Abstract
The recently cloned human chemoattractant receptor-like (CMKRL)1, which is expressed in vivo in CD4-positive immune cells, has structural homology with the two chemokine receptors C-C chemokine receptor (CCR)5 and C-X-C chemokine receptor (CXCR)4, which serve as the major coreceptors necessary for fusion of the HIV-1 envelope with target cells. In view of the structural similarity, CMKRL1 was tested for its possible function as another HIV-1 coreceptor after stable expression in murine fibroblasts bearing the human CD4 receptor. The cells were infected with 10 primary clinical isolates of HIV-1, and entry was monitored by semiquantitative PCR of viral DNA. The efficiency of the entry was compared with the entry taking place in CD4-positive cells expressing either CCR5 or CXCR4. Seven of the isolates used CMKRL1 for viral entry; they were mainly of the syncytium-inducing phenotype and also used CXCR4. Entry efficiency was higher with CMKRL1 than with CXCR4 for more than half of these isolates. Three of the ten isolates did not use CMKRL1; instead, entry was mediated by both CCR5 and CXCR4. The experiments thus indicate that CMKRL1 functions as a coreceptor for the entry of HIV-1 into CD4-positive cells. In the course of this study, leukotriene B4 was shown to be the natural ligand for this receptor (now designated BLTR), which therefore represents a novel type of HIV-1 coreceptor along with the previously identified chemokine receptors. BLTR belongs to the same general chemoattractant receptor family as the chemokine receptors but is structurally more distant from them than are any of the previously described HIV-1 coreceptors.
During infection, HIV-1 generally fuses with and enters its target cell through a series of events involving two classes of cell membrane receptors. It is well established that the virus associates with the CD4 receptor (1–3), which induces conformational changes in the glycoprotein envelope (4, 5) allowing the virus to bind to a second, heptahelix-type cell-surface receptor. This coreceptor triggers the necessary fusion of the envelope with the cell membrane, facilitating viral entry. Two major coreceptors have been identified, both belonging to the chemokine family of G-protein-coupled receptors: C-C chemokine receptor (CCR)5, which is primarily involved in the infection of the so-called M-tropic, or non-syncytium-inducing (NSI), viral strains (6–10), and C-X-C chemokine receptor (CXCR)4, which is required for the fusion of primarily T-tropic, or syncytium-inducing (SI), strains (11).
A key observation leading to the recent discovery of the viral entry cofactors was the finding that certain β-chemokines have a strong suppressive effect on the HIV-1 infection in vitro (12). Analysis of viral isolates obtained sequentially from infected individuals has shown that there is a loss of the sensitivity to inhibition by β-chemokines, along with a shift of the virus from NSI to SI phenotype (13–17). This finding suggests that, with progression of the infection, there is a shift in chemokine receptor usage from CCR5 to CXCR4. These results are in accordance with the findings that dual-tropic virus strains use both types of coreceptors, and that additional coreceptors exist for certain subsets of primary viruses, in addition to their primary usage of CCR5 or CXCR4 (9, 10, 18–20). Hence, the usage of the two major coreceptors may be viewed as extremes in an adaptation process, along which the virus expands its receptor usage to include several different coreceptors. A region in the gp120 envelope glycoprotein that has been shown to be a major determinant in chemokine sensitivity is the third hypervariable loop, or V3 loop (21–23). In view of the varying coreceptor usage, it is important to search for additional G-protein-coupled receptors that might facilitate the entry of various primary HIV-1 isolates into suitable target cells during the course of the infection.
Within the superfamily of G-protein-coupled, seven-transmembrane receptors, the chemokine receptors form a structurally related group that belongs to the subfamily of leukocyte chemoattractant receptors. This subfamily also includes receptors for the so-called classical chemoattractants, such as leukotriene B4, platelet-activating factor, and the anaphylatoxins (24). Recently, a novel chemoattractant-like receptor, CMKRL1, was cloned (25). The natural ligand was subsequently shown to be leukotriene B4 (26, 27). Although the leukotrienes themselves have been known for more than two decades (28), no leukotriene receptor has been cloned previously. The leukotriene B4 receptor, or BLTR, is widely expressed in the immune system (25, 27), including thymus, spleen, lymph nodes, and peripheral blood mononuclear cells (PBMC), and it is ≈30% identical with CCR5 and CXCR4 which, in turn, exhibit the same degree of homology when compared with each other.
