Abstract
Facultative heterochromatin is a cytological manifestation of epigenetic mechanisms that regulate gene expression. Constitutive heterochromatin is marked by distinctive histone H3 methylation and the presence of HP1 proteins, but the chromatin modifications of facultative heterochromatin are less clear. We have examined histone modifications and HP1 in the facultative heterochromatin of nucleated erythrocytes and show that mouse and chicken erythrocytes have different mechanisms of heterochromatin formation. Mouse embryonic erythrocytes have abundant HP1, increased tri-methylation of H3 at K9 and loss of H3 tri-methylation at K27. In contrast, we show that HP1 proteins are lost during the differentiation of chicken erythrocytes, and that H3 tri-methylation at both K9 and K27 is reduced. This coincides with the appearance of the variant linker histone H5. HP1s are also absent from erythrocytes of Xenopus and zebrafish. Our data show that in the same cell lineage there are different mechanisms for forming facultative heterochromatin in vertebrates. To our knowledge, this is the first report of cell types that lack HP1s and that have gross changes in the levels of histone modifications.
Keywords: differentiation/erythrocyte/gene expression/heterochromatin/histone modification
Introduction
Chromatin regulates critical cellular processes such as transcription, DNA replication and repair. Although chromatin packaging at the level of the nucleosome is well understood, it is not clear how further levels of packaging occur. DNA binding dyes stain some chromatin domains within nuclei more intensely than others, differentiating chromatin into heterochromatin and euchromatin respectively. Euchromatin generally corresponds to active (or potentially active) gene regions, while heterochromatin is normally transcriptionally silent. Constitutive heterochromatin describes large segments of the genome, primarily arrays of tandemly repeated (satellite) sequences, which are packaged in a permanently inactive form that is thought to be more compact than euchromatin (Gilbert and Allan, 2001). In mammalian cells constitutive heterochromatin is principally found at centromeric and peri-centromeric regions.
The cytological division of chromatin into euchromatin and heterochromatin does not readily lend itself to a direct molecular definition. However, there are different chromatin-associated biochemical marks important in distinguishing the heterochromatic state from euchromatin. These include: DNA methylation, histone methylation and the absence of histone acetylation (reviewed in Richards and Elgin, 2002). In addition, heterochromatin protein 1 (HP1) proteins, a class of multifunctional chromatin-associated adapter proteins, are present at blocks of constitutive heterochromatin in diverse eukaryotes, where they are thought to be important for regulating heterochromatin-mediated silencing and chromosome structure (Ekwall et al., 1995; Kellum et al., 1995; Yamaguchi et al., 1998). There are three HP1 proteins (α, β and γ) in mammals. HP1α and β are concentrated at pericentric heterochromatin, although HP1β can also be seen at more diffuse nucleoplasmic sites, whereas HP1γ is predominantly localized in euchromatin (Minc et al., 2000; Nielsen et al., 2001). This is a similar range of distribution patterns to those reported for the three HP1 proteins in Drosophila (Smothers and Henikoff, 2001).
Analysis of HP1 structure reveals three functional domains; an N-terminal chromodomain (CD), a central hinge domain (HD) and a C-terminal chromoshadow domain (CSD). Dimerization and interaction of HP1s with other chromosomal proteins is thought to occur through the CSD (Brasher et al., 2000; Smothers and Henikoff, 2000). The CD binds to histone H3 methylated at K9 (metH3-K9) (Bannister et al., 2001; Lachner et al., 2001; Jacobs and Khorasanizadeh, 2002; Nielsen et al., 2002). HP1α may also bind to DNA and linker histones through the HD (Nielsen et al., 2001; Meehan et al., 2003). The HD may also be involved in targeting HP1 to heterochromatin through an RNA binding activity (Muchardt et al., 2002).
Facultative heterochromatin is defined as euchromatic regions that become packaged into a compact heterochromatic-like form in a developmentally regulated manner. Facultative heterochromatin is not characterized by repetitive sequences, so at the DNA sequence level it is entirely different from constitutive heterochromatin. However, facultative heterochromatin has many of the same molecular signatures as constitutive heterochromatin at the nucleosome level. Histone hypoacetylation and H3-K9 methylation occur during formation of the inactive X chromosome (Xi) in somatic cells of female mammals (Jeppesen and Turner, 1993; Peters et al., 2002) and the X chromosome that is silenced within the XY body in spermatogenesis is also enriched in metH3-K9 (Cowell et al., 2002), but is not depleted in H4 acetylation (Armstrong et al., 1997). In insects, both the bithorax complex in Drosophila and the paternal heterochromatic chromosomes in male mealy bugs are also enriched in metH3-K9 (Cowell et al., 2002).
HP1β and γ, but not HP1α, also concentrate on the mammalian XY body during pachytene (Cowell et al., 2002; Metzler-Guillemain et al., 2003), and on the condensed chromosome set in mealy bugs (Cowell et al., 2002). The presence of HP1s at these types of facultative heterochromatin suggests a mechanism of formation similar to that of constitutive heterochromatin. However, HP1s do not accumulate on the Xi (Peters et al., 2002), suggesting there are HP1-independent pathways to the formation of facultative heterochromatin. Instead, blocks of facultative heterochromatin can be enriched in variant histones and non-histone proteins. For example, the mammalian inactive X of somatic and germinal cells contains variant histones macroH2A.1 and 2 (Costanzi and Pehrson, 1998; Hoyer-Fender et al., 2000; Chadwick and Willard, 2001), and Brca1 (Ganesan et al., 2002).
