Whereas the sec35-1 strain accumulated the p1 form of CPY, as shown previously (Wuestehube et al., 1996), the expression of Sly1-20p or Ypt1p was capable of suppressing the ER to Golgi defect (Fig. 4), resulting in the procession of the majority of CPY to its mature form. Nevertheless, since a significant amount of the p1 form of CPY did accumulate, neither of these proteins was capable of full suppression of the sec35-1 defect. A plasmid bearing the wild-type SEC35 gene, however, was capable of completely restoring the secretory block and all CPY was found in the mature form.
Figure 4.
Suppression of the ER to Golgi transport defect of sec35-1 by expression of SLY1-20 or high levels of YPT1. Wild-type and sec18-1 strains, as well as the sec35-1 strain with and without the indicated plasmids, were grown at 21°C to mid-logarithmic phase and then shifted to 39°C for 45 min. Cells were labeled at 39°C for 10 min with Tran35S-label and then chased for 20 min at the same temperature. CPY was immunoprecipitated from each strain, resolved by SDS-7%PAGE, and then visualized by autoradiography. The migration of the ER precursor (p1) and mature (m) forms of CPY are indicated on the left.
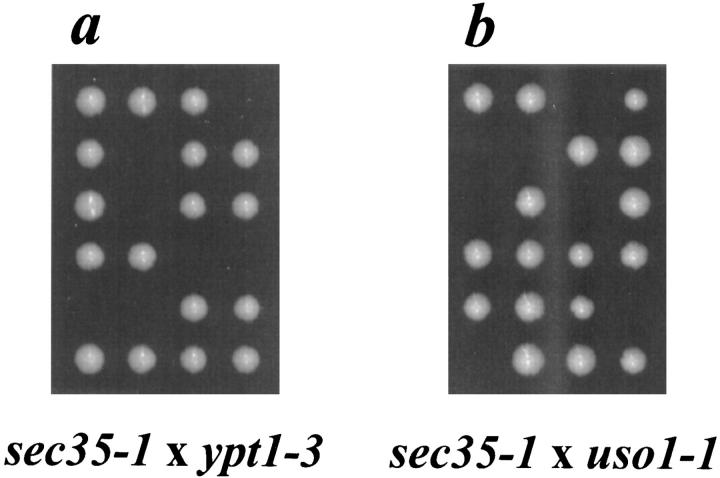
sec35-1 Displays a Synthetic Interaction with ypt1-3 and uso1-1
To further define the genetic interactions of SEC35 we tested whether the sec35-1 strain displayed a synthetic lethal interaction with mutant alleles of other known genes in the secretory pathway. Thus, the sec35-1 strain was mated individually to several strains each containing a temperature-sensitive mutation in one of the known secretory genes and the resulting diploids were sporulated and dissected. Due to the suppression of sec35-1 by YPT1, we first tested whether the mutant alleles of the two genes would display a synthetic interaction. Incubation of dissected tetrads from the diploid strain heterozygous for both sec35-1 and ypt1-3 at 30°C, a temperature permissive for both mutations, resulted in a pattern of viable spores consistent with a synthetic lethal interaction between the two alleles (Fig. 5 a): tetrads consisting of four viable spores segregated 0+:4− for temperature sensitivity (parental ditypes), tetrads consisting of two viable spores segregated 2+:0− for temperature sensitivity (nonparental ditypes), and tetrads consisting of three viable spores segregated 1+:2− for temperature-sensitivity (tetratypes). In all cases the inviable progeny must be the haploid segregant bearing both the sec35-1 and the ypt1-3 mutation.
Figure 5.
The sec35-1 allele exhibits a synthetic interaction with ypt1-3 and uso1-1. The sec35-1 strain was mated to a strain bearing the ypt1-3 allele (a) or the uso1-1 allele (b), and the resulting diploid strains were sporulated, dissected on YPD plates, and then incubated at 30°C for 3 d. Six representative tetrads of each dissection are shown.
Although we failed to see a genetic interaction between SEC35 and USO1 through multicopy suppression analysis, we tested whether mutations in the two genes displayed a synthetic interaction. Analysis of tetrads from a diploid strain heterozygous for both sec35-1 and uso1-1 revealed a pattern of viability identical to that seen with the segregants from the diploid heterozygous for both sec35-1 and ypt1-3 (Fig. 5 b). This synthetic lethal interaction is the first genetic interaction that has been observed between SEC35 and USO1. Interestingly, when tetrads dissected from diploid strains heterozygous for both sec35-1 and ypt1-3 or sec35-1 and uso1-1 were incubated at 21°C, all spores were viable (data not shown). The interaction between sec35-1 and both ypt1-3 and uso1-1 is best described as a conditional synthetic lethality.
Localization of Sec35p
To initiate a biochemical analysis of Sec35p, recombinant Sec35p was used to generate anti-Sec35p antibodies in rabbits. The polyclonal serum was affinity purified on bacterially expressed His6-Sec35p and used to probe total yeast protein extracts. As shown in Fig. 6, the affinity-purified anti-Sec35p antibody specifically recognizes a protein of ∼35 kD, close to the predicted mol wt of 31.8 kD. This band was absent in a strain deleted for SEC35 (expressing SLY1-20 from a CEN plasmid to suppress the sec35Δ growth defect), and is thus specific for Sec35p. As expected, Sec35p is highly overexpressed in a strain bearing a 2-μm SEC35 plasmid. In the temperature-sensitive sec35-1 strain incubated at either the permissive or the restrictive temperatures, the truncated 24-kD protein predicted from the sequence of the sec35-1 allele is not detected, indicating that the mutation greatly decreases the stability of the protein. However, upon longer exposure of the immunoblot the fragment of Sec35p was visible, migrating with an apparent mol wt of 27 kD. This truncated Sec35p is more abundant at the permissive temperature than at the restrictive temperature (data not shown).