In the present study, we have been able to show that CMKRL1/BLTR supports the entry of select primary isolates of mainly the SI phenotype into CD4-positive murine host cells. Thus, an essentially new class of coreceptors that is used and even required for the cellular entry of HIV-1 has been identified.
MATERIALS AND METHODS
Receptor-Expressing Cells.
NIH 3T3 cells (murine fibroblasts) were transfected with cDNA encoding various human receptors and selected to obtain homogenous, stably expressing cell lines verified by Northern blot hybridization of the corresponding receptor message. The experiments focused on the human leukotriene receptor CMKRL1, clone Lyme21–9 (25), permanently transfected into NIH 3T3.CD4 cells. Cells expressing CD4 alone or in combination with CCR5 or CXCR4 (kindly provided by Dan Littman, Skirball Institute for BioMolecular Medicine, New York University Medical Center, New York) were established, as previously described (29, 30). The receptor-expressing cells served as targets for HIV-1 infectivity assays.
Virus Isolates.
The primary virus isolates from HIV-infected individuals were recovered from phytohemagglutinin (PHA)-activated (Murex Diagnostics, Norcross, GA) PBMC or CD8-depleted PBMC that were cocultivated with PHA-stimulated PBMC obtained from two healthy blood donors. The PBMC were separated by Ficoll (Pharmacia) gradient centrifugation. To improve the efficiency of HIV replication, CD8-positive cells were depleted from the patient’s PBMC by immunomagnetic beads (Dynabeads M-450 CD8) according to the manufacturer’s (Dynal, Great Neck, NY) protocol. Freshly isolated PBMC or CD8-depleted PBMC (2 × 106 cells) were activated with PHA (1 μg/ml) for 3 days and cocultivated with PHA blasts from the two normal donors at 1:3 ratio. The cultures were maintained in complete RPMI 1640 medium (BioWhittaker) supplemented with 10% fetal calf serum (Gemini Biological Products, Calabasas, CA) and 10 ng/ml recombinant human IL-2 (R & D Systems). The cultures were evaluated for the presence of HIV-1 p24 core antigen by a commercially available ELISA (DuPont/NEN) for a total of 4 weeks. At the time of positive virus cultures, stocks were generated by expanding the isolates in PHA-stimulated donor PBMC, after which the viruses were filter-sterilized, aliquotted, and cryopreserved at −150°C. After the infection experiments, virus phenotype was determined by using the MT-2 cell assay (31). Briefly, MT-2 cells and PHA-activated PBMC from healthy donors were infected with the primary isolates in parallel (16–200 ng per 0.5 × 106 cells). MT-2 cell cultures were monitored microscopically for syncytium formation and p24 antigen production. Isolates producing syncytia and p24 antigen in the culture were classified as MT-2 positive with an SI phenotype, whereas virus classified as MT-2 negative with an NSI phenotype replicated in PBMC but did not induce syncytia or p24 production in MT-2 cells. The clinical HIV-1 isolates, the patient’s CD4-positive T cell number, the virus phenotype, and p24 levels in the original virus stock are given in Table 1. The prototypic M-tropic isolate, BaL (32), and the T-tropic isolate, IIIB (33), propagated extensively in the neoplastic T cell line H9, were used in control experiments.
Table 1.
Primary and laboratory isolates of HIV-1 used in the infection experiments: Patients and characteristics of the isolates
| Patient | CD4 count,* cells/mm3 | Virus isolate | Virus phenotype† | p24 core antigen,‡ ng/ml |
|---|---|---|---|---|
| A | 400 | DS pbmc CD8- | SI | 150 |
| B | 354 | OR pbmc CD8- | SI | 262 |
| C | 79 | 5774 PBMC | SI | 68 |
| D | — | G3 | NSI | 120 |
| E | 923 | OT pbmc CD8- | SI | 110 |
| F | 411 | P001 pbmc CD8- | SI | 24 |
| G | 386 | L002 PBMC | SI | 336 |
| H | — | 22069-05 | SI | 312 |
| J | — | JV 1083 | NSI | 16 |
| K | — | 571 | SI | 170 |
| — | — | IIIB | SI | 237 |
| — | — | BaL | NSI | 688 |
Primary virus isolates from 10 patients, as well as two laboratory-adapted model isolates, tested on CD4-positive mouse fibroblasts (NIH 3T3) expressing various types of human chemotactic receptors. Primary isolates were grown by coculturing with stimulated human PBMC (in the four isolates designed “pbmc CD8-,” PBMC were depleted of CD8-positive cells by using DynaBeads). G3 and JV 1083 are primary isolates from Nigerian clade G/A. HIV-1 BaL was passaged several times exclusively in primary adherent macrophage cultures derived from peripheral blood. HIV-1 IIIB is a long-term, laboratory-passed isolate grown in H9 cells.