At the bithorax complex in Drosophila, polycomb group (PcG) proteins establish a repressed chromatin state in order to regulate the pattern of Hox gene expression during development (reviewed by Orlando, 2003). Some PcG proteins contain a chromodomain, similar to that found in HP1s, that has been shown to bind to tri-metH3-K27 (Cao et al., 2002; Czermin et al., 2002). Recent evidence has implicated PcG proteins and metH3-K27 in the initiation of mammalian X chromosome inactivation (Wang et al., 2001; Plath et al., 2003; Silva et al., 2003). Therefore, while the role of HP1s in formation of constitutive heterochromatin seems almost universal, there appear to be many routes to the formation of facultative heterochromatin.
The facultative heterochromatin formed in the nuclei of terminally differentiated erythrocytes of chicken has been used as a model system to study the developmentally regulated condensation and repression of chromatin (Weintraub, 1984). In chickens this correlates with the expression of the variant linker histone H5 that is able to condense the chromatin fibre (Bergman et al., 1988), and with the expression of a serpin-like protein called MENT (Grigoryev et al., 1999). Xenopus nucleated erythrocytes have the replacement linker histone H10, the accumulation of which also coincides with cessation of proliferation and the compaction of chromatin (Koutzamani et al., 2002), and fish erythrocytes contain similar, although less well characterized, replacement linker histones. Likewise mouse embryonic erythrocytes are nucleated and have condensed chromatin.
To determine whether HP1s and histone modifications play a role in these forms of facultative heterochromatin we have examined the expression of HP1 proteins and the presence of histone H3 K4, 9 and 27 methylation in mouse and chicken erythrocyte nuclei. Our data indicate that although centromeric heterochromatin is universally associated with tri-methylated histone H3 K9 and HP1 proteins, facultative heterochromatin is formed and maintained by different mechanisms. We find elevated levels of metH3-K9 and abundant HP1 in the nuclei of mouse erythrocytes, and an absence of metH3-K27. In contrast there is a total absence of HP1s from adult chicken, frog and fish nucleated erythrocytes, and decreased levels of metH3-K9. Hence there must be an HP1-independent pathway for the formation of heterochromatin during erythrocyte differentiation in these vertebrates. HP1 levels decrease during the differentiation of chicken embryonic erythrocytes as the levels of H5 increase, suggesting that H5 might replace the role of HP1s. To our knowledge, this is the first report of cell types that lack HP1s and that have gross changes in the levels of histone modifications.
Results
Chromatin association and localization of HP1 isoforms in mammalian and chicken cells
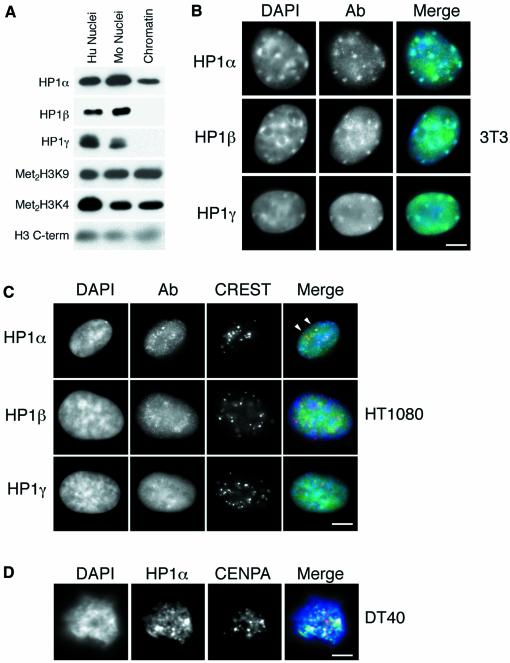
Differential localizations of HP1 isoforms in mouse cells have been reported (Minc et al., 2000; Nielsen et al., 2001). This suggests that despite their sequence similarities, HP1s α, β and γ interact with chromatin in different ways. To examine this we isolated the proteins bound to higher order chromatin fibres from mouse cells under physiological conditions (Gilbert and Allan, 2001). Immunoblotting using monoclonal antibodies specific to each HP1 isoform (Nielsen et al., 1999) demonstrates that most HP1α is stably associated with the chromatin fibres, while HP1β and γ are readily lost during purification (Figure 1A). This suggests that HP1β and γ have a lower affinity for the chromatin fibre than does HP1α, which is surprising given the similar exchange kinetics of all three HP1 isoforms at either euchromatin or heterochromatin measured by fluorescence recovery after photobleaching (Cheutin et al., 2003). HP1α has previously been shown to remain tightly bound to chromatin during salt extraction (Remboutsika et al., 1999).
Fig. 1. Distribution of HP1 isoforms in human, mouse and chicken cells. (A) Immunoblotting of proteins isolated from whole human (hu) or mouse (mo) nuclei, or from mouse chromatin using antisera that detect HP1s α, β and γ, di-methyl H3-K9 (met2H3-K9), and di-methyl H3-K4 (met2H3-K4). An antibody that recognizes H3 is used as a loading control. (B) Immunofluorescence of mouse 3T3 nuclei fixed with pFa using antibodies that detect HP1s α, β and γ (green). DNA was counterstained with DAPI (blue) to highlight the foci of heterochromatin. (C) Co-immunostaining of human HT1080 cells using antibodies that detect HP1s α, β and γ (green) and with CREST antiserum, which detects centromeric antigens (red). DNA was counterstained with DAPI (blue). Arrowheads indicate where foci of HP1α and CREST staining are coincident. (D) Co-immunostaining of chicken DT40 cells with antibody recognizing HP1α (green) and CENP-A (red). Scale bars, 5 µm.
We compared the nuclear distribution of the three HP1 isoforms in mouse and human tissue culture cells by immunofluorescence (Figure 1B and C). As reported previously (Minc et al., 2000; Nielsen et al., 2001), HP1α is concentrated at constitutive heterochromatin, indicated by DAPI bright regions, in mouse cells fixed with paraformaldehyde (pFa) (Figure 1B). Consistent with the strong biochemical association of HP1α with chromatin (Figure 1A), this staining pattern persists in cells fixed with methanol:acetic acid (data not shown). However, the diffuse nucleoplasmic or euchromatin-associated pool of HP1α is lost after extraction of mouse nuclei with methanol:acetic acid. This suggests that the mode of binding of HP1α in heterochromatin differs from that at euchromatic locations, and is consistent with the differing dynamics of HP1s at heterochromatic and euchromatic sites (Cheutin et al., 2003; Festenstein et al., 2003).