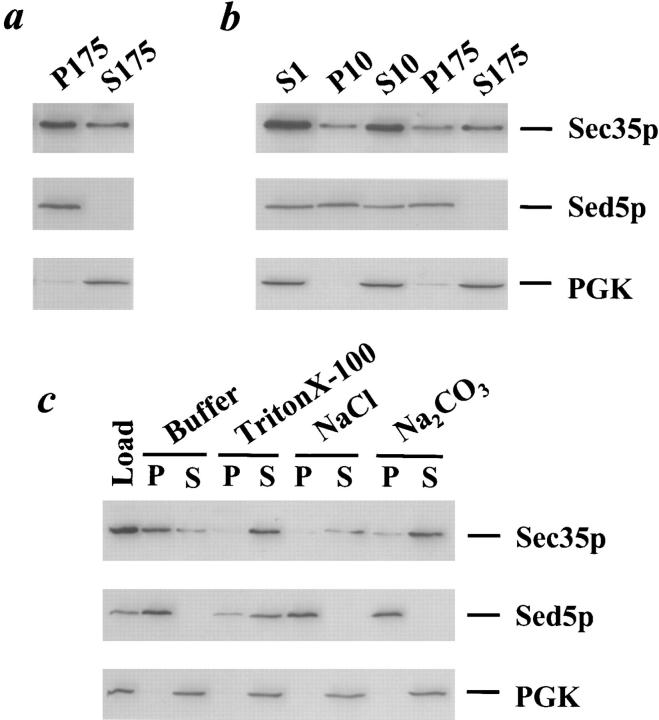
We next tested whether there existed a membrane-associated pool of Sec35p, as might be predicted for a protein required for membrane traffic. Wild-type yeast were lysed with glass beads and centrifuged at 1,000 g for 3 min to remove unbroken cells, yielding the low-speed supernatant S1. The S1 supernatant was then centrifuged at 175,000 g for 60 min and separated into pellet (P175) and supernatant (S175) fractions that should contain the membrane-bound and soluble proteins, respectively. The P175 and S175 fractions were resolved by SDS-PAGE and immunoblotted with affinity-purified antibodies against Sec35p, the cis-Golgi protein Sed5p, and the cytosolic protein PGK. As shown in Fig. 7 a, approximately two-thirds of Sec35p is found in the P175 fraction, and thus a significant proportion of the protein may be membrane-associated. As expected of an integral membrane protein, Sed5p localizes to the P175 fraction, whereas the cytosolic protein PGK remains in the supernatant fraction. Although we consistently observe a fraction of Sec35p in the P175 pellet, the proportion of pelletable protein is variable, ranging from about one-third to two-thirds of the total Sec35p (data not shown).
Figure 7.
Sec35p is a peripheral membrane protein. (a) Sec35p partitions into soluble and membrane-associated pools. A wild-type strain was grown to logarithmic phase, lysed with glass beads, and then centrifuged at 1,000 g for 3 min to generate the S1 supernatant fraction. The S1 supernatant was centrifuged at 175,000 g to obtain the S175 supernatant and P175 pellet fractions. (b) The membrane-associated portion of Sec35p fractionates similarly to the Golgi protein, Sed5p. The S1 supernatant was centrifuged at 10,000 g to generate the S10 supernatant and P10 pellet fractions, and the S10 was subsequently centrifuged at 175,000 g to obtain the S175 supernatant and P175 pellet fractions. (c) Sec35p behaves as a peripheral membrane protein. The S1 supernatant was incubated with buffer 88, 1% Triton X-100, 1 M NaCl, or 100 mM Na2CO3, pH 11, centrifuged at 175,000 g, and separated into supernatant and pellet fractions. (a–c) Aliquots of each fraction were separated by SDS-12%PAGE and immunoblotted with affinity-purified anti-Sec35p (top panel), anti-Sed5p (center panel), or anti-PGK (bottom panel) antibodies. The samples loaded in each lane were derived from equivalent amounts of starting material.
The localization of Sec35p was then examined through subcellular fractionation. The S1 fraction described above was centrifuged at 10,000 g for 15 min, yielding the P10 pellet and S10 supernatant fractions, and the S10 was further centrifuged at 175,000 g for 60 min to generate the P175 pellet and S175 supernatant fractions. With this series of differential centrifugation steps, all organelles of the secretory pathway should be contained in one or more of the pellet fractions: the ER and vacuole are located primarily in the P10 fraction, whereas the Golgi partitions between the P10 and P175 fractions (Walworth et al., 1989; Nakayama et al., 1992). To determine the localization of Sec35p, the supernatant and pellet fractions were resolved by SDS-PAGE and immunoblotted with affinity-purified antibodies. As shown in Fig. 7 b, although a portion of Sec35p once again remains in the S175, the membrane-associated pool of Sec35p partitions between the P10 and P175 fractions. The fractionation pattern of the membrane-associated Sec35p is similar to that of the cis-Golgi marker Sed5p, which also localizes to the P10 and P175 fractions. Nonetheless, because of the large pool of soluble Sec35p and the labile nature of its membrane association in vitro (data not shown), we have been unable to unequivocally determine its precise subcellular localization.