Absolute counts of CD4-positive lymphocytes in patients were measured at the time of initial blood draw.
Phenotype was classified in MT-2 assays after the infection.
Viral titers were evaluated in vitro by using p24 core antigen ELISA.
Infection Experiments.
NIH 3T3 cells expressing CD4 alone or in combination with one of the test receptors were initially grown for at least two passages before 105 or 2 × 105 cells were seeded in 6-well plates. On the following day, they were infected with the various primary isolates (10–75 ng of p24 antigen) or the laboratory isolates, BaL or IIIB (10 ng of p24 antigen), for 2 h at 37°C. The cells were then washed three times with PBS and resuspended in complete culture medium (DMEM containing 10% bovine serum) and grown for another 16 h. They were harvested, washed several times in cell medium, and pelleted by centrifugation. The supernatant was removed and the pellet stored until further analysis at −20°C.
PCR Amplification of HIV-1 DNA.
The infected cell pellet was dissolved at room temperature in 100 ml of K buffer containing 100 μg/ml proteinase K, followed by incubation at 55°C for 60 min, after which the protease was inactivated at 95°C for 10 min (34). Ten microliters of the suspension was used as template in 50-μl PCR for semiquantitative amplification of the gag region of HIV-1 by using recombinant Taq DNA polymerase (GIBCO/BRL) in 35 cycles, each consisting of 94°C 45-sec denaturing (pseudo-hot start), 55°C 1-min annealing, 72°C 1-min extension, and 72°C 7-min final extension. After every sixteenth PCR sample, a negative control was included (K buffer instead of pellet suspension as template), followed by a positive control [10 ng of pHXB2, a full-length proviral clone derived from the IIIB isolate (35, 36)]. The sense primer, SK 38 (5′-AGTAGCAACCCTCTATTGTGTGCA-3′), corresponded to nucleotides 1030–1053 of the pHXB2 sequence, and the antisense primer, SK 39 (5′-ACATTTGCATGGCTGCTTGATGT-3′), corresponded to nucleotides 1367–1389. The 360-bp PCR product was separated by 1.5% agarose gel electrophoresis (10 μl per lane), transferred to a nitrocellulose membrane filter, and hybridized with a 32P-labeled internal probe (5′-TGGCTCCTTCTGATAATGCTGA-3′), which corresponds to nucleotides 1305–1326 in the pHXB2 sequence. The filters were exposed to Kodak X-Omat AR film for 24 h.
Computer-Assisted Densitometry.
The intensity of the hybridization signals from the bands in the autoradiograms was measured by using National Institutes of Health Image software version 1.61, after image acquisition with a charge-coupled device camera. The optical densities of the individual bands were recorded, and the film background as well as the respective biological background (electrophoretic bands obtained after infection of cells expressing CD4 alone) were then subtracted. Comparison was made only between values obtained in the same set of experiments, on the basis of simultaneous PCR amplifications and lined up in the same autoradiogram. Results from repeated experiments were averaged.
RESULTS
Entry of HIV-1 into the murine host cell model was estimated in terms of the amount of PCR-amplified virus DNA measured by computer-assisted densitometry in autoradiograms from Southern blots after cellular uptake and reverse transcription of viral RNA (Fig. 1). To adjust the infectious dose so that little or no virus entry occurred in control cells expressing the CD4 receptor alone (NIH 3T3.CD4 cells), pilot titration experiments were performed by using BaL at 1, 5, 10, 15, and 20 ng of p24 antigen. The optimal dose was found to be 10 ng, which resulted in a negligible entry of the virus (corresponding to a mean optical density in the autoradiograms that yielded pixel values between 0 and 15, on a scale of 0 to 255). In all experiments testing primary isolates, this dose of IIIB or BaL was used as a positive control after infection of NIH 3T3.CD4 cells coexpressing their respective coreceptor, CXCR4 or CCR5 (Fig. 1). As shown in Fig. 2, coexpression of either of these receptors increased the entry of the corresponding virus strain into the cells to a mean optical density of 200 pixels. The graph also shows that the “cross-entry” of BaL into cells expressing CXCR4, or IIIB in the presence of CCR5, was insignificant.