HP1β and γ are also detected at sites of heterochromatin, but there is also a large pool of these proteins more diffusely distributed throughout the nucleoplasm and HP1γ is predominantly found at euchromatic sites (Figure 1B).
The distribution of HP1 isoforms in human cells has not been well described. Compared with mouse cells, human cells have smaller blocks of constitutive heterochromatin that are not easily distinguished cytologically. Much constitutive heterochromatin in human cells is pericentromeric; therefore, we compared the distribution of HP1 isoforms with centromeric antigens detected with CREST antisera. HP1α shows a concentration at centromeric sites in human cells, coincident with foci of CREST staining (arrowheads in Figure 1C). No concentration of HP1β or γ could be detected at centromeric domains (detected by CREST) and there are clear non-centromeric foci of these HP1 isoforms in human cells (Figure 1C).
On chicken chromosomes, blocks of constitutive heterochromatin are found at centromeric and telomeric regions of the macrochromosomes, at centromeric regions of the microchromosomes, and also on the Z and W sex chromosomes (Schmid et al., 1989). In chicken DT40 cells the most prominent foci of HP1α staining are close to centromeres (detected by antibody recognizing the centromere protein CENP-A) (Figure 1D).
metH3-K9 and HP1 distribution in mammalian and chicken nuclei
Binding of HP1 proteins to metH3-K9 is important for their recruitment to chromatin (Bannister et al., 2001; Lachner et al., 2001; Jacobs and Khorasanizadeh, 2002; Nielsen et al., 2002). We assessed how the distribution of HP1s compares with that of metH3-K9 using two different antibodies. The first was raised against a linear peptide, recognizes di-methylH3-K9 (met2H3-K9) and detects facultative heterochromatin of the Xi (Boggs et al., 2002). met2H3-K9 is distributed diffusely in mouse and human nuclei with no concentration at the murine heterochromatic foci detected by DAPI staining (Figure 2A), nor the centromeric domains detected by CREST sera in male human cells (Figure 2B). The second antibody was raised against a branched di-methylH3-K9 peptide and is thought to predominantly recognize tri-methylH3-K9 (met3H3-K9) (Peters et al., 2001). Antigens detected by this antibody are concentrated at mouse pericentric heterochromatin (Figure 2A) and foci in human nuclei, some of which are coincident with CREST staining (arrows in Figure 2B). Other foci do not correspond to centromeric regions detected by CREST (arrowheads in Figure 2B) and may represent other blocks of heterochromatin in the human genome. met3H3-K27 has recently been associated with PcG protein recruitment in the X-inactivation process (Silva et al., 2003). Western blot analysis indicated that male mouse 3T3 cells have this modification (in the absence of Xi) (Figure 6A), prompting us to study its distribution pattern (Figure 2A). As for met2H3-K9, it is nuclear diffuse and does not appear to be associated with any specific cellular structures.
Fig. 2. Distribution of metH3-K9 in human, mouse and chicken cells. (A) Immunostaining of mouse 3T3 with antisera (green) raised against a linear peptide dimethylated at K9 of H3 (met2H3-K9 linear; Boggs et al., 2002), or against a branched di-methylated peptide (met3H3-K9 branched, but which detects tri-methylated H3-K9; Peters et al., 2001) or against a linear peptide tri-methylated at K27 (Silva et al., 2003). DNA was counterstained with DAPI (blue). (B) Co-immunostaining of human HT1080 cells with metH3-K9 linear and branched antibodies (green) and CREST serum (red). DNA was counterstained with DAPI (blue). Arrows indicate sites of co-staining. Arrow heads indicate foci of met3H3-K9 that do not correspond to centromeres detected by CREST. (C) Co-localization of HP1α with tri-methyl H3-K9 in mouse 3T3, human HT1080 and chicken DT40 cells. Scale bars, 5 µm.
Fig. 6. HP1α and methylated H3 in mouse erythrocytes. (A) Western blot of proteins from mouse 3T3, liver and embryonic erythrocyte nuclei using antibodies detecting HP1α, met3H3-K9 and met3H3-K27. H3 antibody is used as a loading control. The graph to the right shows the quantification of these blots. (B) Immunostaining of HP1α and met3H3-K9 (green) in mouse embryonic nucleated erythrocytes. DNA was counterstained with DAPI (blue). (C) Female C127 cells: immunostaining for met3H3-K27 (red), RNA FISH for Xist (green) and double immunostaining for met3H3-K27 (red) and RNA FISH for Xist (green). Mouse embryonic nucleated erythrocytes: RNA FISH for Xist (green) followed by DNA FISH for X chromosome (red), and immunostaining for met3H3-K27 (red) followed by DNA FISH for X chromosome (green). Scale bar, 5 µm.
To assess the correspondence between met3H3-K9 and HP1α distribution, we carried out double staining on mouse and human nuclei. While there is a strong correspondence between met3H3-K9 and HP1 staining at the foci of pericentric heterochromatin detected by DAPI staining of mouse cells, there is little coincidence between the two antibody staining patterns outside of these regions in mouse and human cells (Figure 2C). This suggests that HP1α and met3H3-K9 are not interdependent outside of centromeric heterochromatin. Likewise, HP1α and me2H3-K9 are not coincident in human, mouse and chicken nuclei (Supplementary figure 1, available at The EMBO Journal Online).