Finally, to determine whether the membrane association of Sec35p is as an integral or peripheral membrane protein, we attempted to extract the protein from the membrane using detergent, high salt, or high pH. Although both integral and peripheral membrane proteins should be extracted by detergent that solubilizes the membranes, the hallmark of a peripheral membrane protein is the ability to be extracted by salt or high pH, conditions which often disrupt protein–protein interactions. The S1 supernatant was thus incubated for 30 min on ice in either buffer, 1% Triton X-100, 1 M sodium chloride, or 100 mM sodium carbonate, pH 11, centrifuged at 175,000 g for 60 min, and then separated into pellet and supernatant fractions. The fractions were then resolved by SDS-PAGE and immunoblotted with antibodies against Sec35p, Sed5p, and PGK. Whereas Sed5p and PGK behaved as expected for an integral membrane protein or a cytosolic protein, respectively (Fig. 7 c), Sec35p was extractable by Triton X-100, NaCl, and Na2CO3 and thus behaves as a peripheral membrane protein.
Sec35p Is Required for ER to Golgi Vesicle Docking
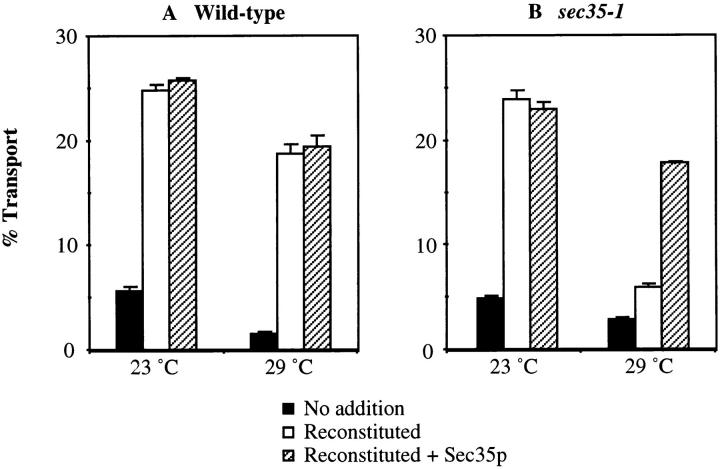
Our genetic and biochemical experiments suggest that Sec35p functions during the docking stage of ER to Golgi transport perhaps through peripheral association with membranes. To directly test this idea, we used a cell-free assay that measures distinct stages of ER to Golgi transport by following the radioactively labeled precursor of the secretory protein α-factor, glycosylated pro-α-factor ([35S]gp-α-factor). Transport in semi-intact yeast cells may be reproduced in vitro with washed membranes, purified COPII proteins, Uso1p, a protein complex called LMA1 (Xu et al., 1997), and ATP (Barlowe, 1997). COPII-coated vesicles that have budded from the ER are freely diffusible and then dock to Golgi membranes upon addition of Uso1p. Sec18p and LMA1 are not required for the docking phase, but are required for efficient fusion of vesicles (Barlowe, 1997). These soluble proteins represent some, but not all, of the genetically defined components for fusion of ER-derived vesicles. Other proteins that are peripherally bound to membranes (such as Sly1p and Ypt1p) or are integral membrane proteins (such as Sed5p, Bet1p and Bos1p) have required schemes designed to selectively inactivate their function in vitro such that reconstitution experiments can be performed (Cao et al., 1998). To inactivate Sec35p in vitro, we prepared membranes from the temperature-sensitive sec35-1 strain and measured reconstituted ER to Golgi transport at restrictive temperatures.
In Fig. 8, semi-intact yeast cell membranes prepared from a wild-type strain and the sec35-1 strain were incubated with purified transport factors at 23° or 29°C. Wild-type membranes show a modest decrease in transport at 29°C, whereas transport in the sec35-1 semi-intact membranes is reduced to near background levels (Fig. 8, A and B, compare open bars). This inhibition is specific for Sec35p function because the addition of purified His6-Sec35p (1.2 μg) restored transport in mutant membranes to near wild-type levels at 29°C (Fig. 8, A and B, compare hatched bars). The restoration of transport was maximal with 1.2 μg of His6-Sec35p, although stimulation could be observed with as little as 0.24 μg (data not shown). Because no stimulation of transport was observed when His6-Sec35p was added to wild-type reactions, it is likely that ample levels of Sec35p are present on washed membranes. However, we have established a specific requirement for Sec35p in cell-free transport of [35S]gp-α-factor from the ER to the Golgi when membranes are prepared from the sec35-1 strain and reactions performed at 29°C.
Figure 8.
Sec35p is required for ER to Golgi transport in vitro. Semi-intact yeast cell membranes prepared from wild-type (A) or sec35-1 (B) strains were incubated at 23° or 29°C for 60 min. Reactions contained an ATP regeneration system alone (No addition, solid bars), COPII, Uso1p, and LMA1 (Reconstituted, open bars), or COPII, Uso1p, LMA1 and His6-Sec35p (Reconstituted + Sec35p, hatched bars). Outer-chain modified forms of [35S]gp-α-factor were immunoprecipitated with anti–α1,6-mannose–specific antibodies and the percent transport reflects the amount of total [35S]gp-α-factor that has acquired Golgi-specific outer-chain modifications.