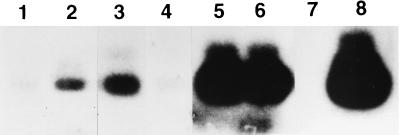
Figure 1.
Examples from a single autoradiogram compiled from a typical infection experiment on NIH 3T3 cells with isolate P001 pbmc CD8- (see Table 1; the mean value from all four experiments is shown in Fig. 3), together with controls. Positive bands are of 360-bp size. Lane 1 shows the minimal background entry of virus into cells expressing CD4 alone; all other lanes show viral entry into CD4-positive cells coexpressing other receptors: lane 2, CMKRL1/BLTR; lane 3, CXCR4; lane 4, CCR5. Lanes 5 and 6 show the biological controls, where CD4-positive cells expressing CXCR4 or CCR5 are infected with IIIB or BaL, respectively. Lane 7 shows the negative PCR control (K buffer instead of infected cell pellet suspension), lane 8 shows the positive PCR control (pHXB2 plasmid serving as template).
Figure 2.
Infection experiments of CD4-positive NIH 3T3 cells (murine fibroblasts), coexpressing three different human chemotactic receptors (CMKRL1/BLTR, CXCR4, and CCR5), with the two laboratory-adapted model isolates, IIIB and BaL (see Table 1). After 16-h exposure to the various isolates, viral cDNA was amplified in semiquantitative PCR followed by hybridization of Southern blots with a 32P-labeled, 22-mer oligonucleotide probe internal to the pHXB2 viral sequence. The graph illustrates computer-assisted densitometry of the autoradiograms and is expressed as relative density (number of pixels) of the hybridizing bands after subtraction of the film background and of any positive signal from control cells expressing CD4 alone. Bars show averages from two to four experiments, the standard deviation being less than 20% of the mean absolute pixel values.
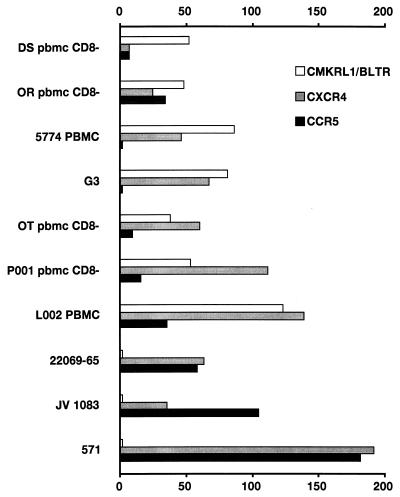
Pilot experiments were also carried out with the primary isolates to adjust the dose levels to a negligible nonspecific entry of virus into the CD4-positive control cells (Fig. 1), and subsequent experiments were carried out at the 40-ng dose. There was an approximately 50-fold increase in the cellular entry of patient isolates A–G when testing cells expressing CMKRL1 compared with the CD4-positive controls (Fig. 3). All isolates (except DS pbmc CD8-) also entered cells that expressed CXCR4 instead. In the case of isolates A–D, the entry into cells expressing CMKRL1 was even higher than the entry into cells equipped with CXCR4. A particularly high level of viral entry into cells expressing CMKRL1 was observed when the titer of the isolate L002 had been increased by passaging in PBMC (Fig. 3).
Figure 3.
Infection experiments of CD4-positive NIH 3T3 cells (murine fibroblasts), coexpressing three different human chemotactic receptors (CMKRL1/BLTR, CXCR4, and CCR5), with 10 clinical isolates of HIV-1 (see Table 1). For further technical details, see Fig. 2.
In accordance with the mainly SI characteristics of isolates A-G, the use of CCR5 in most instances (except for isolate OR pbmc CD8-) was lower than that of CXCR4 (Fig. 3). This is in contrast to isolates H–K, which readily entered both CD4-positive cells expressing either CXCR4 or CCR5. None of these isolates used CMKRL1 for their entry (Fig. 3). When comparing Figs. 2 and 3, it is evident that in most instances the primary virus isolates used the major coreceptors, CXCR4 and CCR5, less efficiently than the laboratory-adapted isolates used the same coreceptors.