In chicken DT40 cells, foci of met3H3-K9 and HP1α staining are coincident, resembling the pattern seen in mouse cells (Figure 2C).
HP1 and H3 methylation in the compact facultative heterochromatin of vertebrate erythrocytes
Whilst a relationship between HP1 proteins and the formation of constitutive heterochromatin is well established, it is not clear what role HP1s may have in the formation of compact facultative heterochromatin. Chromatin compaction and a widespread silencing of gene expression occur during the terminal differentation of erythrocytes (Rowley and Radcliffe, 1988). The genome-wide compaction of chromatin within nucleated erythrocytes is apparent in the intense DAPI-staining and reduced nuclear volume of mouse and chicken erythrocytes in comparison with mouse liver and chicken pre-B cells (Figure 3). The chromatin condensation within Xenopus and zebrafish erythrocyte nuclei is less apparent.
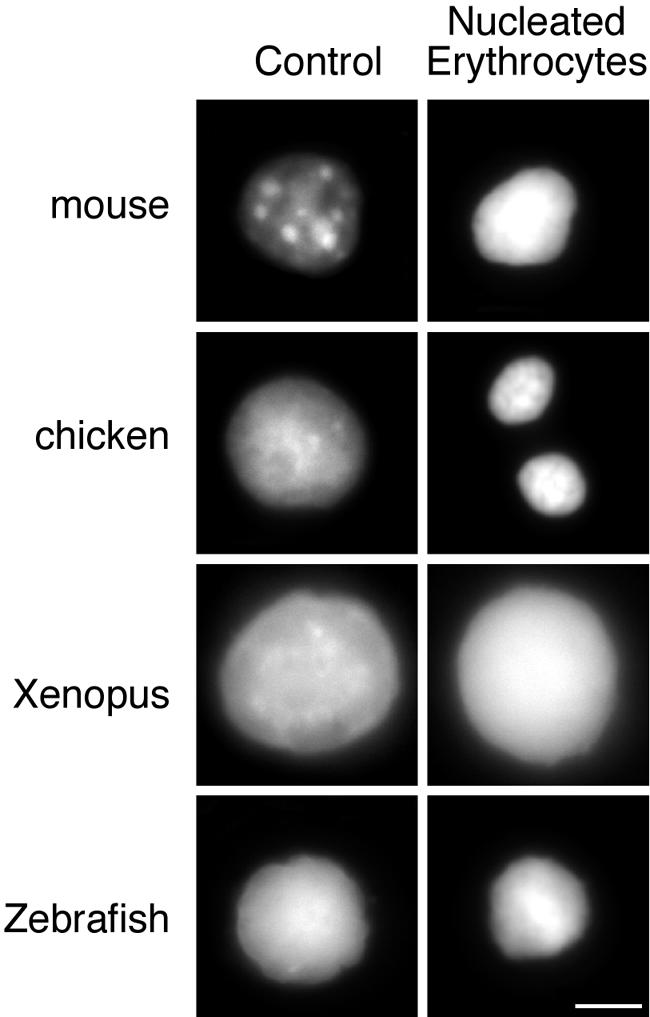
Fig. 3. Chromatin condensation in vertebrate nucleated erythrocytes. DAPI staining of mouse, chicken, Xenopus and zebrafish nucleated erythrocytes (right hand panel) contrasted with the staining pattern of nuclei from control tissues (mouse, Xenopus and zebrafish liver) and chicken DT40 cells. Scale bar, 5 µm.
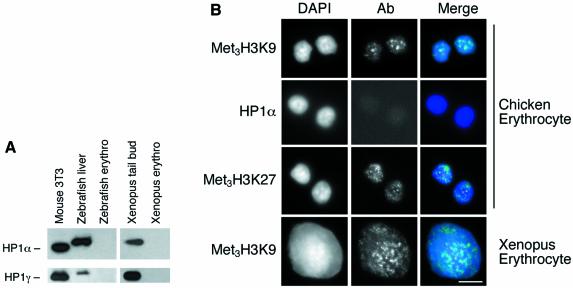
To establish whether there is a role for HP1s and histone modifications in the chromatin condensation in chicken erythrocytes, we analysed chromatin from DT40 (pre-B) chicken cells and from nucleated adult chicken erythrocytes by western blot. Similar levels of H3 dimethylated at K9 (Figure 4A) were found in the chromatin of mouse 3T3 cells, chicken DT40 cells and chicken erythrocytes. In contrast, we found decreased levels of met3H3-K9 (antisera raised against a linear tri-methylated H3-K9 peptide) in chicken erythrocyte chromatin in comparison with DT40 chromatin (Figure 4A and B). This suggested that there might also be reduced levels of HP1 proteins in chicken erythrocytes. In fact, we were unable to detect any HP1 isoforms in adult chicken erythrocytes (Figure 4A and B) by western blotting. Recently, it has been shown that chromodomain containing PcG proteins can bind to met3H3-K27 (Cao et al., 2002; Czermin et al., 2002), leading to the possibility that heterochromatin formation in chicken erythrocytes is mediated by this H3 modification and binding of a chicken PcG protein. However, western blotting with an antibody against met3H3-K27 showed that levels of this histone modification are very low in chicken erythrocytes (Figure 4A and B).
Fig. 4. Loss of HP1s from chicken erythrocytes. (A) Western blot of mouse 3T3, chicken DT40 and adult chicken erythrocyte nuclear proteins with antibodies detecting HP1 isoforms, di- (met2H3-K9) and tri- (met3H3-K9) methylated H3 K9, and H3 methylated at K4 and K27. Antibody detecting H3 is used as a control. (B) Quantification of HP1α, met2H3-K4, met3H3-K9 and met3H3-K27 levels in mouse, DT40 and chicken erythrocyte nuclei. Signals were normalized with respect to total histone H3. (C) Coomassie-stained gel of mouse 3T3, chicken DT40, chicken day 10 embryonic erythrocytes and adult chicken erythrocytes. Histone H5 is indicated by an arrow. Western blots of equivalent samples probed with antibodies against H5 and HP1α are shown below.