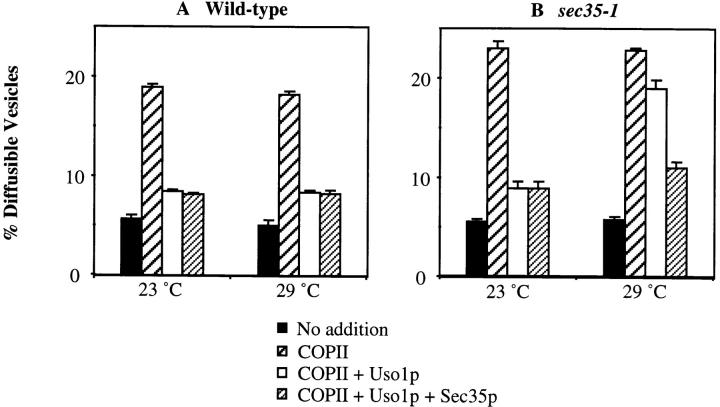
To further characterize Sec35p function in distinct transport stages, we assayed vesicle docking in mutant sec35-1 semi-intact cells. For this assay, docking is measured as the amount of COPII-generated vesicles that attach to Golgi membranes in the presence of Uso1p. The docked vesicles sediment with semi-intact cells after a brief centrifugation step (Barlowe, 1997). As seen in Fig. 9, wild-type and sec35-1 semi-intact membranes bud nearly equivalent amounts of ER-derived vesicles when purified COPII proteins are added at 23°and 29°C (Fig. 9, A and B, wide hatched bars). Uso1p addition docked this intermediate and reduced the level of freely diffusible vesicles in wild-type reactions but docking was not efficient in reactions containing sec35-1 membranes at the restrictive temperature (Fig. 9, A and B, compare open bars). Finally, the addition of purified His6-Sec35p restored docking efficiency in sec35-1 membranes at 29°C and did not alter the level of docking in wild-type reactions (Fig. 9, A and B, narrow hatched bars). In summary, ER to Golgi transport in sec35-1 membranes is prevented because vesicles fail to dock.
Figure 9.
Sec35p is required for vesicle docking. Semi-intact yeast cell membranes prepared from wild-type or sec35-1 strains were incubated at 23° or 29°C for 30 min. Reactions contained an ATP regeneration system alone (No addition, solid bars), COPII (COPII, wide hatched bars), COPII and Uso1p (COPII + Uso1p, open bars), or COPII, Uso1p, and His6-Sec35p (COPII, Uso1p + Sec35p, narrow hatched bars). Freely diffusible vesicles containing [35S]gp-α-factor were separated from semi-intact membranes by centrifugation at 18,000 g for 3 min. The percent diffusible vesicles reflects the amount of total concanavalin A precipitable [35S]gp-α-factor contained in the 18,000 g supernatant fluid.
Discussion
sec35-1 is a novel secretory mutant identified in a genetic screen for temperature-sensitive mutants that accumulate unprocessed α-factor internally (Wuestehube et al., 1996). The sec35-1 strain is defective in the transport of both soluble and membrane proteins from the ER to the Golgi apparatus. In addition, numerous vesicles accumulate at the restrictive temperature, suggesting that there is a defect in vesicle docking or fusion (Wuestehube et al., 1996). We have cloned SEC35 and demonstrated that it encodes a novel cytosolic protein of 32 kD. The cloned gene can suppress the temperature-sensitive growth defect, as well as the ER to Golgi secretory defect, of the sec35-1 strain; it is authentic SEC35 as shown by integrative mapping.
SEC35 displays a strong genetic interaction with two genes whose protein products are involved in the regulation of vesicle docking: the small GTP-binding protein YPT1 and the t-SNARE–associated protein SLY1. The temperature-sensitive growth defect of the sec35-1 strain is suppressed by either high levels of YPT1 or low levels of a dominant allele of SLY1, termed SLY1-20. Suppression of the growth defect is due to a restoration of secretory pathway function, since a vacuolar protein, CPY, which accumulates in its ER-modified form in the sec35-1 strain, is transported to the vacuole in the presence of SLY1-20 or high-copy YPT1. Neither gene can completely replace the function of SEC35, however, since only partial restoration is obtained.
High levels of YPT1 or low levels of SLY1-20 can also rescue the cold-sensitive lethality of the SEC35 deletion strain. This suppression of the sec35Δ strain indicates that both Ypt1p and Sly1p function downstream of, or in a parallel pathway with, Sec35p. However, overexpression of SEC35 fails to suppress the temperature-sensitive growth defect of either the ypt1-3 or the sly1ts strains, contrary to what might be expected if Sec35p operates in a parallel pathway. Thus, it seems more likely that Ypt1p and Sly1p function downstream of Sec35p. Furthermore, since Ypt1p acts upstream of the SNAREs and Sly1p to regulate SNARE complex assembly (Søgaard et al., 1994; Lupashin and Waters, 1997), Sec35p is likely to impinge on this process as well.
The interpretation that Sec35p is involved in regulating SNARE complex assembly is supported by the result that overexpression of a number of v-SNAREs involved in docking vesicles to the cis-Golgi (Sec22p, Bet1p, and Ykt6p) are capable of suppressing the temperature-sensitive growth defect of the sec35-1 strain, albeit to a lesser extent than YPT1 or SLY1-20. This suppression may be due to a mass action effect, in which increasing the amount of one v-SNARE can drive v/t-SNARE complex formation to an extent sufficient to allow a low level of growth. Intriguingly, the sec35-1 strain can also be suppressed by overexpression of one of the Golgi to plasma membrane v-SNAREs, SNC2. This suppression pattern suggests that Sec35p may function at more than one step in the secretory pathway. However, overexpression of the Golgi to plasma membrane Ypt1p-like GTPase, SEC4, is unable to suppress sec35-1. Attempts to address the effect of Sec35p at multiple stages by preloading the secretory pathway of the sec35-1 strain with labeled CPY and shifting to the nonpermissive temperature were inconclusive, since the temperature-sensitive defect of the sec35-1 strain is slow relative to the rate of secretion (data not shown).
Multicopy suppression analysis failed to identify a genetic interaction between SEC35 and the third gene whose protein product has a role in the regulation of SNARE complex assembly, USO1. However, mutations in SEC35, USO1, and YPT1 exhibit a very similar pattern of suppression since each is suppressed by both SLY1-20 and ER to Golgi v-SNAREs (Sapperstein et al., 1996). In addition, the three genes are related through synthetic lethal interactions: the sec35-1 allele is conditionally lethal in combination with either uso1-1 or ypt1-3, and uso1-1 displays a synthetic lethal interaction with ypt1-3 (Sapperstein et al., 1996). These data suggest that the functions of Sec35p, Uso1p, and Ypt1p in ER to Golgi transport are intimately associated.