Consistent with the above findings on the clinical isolates, the SI type of laboratory isolate, IIIB, also showed significant entry into cells expressing CMKRL1 (Fig. 2), in addition to the entry into cells transfected with CXCR4. BaL, on the other hand, which was efficiently taken up into CD4-positive cells expressing CCR5, did not enter CMKRL1-expressing cells to any significant degree (Fig. 2).
DISCUSSION
In the present study, we have used a cell model in which various human chemotactic receptors were stably expressed, together with human CD4, in otherwise nonpermissive mouse fibroblasts. Infection experiments were carried out with 10 primary isolates of HIV-1, followed by PCR amplification of viral cDNA. They suggest that a new type of chemotactic cell surface receptor—the leukotriene B4 receptor, BLTR—is required for the entry of certain, primarily SI-type, clinical isolates into CD4-positive target cells. In accordance with their SI phenotype, the isolates also efficiently use the major chemokine receptor, CXCR4, rather than CCR5.
Several types of target cells have been used in previous infection experiments. Among them, NIH 3T3 mouse fibroblasts (7, 8, 11) are often used, as they are nonpermissive because of the lack of necessary membrane receptors. Moreover, even when murine cells are transfected with human CD4, they still resist infection with all tested strains of HIV-1 (7). Accordingly, we could minimize any entry of test isolates into such cells when nonspecific uptake of virus particles was eliminated, by properly reducing infectious dose or exposure time in the infection assays. We could confirm that prototypic isolates of M- or T-tropic virus strains efficiently enter the CD4-positive murine cells coexpressing human CCR5 (7) or CXCR4 (11). These reactions were therefore used as positive biological controls in the experiments with clinical test isolates.
PCR-based amplification of viral cDNA is a convenient way to evaluate cellular entry and intracellular reverse transcription of HIV-1 (21). In calibration trials with pHXB2, a full-length clone derived from HIV-1 IIIB (35, 36) which we used throughout as internal control in the PCR reactions, the method gave a semiquantitative reflection of the number of viral DNA copies present in the host cells after infection in vitro (21). Hence, we expressed the degree of infection with the clinical isolates as the amount of increase of viral cDNA in cells stably expressing the various test receptors beyond the level in control cells expressing CD4 alone.
The presently identified new HIV coreceptor was originally cloned as an “orphan” G-protein linked, heptahelix receptor with structural similarities to receptors in the chemoattractant family and provisionally named CMKRL1 (25). At the time when the molecular genetic features of this receptor were submitted for publication, the two chemoattractant receptors, CXCR4 (then an “orphan” receptor, too) and CCR5, had just been identified as HIV coreceptors (6–11). It was striking that all three receptors showed some 30% overall sequence similarity. Moreover, the distribution of CMKRL1 in cells and tissues of the immune system (25) would make this new receptor a suitable target for HIV-1. With these findings in mind, we set out to investigate whether CMKRL1 could function as coreceptor for HIV-1. This approach was further stimulated by observations indicating that additional coreceptors seemed to exist which, in concert with either of the two major coreceptors, could promote cellular entry of some HIV-1 strains (9, 10). In the course of our infection experiments, leukotriene B4 was identified in separate studies (26, 27) as the natural ligand for CMKRL1; thus the first member of the leukotriene receptor family has been cloned.
The role of CCR5 and CXCR4 as the major HIV coreceptors and their interaction with M- and T-tropic virus strains, respectively, is well established (6–11). Dual-tropic isolates use both receptors (10). Some M-tropic and dual-tropic strains can also use other receptors, such as CCR2b and CCR3 (9, 10, 37). More recently, additional β-chemokine receptors have been shown to support HIV-1 entry (18, 19). This finding suggests that in the course of disease progression, the viruses expand their receptor usage to include also secondary coreceptors, providing the viruses with new cell populations to target (38).
Our experiments introduce the recently cloned leukotriene B4 receptor BLTR, originally designated CMKRL1 (25–27), as a new type of HIV-1 coreceptor for select virus strains. We used primary clinical isolates because they should reflect more closely the actual mode of viral usage at a given disease stage, compared with isolates that have been passaged and adapted in immortalized cell lines. The murine target cell model reflects a limited, albeit crucial, phase in the virus life span, namely the cellular entry and subsequent retrograde transcription. Preliminary data from a different infection model, consisting of human astrocytes coexpressing CD4 and CMKRL1, have indicated that the latter receptor supports the infection of yet another SI-type isolate (designated LW), as reflected in p24 ELISA (unpublished observations).