To examine how these altered levels of HP1s and histone modifications are regulated during development we analysed the chromatin from 10-day embryonic chicken erythrocytes by western blot and Coomassie-stained gel (Figure 4C). During erythrocyte maturation the level of H5 increases (Ruiz-Carrillo et al., 1974) while the level of HP1α decreases. In some adult chicken erythrocyte preparations trace amounts of HP1α are present due to white-cell contamination.
We also detected no HP1α or γ in pycnotic chromatin from frog and fish nucleated erythrocytes (Figure 5A) (we were unable to detect Xenopus or zebrafish HP1β isoforms using available antibodies). HP1α was also undetectable in adult chicken erythrocytes by immunofluorescence (Figure 5B), but the distribution of met3H3-K9 in chicken erythrocytes and Xenopus erythrocytes is similar to that in other somatic cells (e.g. DT40 chicken cells in Figure 1C). This indicates that the distribution of this histone modification is unaffected by the absence of HP1α and suggests that chromatin condensation in non-mammalian vertebrate erythrocytes occurs without HP1s. Moreover, HP1s must also have been lost from the constitutive heterochromatin in these cells (Figures 1 and 5), and so cannot be essential for their survival (Filesi et al., 2002). met3H3-K27 in chicken erythrocytes was present at low levels by western blot (Figure 4A) and has a diffuse distribution pattern by immunofluorescence (Figure 5B). These nuclei were obtained from female chicken cells that carry ZW sex chromosomes and so would not exhibit any dosage compensation, unlike male cells, which carry ZZ sex chromosomes. Therefore, male cells might inactivate one chromosome (McQueen et al., 2001) possibly by tri-methylation at H3-K27 (Figure 6C).
Fig. 5. Absence of HP1α from chicken, fish and Xenopus erythrocytes. (A) Western blot of proteins from fish liver and nucleated erythrocytes, and from Xenopus tail bud tadpole nuclei and erythrocytes, using antibodies raised against Xenopus HP1α and HP1γ. HP1β isoforms could not be detected in Xenopus or zebrafish cells using available antibodies. (B) Immunostaining of chicken and Xenopus nuclei with met3H3-K9 or met3H3-K27 antisera, or HP1α antibody (green) and DNA (blue). Scale bar, 5 µm.
HP1 and H3 methylation in mouse nucleated embryonic erythrocytes
To assess whether the loss of HP1s from erythrocytes is restricted to avian and fish species or whether it also occurs during mammalian erythrocyte differentiation, we examined nucleated erythrocytes from early mouse embryos. During mouse embryogenesis non-enucleated erythrocytes express embryonic haemoglobins, before the switch to expression of foetal haemoglobins at ∼12 days post-coitum (d.p.c.). The chromatin in erythrocytes from 12.5 d.p.c. mouse embryos appears to be highly compacted compared with that in mouse foetal liver cells (Figure 3). However, western blotting shows that in contrast to non-mammalian vertebrate erythrocytes, mouse embryonic erythrocytes have elevated levels of HP1α compared with liver cells (Figure 6A). This is accompanied by increased levels of met3H3-K9 and a dramatic loss of tri-methylation at H3-K27 (Figure 6A). met3H3-K27 has been shown to be present during X inactivation (Silva et al., 2003). In male 3T3 cells it has a nuclear diffuse distribution pattern (Figure 2A), whilst in female mouse C127 epithelial cells it is nuclear diffuse with a very clear accumulation on the inactive X chromosome coincident with Xist staining (Figure 6C). However, by immunofluorescence we were unable to detect met3H3-K27 staining in mouse erythrocytes from embryos (confirmed to be female by X chromosome painting), and we were also unable to detect Xist by RNA fluorescence in situ hybridization (FISH) (Figure 6C). This suggests that facultative heterochromatin formation in murine embryonic erythrocytes may be mediated by HP1, and that PcG family members are unlikely to be involved.
The presence of elevated levels of HP1α and met3H3-K9 in mouse erythrocytes suggests that there might be a spreading of HP1 from the normal sites of constitutive heterochromatin (Figure 2C) to a more global distribution across the genome. However, immunofluorescence indicates that HP1α is still predominantly localized to blocks of heterochromatin in mouse erythrocytes whilst the distribution of met3H3-K9 is more diffuse (Figure 6B). Thus, a redistribution of HP1α to sites throughout the genome is unlikely to be solely responsible for the formation of facultative heterochromatin in these cells.
Discussion
Initial progress in understanding heterochromatin structure came from the analysis of constitutive heterochromatin, for example, in screens for suppressors of position effect variegation in flies. HP1 was identified in this way, and HP1 proteins have now been shown to be important in the formation and propagation of constitutive heterochromatin structure in many organisms. HP1 proteins have been implicated in some forms of facultative heterochromatin (XY body and mealy bug chromosome condensation), but not in others (somatic X inactivation) (Cowell et al., 2002; Peters et al., 2002; Metzler-Guillemain et al., 2003). In addition, differently methylated forms of H3 have been seen within facultative heterochromatin. SET domain-containing PcG group proteins in Drosophila have been shown to be H3 K27 histone methyltransferases (HMTases). The chromodomain of other PcG proteins bind this methylated form of H3 (Cao et al., 2002; Czermin et al., 2002). However, the bithorax complex that is repressed by PcG complexes and the condensed chromosome set in mealy bugs, are also enriched in meH3-K9 (Cowell et al., 2002). H3 becomes transiently tri-methylated at K27 during the initiation of X inactivation in embryonic stem cells (Plath et al., 2003; Silva et al., 2003). This mark may be supplemented by metH3-K9 at later stages of inactivation (Boggs et al., 2002), although we have clearly shown there is a concentration of met3H3-K27 on the inactive X in terminally differentiated somatic mouse cells (Figure 6C). Therefore, there appear to be many mechanisms through which facultative heterochromatin is formed, and there is a need to study this in a variety of systems.