At the ER to Golgi step of the secretory pathway it is likely that v/t-SNARE complex formation occurs during the docking phase of the reaction, before membrane fusion. This interpretation is based on the observation that Sec18p (NSF), which is required to disassemble v/t-SNARE complexes (Söllner et al., 1993a ,b) allows docked vesicles to fuse in vitro (Barlowe, 1997). Because our genetic analysis indicates that Sec35p functions the docking step, we tested this hypothesis directly using an in vitro assay that monitors individual stages of ER to Golgi transport. In this cell-free assay, vesicle budding from washed semi-intact cells requires the addition of COPII components, whereas docking is dependent upon both COPII proteins and Uso1p. Membranes isolated from the sec35-1 strain and incubated at the restrictive temperature were found to be competent for vesicle formation but defective in Uso1p-dependent vesicle docking. Thus, Sec35p appears to be required for the docking of ER-derived vesicles.
Sec35p was not identified as one of the cytosolic components required for vesicle docking in wild-type semi-intact cells (Barlowe, 1997). In addition, purified Sec35p does not stimulate docking in wild-type cells and thus sufficient Sec35p is present in the cytosol-free membranes to provide its essential function. These results indicate that Sec35p is associated with membranes. Indeed, we find that a significant portion of Sec35p behaves as a peripheral membrane protein, potentially associating with the Golgi. The above in vitro data from wild-type cells implies that the membrane-associated pool of Sec35p may be the functional form of this protein since neither the removal of soluble Sec35p nor the addition of recombinant Sec35p affects docking. It is possible, however, that either the soluble fraction of Sec35p is not removed with the washes or that very little Sec35p, soluble or membrane-associated, is required for function.
Recent results from a number of laboratories suggest the following outline for ER to Golgi transport. First, vesicle budding occurs under the direction of COPII proteins resulting in a freely diffusible vesicle (Barlowe et al., 1994). Docking then occurs in two stages: in the first stage, the vesicle is “tethered” to the Golgi in a reaction that requires Uso1p and Ypt1p (Cao et al., 1998) while the second stage may be a SNARE-dependent event characterized by an assembled v/t-SNARE complex spanning the vesicle and target membrane, as proposed in the original SNARE hypothesis (Söllner et al., 1993b ). In this case, the role of Uso1p and Ypt1p in the regulation of SNARE-complex assembly (Sapperstein et al., 1996; Lupashin and Waters, 1997) may equate to a dependence on the tethered state for SNARE-dependent docking. After docking, Sec18p would act to disassemble the v/t-SNARE complex and allow the reaction to proceed towards fusion. What is perhaps least clear in this scenario for ER to Golgi transport is the exact role of Sec18p in facilitating progression towards fusion. For example, it is possible that Sec18p acts to disassemble v/t-SNARE complexes within (not across) the vesicle and/or target membranes of the tethered intermediate. This disassembly would allow the freed v- and t-SNAREs to enter into new v/t-SNARE complexes that span the vesicle and target membranes. Fusion could then ensue without a further requirement for Sec18p, as suggested by the recent work on vacuole fusion in yeast (Mayer et al., 1996; Mayer and Wickner, 1997) and the reconstitution of membrane fusion with SNAREs alone (Weber et al., 1998). The essential difference in the models is that the first (tethering → SNARE-dependent docking → Sec18p function → fusion) a vesicle-target membrane-spanning v/t-SNARE complex is disassembled by Sec18p, allowing fusion, whereas in the second model (tethering → Sec18p function → SNARE- dependent docking → fusion) the vesicle-target membrane-spanning v/t-SNARE complex is, in effect, generated by Sec18p and proceeds spontaneously to fusion. Regardless of the precise role of Sec18p in generating or consuming a SNARE-docked state, our genetic and biochemical results clearly place Sec35p as acting in the Uso1p-dependent docking phase of ER to Golgi vesicle docking.
Acknowledgments
This work was supported by grants from the National Institutes of Health (NIH) (GM48753 to M.G. Waters and GM52549 to C. Barlowe), the Pew Scholars Program in the Biomedical Science to C. Barlowe, and a fellowship from the Lucille P. Markey Charitable Trust to M.G. Waters. S.M. VanRheenen was supported by an NIH training grant (GM07312).
Abbreviations used in this paper
- CEN
centromere
- COP
coat protein
- CPY
carboxypeptidase Y
- gp-α-factor
glycosylated pro-α-factor
- GST
glutathione-S-transferase
- NSF
N-ethylmaleimide–sensitive fusion protein
- ORF
open reading frame
- PGK
phosphoglycerate kinase
- SC
synthetic complete medium
- SNAP
soluble NSF–attachment protein
- t-SNARE
target membrane–SNAP receptor
- v-SNARE
vesicle-SNAP receptor
- YPD
rich medium
Footnotes
We are very grateful to R. Schekman (University of California, Berkeley, CA) for the sec35 strain. We thank S. Emr, S. Ferro-Novick, J. Gerst, C. Kaiser, P. Novick, H. Pelham, M. Rose (Princeton University, Princeton, NJ), R. Schekman, H.D. Schmitt and members of their laboratories for generously supplying reagents and strains, and D. Hasara (Princeton University) for expert assistance in antibody production. We are particularly grateful to S. Harris, K. Paul, and S. Sapperstein (all three from Princeton University) for critical reading of the manuscript and to all members of the laboratories for helpful discussion.