The pathophysiological significance of the new coreceptor CMKRL1/BLTR is emphasized by the fact that, like previously identified coreceptors, it is widely distributed in the human immune system (25, 27), where it is expressed in CD4-positive cells (39–41). This receptor is unusual in that the major coreceptor “partner” is CXCR4 rather than CCR5. Accordingly, the virus isolates using CMKRL1/BLTR turned out to be of the SI phenotype (except the G3 isolate, which is of a different genotype and uncharacteristically used CXCR4, although it was classified as NSI phenotype). Also, the laboratory-adapted, T-tropic protoype virus strain IIIB was taken up into cells expressing CMKRL1/BLTR. The three isolates that did not use this receptor all behaved as dual-tropic isolates (10). In its coreceptor function, BLTR could be reminiscent of another coreceptor, CCR2b, which also mediates entry of select HIV-1 isolates. There is reason to believe that CCR2b may be important in late stages of the infection, as evidenced by observations that the mutant V64I seems to be relevant for the rate of progression of AIDS (42). It should be noted that four of the virus isolates tested entered cells expressing BLTR more efficiently than cells expressing CXCR4 (or CCR5).
The interaction between gp120 and CD4 induces conformational changes, which promote the physical association between this complex and the coreceptor (43–45). The search for determinants of HIV entry via the chemokine receptors has revealed a complex picture. All extracellular domains of the receptors seem to play a role in viral entry, although the amino terminus and the second extracellular domain appear to be particularly important (46–50). The fusion activity of the receptor does not require G-protein signaling (51). Because the isolates using BLTR as coreceptor also entered cells expressing CXCR4 (rather than CCR5), it should be important to elucidate any structural similarities between the first two receptors. Thus, Thr-3 and Phe-16 in BLTR (25, 27) correspond to identical residues in CXCR4, whereas Ser and Ile are found in the respective positions of CCR5. Another region of the receptor that stands out is the third transmembrane helix domain (Fig. 4), where eight distinct residues (including the functionally important residue His) are identical between BLTR and CXCR4; only two are identical between CXCR4 and CCR5. In all other transmembrane or extra-cellular domains, no, or maximally four, residues are identical between the first two receptors, whereas in those regions the identities between CXCR4 and CCR5 are always more numerous. Although most of the modeling aimed at elucidating virus–coreceptor interaction has focused on the extracellular domains of the receptor (46–50), proper knowledge of the three-dimensional configuration of the receptor molecule is still lacking, which hampers the detailed mapping of virus-binding epitopes. Thus, paradoxical results have been obtained with chimeric receptor constructs, and there are indications that transmembrane and cytoplasmic domains may also make significant contributions to coreceptor function (49).
Figure 4.
Alignment of deduced amino acid sequence comprising the putative third transmembrane helix region of the three receptors tested, BLTR (originally designated CMKRL1; GenBank/European Molecular Biology Laboratory Data Bank accession no. X98356), CXCR4 (accession no. M99293), and CCR5 (accession no. X91492). The full sequence of CXCR4 is given. For the other two, only the identical amino acid residues are shown, the nonidentical residues being marked as dashes.
Our results on a novel type of HIV-1 coreceptor emphasize that a whole array of membrane receptors may be used by the virus when it adapts over a long period of time. Such receptors may not be found only within a single family (of chemokines), but may involve also other, structurally related but distinct, receptors outside the family. The decisive criterion might be in which cell type, or how extensive among various types of cells, the coreceptor is expressed to serve as a suitable vehicle for the virus when it invades a given population of cells.
Acknowledgments
This work was supported by grants from the Swedish Medical Research Council (Grant 05680), the Crafoord and Segerfalk Foundations in Sweden, the University of Maryland School of Medicine (Grant 02-1-30883), and by funds directed to the Institute of Human Virology from the State of Maryland.