To try to clarify the histone modifications and role of HP1s in the formation of facultative heterochromatin, we used nucleated erythrocytes as a model system. Chicken, frog and fish erythrocytes are normally nucleated, in contrast to adult mammalian erythrocytes, which are enucleated late in differentiation. However, during embryogenesis, mammalian erythrocytes remain nucleated, but during their differentiation there is a dramatic increase in chromatin condensation and a decrease in transcriptional activity (Rowley and Radcliffe, 1988; Grigoryev et al., 1999) (Figure 3).
Abundant HP1 and metH3-K9, but an absence of metH3-K27, in mouse erythrocytes
Analysis of the nucleosome modifications in embryonic erythrocyte chromatin by western blot indicates that there are elevated levels of met3H3-K9 in these cells (in comparison with liver and 3T3 cells) (Figure 6A) and, in contrast, we are unable to detect any tri-methylation of H3-K27 in erythrocytes (Figure 6A and C). Hence, we think it unlikely that PcG proteins are responsible for the chromatin compaction and gene silencing in mouse embryonic erythrocytes. Furthermore, we were unable to detect Xist RNA on the inactive X chromosome. This suggests that the mechanism of global facultative heterochatin formation in these cells is able to obviate the need for both Xist and met3H3-K27. However, coincident with the increased levels of met3H3-K9 in mouse erythrocytes we also detect enhanced levels of HP1α (Figure 6A), suggesting that HP1 could have a role in formation of facultative heterochromatin in these cells. It is suggested that HP1 spreads across heterochromatin through binding to histone H3 methylated at K9 (Bannister et al., 2001; Lachner et al., 2001; Partridge et al., 2002). Furthermore, it has been suggested that HP1 (or its orthologue) is required for the spread of met3H3-K9 itself (Hall et al., 2002). Whilst we see some diffuse re-distribution of met3H3-K9 in mouse erythrocyte nuclei, HP1α is still predominantly concentrated at the foci of constitutive pericentric heterochromatin (Figure 6B). This is consistent with the observation that, in fission yeast, met3H3-K9 can spread from ectopic sites (via the action of the HMTase clr4) in the absence of the HP1 homologue swi6 (Partridge et al., 2002).
Absence of HP1s and reduced H3 methylation of K9 and K27 in chicken erythrocytes
To establish whether similar changes in histone methylation and HP1 are present in the erythrocytes of non-mammalian vertebrates, we analysed nuclei of chicken erythrocytes. DAPI staining reveals the chromatin in these small nuclei to be more compact than that in control (DT40) chicken cells (Figure 3), consistent with electron micrographs of chromatin in avian erythrocytes (Ruiz-Carrillo et al., 1974; Rowley and Radcliffe, 1988). By western blot, levels of H3 tri-methylated at both K9 and K27 are reduced in chicken erythrocytes (Figure 4) and, most strikingly, levels of all three HP1 isoforms are undetectable in adult chicken erythrocytes, as confirmed by immunofluorescence (Figure 5B). We also could not detect HP1α or γ in erythrocytes from Xenopus or zebrafish (Figure 5). Hence, we suggest that there can be no role for HP1 in the formation of compact facultative heterochromatin in erythrocytes of these vertebrates. Levels of di-methylated H3-K9 have been analysed by chromatin immunoprecipitation at the β-globin locus of chicken erythrocytes (Litt et al., 2001). High levels of met2H3-K9 were found over compact chromatin outside of the globin locus and over developmentally inactive globin genes. It was proposed that an insulator acts to prevent the spread of metH3-K9 and accompanying silencing proteins, such as HP1, into the globin locus (Litt et al., 2001). However, the absence of HP1s from chicken erythrocytes suggests that this is not necessary, although we cannot rule out a role for other chromo-box proteins that could bind metH3-K9. For example, fission yeast Chp1 also binds to metH3-K9 (Partridge et al., 2002).
Recently, it was suggested that HP1 function is essential for cell survival in mammalian cells, since injection of HP1 antibodies into cells resulted in cell death (Filesi et al., 2002). Our analyses show that HP1s are not essential for cell survival in some vertebrate erythrocytes, and moreover indicates that the expression of HP1 proteins is developmentally regulated. It will be important to analyse the presence of HP1 isoforms in other differentiated cell types.
Specialized proteins to mediate chromatin condensation in erythrocytes
Our data suggest that HP1 is not involved in the chromatin condensation of chicken, frog and fish erythrocytes. Instead, other specialized chromatin-associated proteins may be recruited to perform this role in erythrocyte differentiation. Currently, there are two such candidate proteins. The serpin-like protein MENT is able to induce large-scale chromatin condensation in vitro and when ectopically expressed (Grigoryev et al., 1999). Its accumulation during avian erythrocyte differentiation strongly correlates with the extent of chromatin condensation (Grigoryev and Woodcock, 1993). The mechanisms of recruitment and action of MENT are unknown, but it is an abundant basic protein that associates with polynucleosomes (Grigoryev et al., 1999).
In avian erythrocytes some H1 is replaced with the variant linker histone H5 (Ruiz-Carrillo et al., 1974). When injected into mammalian cells, H5 can block both transcription and DNA replication (Bergman et al., 1988), and there are two DNA binding sites in the H5 globular domain necessary for binding to nucleosomes (Duggan and Thomas, 2000). The function of linker histones, and their localization with respect to nucleosomes, is poorly understood (Widom, 1998), and the consequences of the presence of the H5 variant on chromatin structure are yet to be determined. However, since we have observed that the decrease in HP1s in chicken erythrocytes during development parallels the increasing levels of H5 (Figure 4C), we speculate that H5 could replace the role of HP1 in chromatin compaction.