Address all correspondence to M. Gerard Waters, Department of Molecular Biology, Princeton University, Princeton, NJ 08544. Tel.: (609) 258-2891. Fax: (609) 258-1701. E-mail: gwaters@molbio.princeton.edu
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Baker D, Hicke L, Rexach M, Schleyer M, Schekman R. Reconstitution of SECgene product-dependent intercompartmental protein transport. Cell. 1988;54:335–344. doi: 10.1016/0092-8674(88)90196-1. [DOI] [PubMed] [Google Scholar]
- Banfield DK, Lewis MJ, Pelham HRB. A SNARE-like protein required for traffic through the Golgi complex. Nature. 1995;375:806–809. doi: 10.1038/375806a0. [DOI] [PubMed] [Google Scholar]
- Barlowe C. Coupled ER to Golgi transport reconstituted with purified cytosolic components. J Cell Biol. 1997;139:1097–1108. doi: 10.1083/jcb.139.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C, Schekman R. SEC12encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature. 1993;365:347–349. doi: 10.1038/365347a0. [DOI] [PubMed] [Google Scholar]
- Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- Barroso M, Nelson DS, Sztul E. Transcytosis-associated protein (TAP)/p115 is a general fusion factor required for binding of vesicles to acceptor membranes. Proc Natl Acad Sci USA. 1995;92:527–531. doi: 10.1073/pnas.92.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger B, Wilson DB, Wolf E, Tonchev T, Milla M, Kim PS. Predicting coiled coils by use of pairwise residue correlations. Proc Natl Acad Sci USA. 1995;92:8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne HR. Do GTPases direct membrane traffic in secretion? . Cell. 1988;53:669–671. doi: 10.1016/0092-8674(88)90081-5. [DOI] [PubMed] [Google Scholar]
- Cao X, Ballew N, Barlowe C. Initial docking of ER-derived vesicles requires Uso1p and Ypt1p but is independent of SNARE proteins. EMBO (Eur Mol Biol Organ) J. 1998;17:2156–2165. doi: 10.1093/emboj/17.8.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dascher C, Ossig R, Gallwitz D, Schmitt HD. Identification and structure of four yeast genes (SLY) that are able to suppress the functional loss of YPT1, a member of the RAS superfamily. Mol Cell Biol. 1991;11:872–885. doi: 10.1128/mcb.11.2.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eakle KA, Bernstein M, Emr SD. Characterization of a component of the yeast secretion machinery: identification of the SEC18gene product. Mol Cell Biol. 1988;8:4098–4109. doi: 10.1128/mcb.8.10.4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elble R. A simple and efficient procedure for transformation of yeasts. BioTechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- Espenshade P, Gimeno RE, Holzmacher E, Teung P, Kaiser CA. Yeast SEC16 gene encodes a multidomain vesicle coat protein that interacts with Sec23p. J Cell Biol. 1995;131:311–324. doi: 10.1083/jcb.131.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro-Novick S, Novick P. The role of GTP-binding proteins in transport along the exocytic pathway. Annu Rev Cell Biol. 1993;9:575–599. doi: 10.1146/annurev.cb.09.110193.003043. [DOI] [PubMed] [Google Scholar]
- Fischer von Mollard G, Nothwehr SF, Stevens TH. The yeast v-SNARE Vti1p mediates two vesicle transport pathways through interactions with the t-SNAREs Sed5p and Pep12p. J Cell Biol. 1997;137:1511–1524. doi: 10.1083/jcb.137.7.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett MD, Zahner JE, Cheney CM, Novick PJ. GDI1encodes a GDP dissociation inhibitor that plays an essential role in the yeast secretory pathway. EMBO (Eur Mol Biol Organ) J. 1994;13:1718–1728. doi: 10.1002/j.1460-2075.1994.tb06436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerst JE, Rodgers L, Riggs M, Wigler M. SNC1, a yeast homolog of the synaptic vesicle-associated membrane protein/synaptobrevin gene family: genetic interactions with the RAS and CAPgenes. Proc Natl Acad Sci USA. 1992;89:4338–4342. doi: 10.1073/pnas.89.10.4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goud B, Salminen A, Walworth NC, Novick PJ. A GTP-binding protein required for secretion rapidly associates with secretory vesicles and the plasma membrane in yeast. Cell. 1988;53:753–768. doi: 10.1016/0092-8674(88)90093-1. [DOI] [PubMed] [Google Scholar]
- Griff IC, Schekman R, Rothman JE, Kaiser CA. The yeast SEC17gene product is functionally equivalent to mammalian α-SNAP protein. J Biol Chem. 1992;267:12106–12115. [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coliwith plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hardwick KG, Pelham HR. SED5encodes a 39-kD integral membrane protein required for vesicular transport between the ER and the Golgi complex. J Cell Biol. 1992;119:513–521. doi: 10.1083/jcb.119.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow, E., and D. Lane. 1988. Antibodies. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 726 pp.