ABBREVIATIONS
- HIV-1
HIV type 1
- CCR
C-C chemokine receptor
- CXCR
C-X-C chemokine receptor
- NSI
non-syncytium-inducing
- SI
syncytium-inducing
- CMKRL1
chemoattractant receptor-like 1
- BLTR
leukotriene B4 receptor
- PBMC
peripheral blood mononuclear cells
- PHA
phytohemagglutinin
References
- 1.Dalgleish A G, Beverly P C L, Clapham P R, Crawford D H, Greaves M F, Weiss R A. Nature (London) 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 2.Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman J C, Montagnier L. Nature (London) 1984;312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 3.Maddon P, Dalgleish A, McDougal J S, Clapham P, Weiss R, Axel R. Cell. 1986;47:333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 4.Sattenau Q J, Zolla-Pazner S, Poignard P. Virology. 1995;206:713–714. doi: 10.1016/s0042-6822(95)80094-8. [DOI] [PubMed] [Google Scholar]
- 5.Chan D C, Fass D, Berger J M, Kim P S. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 6.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 7.Deng H-K, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, et al. Nature (London) 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 8.Dragic T, Litwin V, Allaway G P, Martin S R, Huang X, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, et al. Nature (London) 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 9.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, et al. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 10.Doranz B, J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 11.Feng Y, Broder C C, Kennedy P E, Berger E A. Science. 1996;272:872–877. [Google Scholar]
- 12.Cocchi F, DeVico A L, Garzino-Demo A, Arya S K, Gallo R C, Lusso P. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 13.Zhang L, Huang Y, He T, Cao Y, Ho D D. Nature (London) 1996;383:768. doi: 10.1038/383768a0. [DOI] [PubMed] [Google Scholar]
- 14.Jansson M, Popovic M, Karlsson A, Cocchi F, Rossi P, Albert J, Wigzell H. Proc Natl Acad Sci USA. 1996;93:15382–15387. doi: 10.1073/pnas.93.26.15382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simmons G, Wilkinson D, Reeves J D, Dittmar M T, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, et al. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozak S L, Platt E J, Madani N, Ferro F J, Feden K, Kabat D. J Virol. 1997;71:873–882. doi: 10.1128/jvi.71.2.873-882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao F, Alkhatib G, Peden K W C, Sharma G, Berger E A, Farber J M. J Exp Med. 1997;185:2015–2023. doi: 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng H-K, Unutmaz D, Kewal Ramani V N, Littman D R. Nature (London) 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 20.Loetscher M, Amara A, Oberlin E, Brass N, Legler D F, Loetscher P, D’Apuzzo M, Meese E, Rousset D, Virelizier J-L, et al. Curr Biol. 1997;7:652–660. doi: 10.1016/s0960-9822(06)00292-2. [DOI] [PubMed] [Google Scholar]
- 21.Cocchi F, DeVico A L, Garzino-Demo A, Cara A, Gallo R C, Lusso P. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 22.Oravecz P, Pall M, Norcross M A. J Immunol. 1996;157:1329–1332. [PubMed] [Google Scholar]
- 23.Speck R F, Wehrly K, Platt E J, Atchison R E, Charo I F, Kabat D, Chesebro B, Goldsmith M A. J Virol. 1997;71:7136–7139. doi: 10.1128/jvi.71.9.7136-7139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy P M. Annu Rev Immunol. 1994;12:593–633. doi: 10.1146/annurev.iy.12.040194.003113. [DOI] [PubMed] [Google Scholar]
- 25.Owman C, Nilsson C, Lolait S J. Genomics. 1996;37:187–194. doi: 10.1006/geno.1996.0541. [DOI] [PubMed] [Google Scholar]
- 26.Owman C, Sabirsh A, Boketoft Å, Olde B. Biochem Biophys Res Commun. 1997;240:162–166. doi: 10.1006/bbrc.1997.7628. [DOI] [PubMed] [Google Scholar]
- 27.Yokomizu T, Izumi T, Chang K, Tukawa Y, Shimizu T. Nature (London) 1997;387:620–624. doi: 10.1038/42506. [DOI] [PubMed] [Google Scholar]