Since globin gene expression in chicken and mouse erythrocytes is used as a model system for the study of chromatin structure and gene expression, it is important to understand the molecular basis for the chromatin compaction and gene silencing that occur in these cell types. Examination of histone modifications and HP1 isoforms in these cell types is a first step along this road, and our data show that the conclusions drawn in one species, e.g. chicken, cannot readily be extrapolated to mammals. It will also be important to examine the presence of HP1 proteins and histone modifications in other differentiated cell types.
Materials and methods
Cell lines and tissues
Male mouse NIH 3T3 fibroblasts, female mouse C127 epithelial cells and human HT1080 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% foetal calf serum (FCS). DT40 cells were cultured in RPMI supplemented with 10% FCS and 1% chicken serum. Mouse embryo blood was obtained from 12.5 d.p.c. embryos. Adult chicken blood was isolated from freshly slaughtered female leghorn chickens. Embryonic chicken blood was prepared from 10-day embryos. Zebrafish blood and liver and Xenopus blood were collected from freshly culled animals. Mouse liver was obtained from culled adult mice and Xenopus nuclei were obtained from tail bud tadpoles.
Preparation of nuclei and chromatin
Erythrocyte and tissue culture cell nuclei were prepared by a modification of the method of Gilbert and Allan (2001). The cells were washed in phosphate-buffered saline (PBS) and resuspended in nuclear buffer A (85 mM KCl, 10 mM Tris–HCl, 0.2 mM spermidine, 0.2 mM EDTA, 160 mM sucrose, 250 µM phenylmethylsulfonyl fluoride) on ice. An equal volume of nuclear buffer B (nuclear buffer A supplemented with an appropriate amount of NP-40) was added. The nuclei were pelleted in a benchtop centrifuge (2000 g, 4 min, 4°C) and washed into nuclear buffer A. The concentration of the nuclei was determined by measuring the A260. Mouse and zebrafish liver nuclei were prepared using the above procedure except the cells were first homogenized in nuclear buffer A. Chromatin was prepared as described previously (Gilbert and Allan, 2001). To isolate chromatin-associated proteins the chromatin was fractionated on a sucrose gradient. Peak fractions were dialysed against TE and the proteins were precipitated using acetone and resuspended in SDS loading buffer.
Antibodies
The antibodies used were: mAb HP1α, β and γ [Chemicon; western blotting (wb) 1:1000, immunofluorescence (immuno) 1:500; Nielsen et al., 1999]; mAb histone H5 (from Michael Bustin; wb 1:1000); rabbit polyclonals against xHP1α (affinity purified; wb 1:100) and γ (from Richard Meehan; wb 1:1000); met2H3-K9 (Upstate; wb 1:1000, immuno 1:500; Boggs et al., 2002), met2H3-K4 (Upstate; wb 1:1000, immuno 1:500), met3H3-K9 (wb 1:2000, immuno 1:500), met3H3-K27 (wb 1:1000, immuno 1:100; Silva et al., 2003), 4× met2H3-K9 (immuno 1:500; Peters et al., 2001), C-terminal H3 (wb 1:10 000; Verreault et al., 1996); and CENP-A (from Vinciane Regnier).
Western blotting
Nuclei were suspended in SDS loading buffer, fractionated by SDS–PAGE and transferred to a nylon membrane by semi-dry blotting. The membranes were probed with antibodies using standard techniques and detected by enhanced chemiluminescence.
Immunofluorescence
Mouse NIH 3T3, C127 and human HT1080 were grown on slides and fixed using 4% pFa in PBS. The cells were permeabilized using Triton X-100 in PBS and were sequentially incubated with the primary and secondary antibodies (Jackson Laboratories). All other cells and nuclei were cytospun onto slides and were pFa-fixed and processed as above.
FISH
RNA FISH. Cells were grown on slides and fixed using 4% pFA in PBS. The cells were permeabilized using Triton X-100 in PBS and stored in 70% ethanol at 4°C. The slides were dehydrated using 70, 90, 100% ethanol and air dried. One hundred and fifty nanograms of digoxigenin (DIG)-labelled Xist probe (pGPT16; Duthie et al., 1999), 20 µg of yeast tRNA and 5 µg of sonicated salmon sperm DNA were precipitated together and resuspended in 15 µl of hybridization mix (50% deionized formamide, 10% dextran sulphate, 1% Tween-20 in 2× SSC) and incubated under a sealed coverslip overnight at 37°C. Slides were washed for 3 min in 50% formamide/2× SSC at room temperature, for 3 min in 50% formamide/2× SSC at 37°C, and for 3 min in 2× SSC at room temperature and 4× SSC 0.1% Tween-20. The Xist signal was detected using one layer of FITC-conjugated anti-DIG (raised in sheep) and one layer of FITC-conjugated anti-sheep. Slides were mounted in Vectashield with 1 µg/ml DAPI. For immuno-RNA FISH the cells were subsequently incubated with primary and secondary antibodies.
DNA FISH. Cells were grown on slides and fixed using 4% pFA in PBS. The cells were permeabilized using Triton X-100 in PBS and stored in 70% ethanol. The slides were washed briefly in 2× SSC and incubated with 100 µg/ml RNaseA for 1 h at 37°C. The slides were dehydrated using 70, 90, 100% ethanol and air dried. Slides were denatured in 70% formamide/2× SSC at 70°C for 90 s, transferred to ice-cold 70% ethanol, and then into 90 and 100% ethanol and air dried. Fifteen microlitres of biotin-labelled X-chromosome paint (Cambio) were denatured at 70°C, re-annealed at 37°C for 15 min and hybridized on the slide under a sealed coverslip overnight at 37°C. The slides were washed 4× 3 min in 2× SSC at 45°C, 4× 3 min in 0.1× SSC at 60°C and then 4× SSC 0.1% Tween-20 at room temperature. Biotinylated probes were detected using Texas Red-conjugated avidin, followed by biotinylated anti-avidin and a final layer of Texas Red-conjugated avidin. Slides were mounted in Vectashield with 1 µg/ml DAPI.