- Hicke L, Schekman R. Yeast Sec23p acts in the cytoplasm to promote protein transport from the endoplasmic reticulum to the Golgi complex in vivo and in vitro. . EMBO (Eur Mol Biol Organ) J. 1989;8:1677–1684. doi: 10.1002/j.1460-2075.1989.tb03559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L, Yoshihisa T, Schekman R. Sec23p and a novel 105-kDa protein function as a multimeric complex to promote vesicle budding and protein transport from the endoplasmic reticulum. Mol Cell Biol. 1992;3:667–676. doi: 10.1091/mbc.3.6.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosono R, Hekimi Y, Kamiya Y, Sassa T, Murakami S, Nishiwaki K, Miwa J, Taketo A, Kodaira K-I. The unc-18 gene encodes a novel protein affecting the kinetics of acetylcholine metabolism in the nematode Caenorhabditis elegans. . J Neurochem. 1992;58:1517–1525. doi: 10.1111/j.1471-4159.1992.tb11373.x. [DOI] [PubMed] [Google Scholar]
- Kaiser CA, Schekman R. Distinct sets of SECgenes govern transport vesicle formation and fusion early in the secretory pathway. Cell. 1990;61:723–733. doi: 10.1016/0092-8674(90)90483-u. [DOI] [PubMed] [Google Scholar]
- Lian JP, Stone S, Jiang Y, Lyons P, Ferro-Novick S. Ypt1p implicated in v-SNARE activation. Nature. 1994;372:698–701. doi: 10.1038/372698a0. [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Lupashin VV, Waters MG. t-SNARE activation through transient interaction with a Rab-like guanosine triphosphatase. Science. 1997;276:1255–1258. doi: 10.1126/science.276.5316.1255. [DOI] [PubMed] [Google Scholar]
- Lupashin VV, Hamamoto S, Schekman RW. Biochemical requirements for the targeting and fusion of ER-derived transport vesicles with purified yeast Golgi membranes. J Cell Biol. 1996;132:277–289. doi: 10.1083/jcb.132.3.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupashin VV, Pokrovskaya ID, McNew JA, Waters MG. Characterization of a novel yeast SNARE protein implicated in Golgi retrograde traffic. Mol Biol Cell. 1997;8:2659–2676. doi: 10.1091/mbc.8.12.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini AM, Vissers S, Urrestarazu A, Andre B. Cloning and expression of MEP1 gene encoding an ammonium transporter in Saccharomyces cerevisiae. . EMBO (Eur Mol Biol Organ) J. 1994;13:3456–3463. doi: 10.1002/j.1460-2075.1994.tb06651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, Wickner W. Docking of yeast vacuoles is catalyzed by the Ras-like GTPase Ypt7p after symmetric priming by Sec18p (NSF) J Cell Biol. 1997;136:307–313. doi: 10.1083/jcb.136.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, Wickner W, Haas A. Sec18p (NSF)-driven release of Sec17p (α-SNAP) can precede docking and fusion of yeast vacuoles. Cell. 1996;85:83–94. doi: 10.1016/s0092-8674(00)81084-3. [DOI] [PubMed] [Google Scholar]
- McNew JA, Søgaard M, Lampen NM, Machida S, Ye RR, Lacomis L, Tempst P, Rothman JE, Söllner TH. Ykt6p, a prenylated SNARE essential for ER-Golgi transport. J Biol Chem. 1997;272:17776–17783. doi: 10.1074/jbc.272.28.17776. [DOI] [PubMed] [Google Scholar]
- Miller, J.H. 1972. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor. NY. 466 pp.
- Nakajima H, Hirata A, Ogawa Y, Yonehara T, Yoda K, Yamasaki M. A cytoskeleton-related gene, uso1, is required for intracellular protein transport in Saccharomyces cerevisiae. . J Cell Biol. 1991;113:245–260. doi: 10.1083/jcb.113.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano A, Brada D, Schekman R. A membrane glycoprotein, Sec12p, required for protein transport from the endoplasmic reticulum to the Golgi apparatus in yeast. J Cell Biol. 1988;107:851–863. doi: 10.1083/jcb.107.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano A, Muramatsu M. A novel GTP-binding protein, Sar1p, is involved in transport from the endoplasmic reticulum to the Golgi apparatus. J Cell Biol. 1989;109:2677–2691. doi: 10.1083/jcb.109.6.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Nagasu T, Shimma Y, Kuromitsu J, Jigami Y. OCH1encodes a novel membrane bound mannosyltransferase: outer chain elongation of asparagine-linked oligosaccharides. EMBO (Eur Mol Biol Organ) J. 1992;11:2511–2519. doi: 10.1002/j.1460-2075.1992.tb05316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AP, Ferro-Novick S. Characterization of new mutants in the early part of the yeast secretory pathway isolated by a [3H]mannose suicide selection. J Cell Biol. 1987;105:1587–1594. doi: 10.1083/jcb.105.4.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AP, Shim J, Ferro-Novick S. BET1, BOS, and SEC22are members of a group of interacting yeast genes required for transport from the endoplasmic reticulum to the Golgi complex. Mol Cell Biol. 1990;10:3405–3414. doi: 10.1128/mcb.10.7.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AP, Groesch ME, Ferro-Novick S. Bos1p, a membrane protein required for ER to Golgi transport in yeast, co-purifies with the carrier vesicles and with Bet1p and the ER membrane. EMBO (Eur Mol Biol Organ) J. 1992;11:3609–3617. doi: 10.1002/j.1460-2075.1992.tb05445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi A, Gibson J, Gregor I, Schatz G. Import of proteins into mitochondria. The precursor of cytochrome c1 is processed in two steps, one of them heme-dependent. J Biol Chem. 1982;257:13042–13047. [PubMed] [Google Scholar]
- Ossig R, Dascher C, Trepte HH, Schmitt HD, Gallwitz D. The yeast SLYgene products, suppressors of defects in the essential GTP-binding Ypt1 protein, may act in endoplasmic reticulum-to-Golgi transport. Mol Cell Biol. 1991;11:2980–2993. doi: 10.1128/mcb.11.6.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevsner J, Hsu SC, Braun JE, Calakos N, Ting AE, Bennett MK, Scheller RH. Specificity and regulation of a synaptic vesicle docking complex. Neuron. 1994;13:353–361. doi: 10.1016/0896-6273(94)90352-2. [DOI] [PubMed] [Google Scholar]
- Protopopov V, Govindan B, Novick P, Gerst JE. Homologs of the synaptobrevin/VAMP family of synaptic vesicle proteins function on the late secretory pathway in S. cerevisiae. . Cell. 1993;74:855–861. doi: 10.1016/0092-8674(93)90465-3. [DOI] [PubMed] [Google Scholar]
- Pryer NK, Salama NR, Schekman R, Kaiser CA. Cytosolic Sec13p complex is required for vesicle formation from the endoplasmic reticulum in vitro. . J Cell Biol. 1993;120:865–875. doi: 10.1083/jcb.120.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach MF, Schekman RW. Distinct biochemical requirements for the budding, targeting, and fusion of ER-derived transport vesicles. J Cell Biol. 1991;114:219–229. doi: 10.1083/jcb.114.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M.D. 1995. Modern and post-modern genetics in Saccharomyces cerevisiae. In The Yeasts. A.H. Rose, A.E. Wheals, and J.S. Harrison, editors. Academic Press, San Diego, CA. 69–120.