- 28.Samuelsson B. Trends Pharmacol Sci. 1980;1:227–230. [Google Scholar]
- 29.Landau N R, Littman D R. J Virol. 1992;66:5110–5113. doi: 10.1128/jvi.66.8.5110-5113.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pear W S, Nolan G P, Scott M L, Baltimore D. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karlsson A, Parsmyr K, Sandström E, Fenyö E M, Albert J. J Clin Microbiol. 1994;32:364–370. doi: 10.1128/jcm.32.2.364-370.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gartner S, Markovits P, Markovits D M, Kaplan M H, Gallo R C, Popovic M. Science. 1986;233:215–219. doi: 10.1126/science.3014648. [DOI] [PubMed] [Google Scholar]
- 33.Popovic M, Sarngadharan M G, Read E, Gallo R C. Science. 1984;224:497–500. doi: 10.1126/science.6200935. [DOI] [PubMed] [Google Scholar]
- 34.Kawasaki E S. In: PCR Protocols: A Guide to Methods and Applications. Innis M A, Gelfand D H, Sninsky J J, White T J, editors. New York: Academic; 1990. pp. 146–152. [Google Scholar]
- 35.Fisher A G, Collalti E, Ratner L, Gallo R C, Wong-Staal F. Nature (London) 1985;316:262–265. doi: 10.1038/316262a0. [DOI] [PubMed] [Google Scholar]
- 36.Feinberg M B, Jarrett R F, Aldovini A, Gallo R C, Wong-Staal F. Cell. 1986;46:807–817. doi: 10.1016/0092-8674(86)90062-0. [DOI] [PubMed] [Google Scholar]
- 37.He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hofmann W, Newman W, et al. Nature (London) 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 38.Clapham P R, Weiss R A. Nature (London) 1997;388:230–231. doi: 10.1038/40758. [DOI] [PubMed] [Google Scholar]
- 39.Payan D G, Missirian-Bastian A, Goetzl E J. Proc Natl Acad Sci USA. 1984;81:3501–3505. doi: 10.1073/pnas.81.11.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rola-Pleszczynski M, Bouvrette L, Gingras D, Girard M. J Immunol. 1987;139:513–517. [PubMed] [Google Scholar]
- 41.Zachariae C, Ternowitz T, Larsen C G, Nielsen V, Thestrup-Pedersen K. Arch Dermatol Res. 1988;280:354–357. doi: 10.1007/BF00426613. [DOI] [PubMed] [Google Scholar]
- 42.Smith M W, Dean M, Carrington M, Winkler C, Huttley G A, Lomb D A, Goedert J J, O’Brien T R, Jacobson L P, et al. Science. 1997;277:959–965. doi: 10.1126/science.277.5328.959. [DOI] [PubMed] [Google Scholar]
- 43.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, et al. Nature (London) 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 44.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, More J P. Nature (London) 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 45.Lapham C K, Ouyang J, Chandrasekhar B, Nguyen N Y, Dimitrov D S, Golding H. Science. 1996;274:602–605. doi: 10.1126/science.274.5287.602. [DOI] [PubMed] [Google Scholar]
- 46.Rucker J, Samson M, Doranz B J, Libert F, Berson J F, Smyth R J, Collman R G, Broder C C, Vassart G, Doms R W, et al. Cell. 1996;87:437–446. doi: 10.1016/s0092-8674(00)81364-1. [DOI] [PubMed] [Google Scholar]
- 47.Luz Z, Berson J F, Chen Y, Turner J D, Zhang T, Sharron M, Jenks M H, Wang Z, Kim J, Hoxie J A, et al. Proc Natl Acad Sci USA. 1997;94:6426–6431. doi: 10.1073/pnas.94.12.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alkhatib G, Ahuja S S, Light D, Mummidi S, Berger E A, Ahuja S K. J Biol Chem. 1997;272:19771–19776. doi: 10.1074/jbc.272.32.19771. [DOI] [PubMed] [Google Scholar]
- 49.Alkhatib G, Berger E A, Murphy P M, Pease J E. J Biol Chem. 1997;272:20420–20426. doi: 10.1074/jbc.272.33.20420. [DOI] [PubMed] [Google Scholar]
- 50.Doranz B J, Lu Z H, Rucker J, Zhang T Y, Sharron M, Cen Y H, Wang Z X, Guo H H, Du J G, Accavitti M A, et al. J Virol. 1997;71:6305–6314. doi: 10.1128/jvi.71.9.6305-6314.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alkhatib G, Locati M, Kennedy P E, Murphy P M, Berger E A. Virology. 1997;234:340–348. doi: 10.1006/viro.1997.8673. [DOI] [PubMed] [Google Scholar]