- Rose MD, Novick P, Thomas JH, Botstein D, Fink GR. A Saccharomyces cerevisiaegenomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Rose, M., F. Winston, and P. Hieter. 1990. Methods in Yeast Genetics. A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 198 pp.
- Rossi G, Kolstad K, Stone S, Palluault F, Ferro-Novick S. BET3encodes a novel hydrophilic protein that acts in conjunction with yeast SNAREs. Mol Biol Cell. 1995;6:1769–1780. doi: 10.1091/mbc.6.12.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JE. Mechanisms of intracellular protein transport. Nature. 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- Salama NR, Yeung T, Schekman RW. The Sec13p complex and reconstitution of vesicle budding from the ER with purified cytosolic proteins. EMBO (Eur Mol Biol Organ) J. 1993;12:4073–4082. doi: 10.1002/j.1460-2075.1993.tb06091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapperstein SK, Walter DM, Grosvenor AR, Heuser JE, Waters MG. p115 is a general vesicular transport factor related to the yeast ER-Golgi transport factor Uso1p. Proc Natl Acad Sci USA. 1995;92:522–526. doi: 10.1073/pnas.92.2.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapperstein SK, Lupashin VV, Schmitt HD, Waters MG. Assembly of the ER to Golgi SNARE complex requires Uso1p. J Cell Biol. 1996;132:755–767. doi: 10.1083/jcb.132.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schekman R, Orci L. Coat proteins and vesicle budding. Science. 1996;271:1526–1533. doi: 10.1126/science.271.5255.1526. [DOI] [PubMed] [Google Scholar]
- Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acid as a carrier. CurrGenet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Schulze KL, Littleton JT, Salzberg A, Halachmi N, Stern M, Lev Z, Bellen HJ. rop, a Drosophila homolog of yeast Sec1 and vertebrate n-Sec1/Munc-18 proteins, is a negative regulator of neurotransmitter release in vivo. . Neuron. 1994;13:1099–1108. doi: 10.1016/0896-6273(94)90048-5. [DOI] [PubMed] [Google Scholar]
- Segev N. Mediation of the attachment or fusion step in vesicular transport by the GTP-binding Ypt1 protein. Science. 1991;252:1553–1556. doi: 10.1126/science.1904626. [DOI] [PubMed] [Google Scholar]
- Shaywitz DA, Espenshade PJ, Gimeno RE, Kaiser CA. COPII subunit interactions in the assembly of the vesicle coat. J Biol Chem. 1997;272:25413–25416. doi: 10.1074/jbc.272.41.25413. [DOI] [PubMed] [Google Scholar]
- Shim J, Newman AP, Ferro-Novick S. The BOS1gene encodes an essential 27-kD putative membrane protein that is required for vesicular transport from the ER to the Golgi complex in yeast. J Cell Biol. 1991;113:55–64. doi: 10.1083/jcb.113.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Søgaard M, Tani K, Ye RR, Geromanos S, Tempst P, Kirchhausen T, Rothman JE, Söllner T. A Rab protein is required for the assembly of SNARE complexes in the docking of transport vesicles. Cell. 1994;78:937–948. doi: 10.1016/0092-8674(94)90270-4. [DOI] [PubMed] [Google Scholar]
- Söllner T, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE. A protein assembly-disassembly pathway in vitrothat may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell. 1993a;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- Söllner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993b;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- Stevens T, Esmon B, Schekman R. Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell. 1982;30:439–448. doi: 10.1016/0092-8674(82)90241-0. [DOI] [PubMed] [Google Scholar]
- Walworth NC, Goud B, Ruohola H, Novick PJ. Fractionation of yeast organelles. Methods Cell Biol. 1989;31:335–356. doi: 10.1016/s0091-679x(08)61618-0. [DOI] [PubMed] [Google Scholar]
- Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Söllner T, Rothman JE. SNAREpins: Minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- Wilson DW, Wilcox CA, Flynn GC, Chen E, Kuang WJ, Henzel WJ, Block MR, Ullrich A, Rothman JE. A fusion protein required for vesicle-mediated transport in both mammalian cells and yeast. Nature. 1989;339:355–359. doi: 10.1038/339355a0. [DOI] [PubMed] [Google Scholar]
- Wuestehube LJ, Duden R, Eun A, Hamamoto S, Korn P, Ram R, Schekman R. New mutants of Saccharomyces cerevisiaeaffected in the transport of proteins from the endoplasmic reticulum to the Golgi complex. Genetics. 1996;142:393–406. doi: 10.1093/genetics/142.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Mayer A, Muller E, Wickner W. A heterodimer of thioredoxin and IB 2cooperates with Sec18p (NSF) to promote yeast vacuole inheritance. J Cell Biol. 1997;136:299–306. doi: 10.1083/jcb.136.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]