For RNA/DNA FISH or immuno-DNA FISH the slides were first processed for either RNA FISH or immuno. Coordinates of cells were taken and the slide was then processed for DNA FISH.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Robin Allshire and Richard Meehan for critical reading of the manuscript. We thank Allan Verreault (Cancer Research UK, Clare Hall, UK) for the gift of H3 antibody, Vinciane Regnier (University of Oxford) for the CENP-A antibody, Richard Meehan (University of Edinburgh) for both the Xenopus HP1 antibody and the Xenopus tail bud tadpole nuclei, and Tim Hebbes (University of Portsmouth) for chicken embryonic erythrocytes. W.A.B. is a Centennial Fellow of the James S.McDonnell Foundation and H.S. is funded by the AICR.
References
- Armstrong S.J., Hulten,M.A., Keohane,A.M. and Turner,B.M. (1997) Different strategies of X inactivation in germinal and somatic cells: histone H4 underacetylation does not mark the inactive X chromosome in the mouse germline. Exp. Cell Res., 230, 399–402. [DOI] [PubMed] [Google Scholar]
- Bannister A.J., Zegerman,P., Partridge,J.F., Miska,E.A., Thomas,J.O., Allshire,R.C. and Kouzarides,T. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature, 410, 120–124. [DOI] [PubMed] [Google Scholar]
- Bergman M.G., Wawra,E. and Winge,M. (1988) Chicken histone H5 inhibits transcription and replication when introduced into proliferating cells by microinjection. J. Cell Sci., 91, 201–209. [DOI] [PubMed] [Google Scholar]
- Boggs B.A., Cheung,P., Heard,E., Spector,D.L., Chinault,A.C. and Allis,C.D. (2002) Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat. Genet., 30, 73–76. [DOI] [PubMed] [Google Scholar]
- Brasher S.V. et al. (2000) The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J., 19, 1587–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R., Wang,L., Wang,H., Xia,L., Erdjument-Bromage,H., Tempst,P., Jones,R.S. and Zhang,Y. (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science, 298, 1039–1043. [DOI] [PubMed] [Google Scholar]
- Chadwick B.P. and Willard,H.F. (2001) Histone H2A variants and the inactive X chromosome: identification of a second macroH2A variant. Hum. Mol. Genet., 10, 1101–1113. [DOI] [PubMed] [Google Scholar]
- Cheutin T., McNairn,A.J., Jenuwein,T., Gilbert,D.M., Singh,P.B. and Misteli,T. (2003) Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science, 299, 721–725. [DOI] [PubMed] [Google Scholar]
- Costanzi C. and Pehrson,J.R. (1998) Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature, 393, 599–601. [DOI] [PubMed] [Google Scholar]
- Cowell I.G. et al. (2002) Heterochromatin, HP1 and methylation at lysine 9 of histone H3 in animals. Chromosoma, 111, 22–36. [DOI] [PubMed] [Google Scholar]
- Czermin B., Melfi,R., McCabe,D., Seitz,V., Imhof,A. and Pirrotta,V. (2002) Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell, 111, 185–196. [DOI] [PubMed] [Google Scholar]
- Duggan M.M. and Thomas,J.O. (2000) Two DNA-binding sites on the globular domain of histone H5 are required for binding to both bulk and 5S reconstituted nucleosomes. J. Mol. Biol., 304, 21–33. [DOI] [PubMed] [Google Scholar]
- Duthie S.M., Nesterova,T.B., Formstone,E.J., Keohane,A.M., Turner,B.M., Zakian,S.M. and Brockdorff,N. (1999) Xist RNA exhibits a banded localization on the inactive X chromosome and is excluded from autosomal material in cis. Hum. Mol. Genet., 8, 195–204. [DOI] [PubMed] [Google Scholar]
- Ekwall K., Javerzat J.P., Lorentz,A., Schmidt,H., Cranston,G. and Allshire,R. (1995) The chromodomain protein Swi6: a key component at fission yeast centromeres. Science, 269, 1429–1431. [DOI] [PubMed] [Google Scholar]
- Festenstein R., Pagakis,S.N., Hiragami,K., Lyon,D., Verreault,A., Sekkali,B. and Kioussis D. (2003) Modulation of heterochromatin protein 1 dynamics in primary mammalian cells. Science, 299, 719–721. [DOI] [PubMed] [Google Scholar]
- Filesi I., Cardinale,A., van der Sar,S., Cowell,I.G., Singh,P.B. and Biocca,S. (2002) Loss of heterochromatin protein 1 (HP1) chromodomain function in mammalian cells by intracellular antibodies causes cell death. J. Cell Sci., 115, 1803–1813. [DOI] [PubMed] [Google Scholar]
- Ganesan S. et al. (2002) BRCA1 supports XIST RNA concentration on the inactive X chromosome. Cell, 111, 393–405. [DOI] [PubMed] [Google Scholar]
- Gilbert N. and Allan,J. (2001) Distinctive higher-order chromatin structure at mammalian centromeres. Proc. Natl Acad. Sci. USA, 98, 11949–11954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryev S.A. and Woodcock,C.L. (1993) Stage-specific expression and localization of MENT, a nuclear protein associated with chromatin condensation in terminally differentiating avian erythroid cells. Exp. Cell Res., 206, 335–343. [DOI] [PubMed] [Google Scholar]
- Grigoryev S.A., Bednar,J. and Woodcock,C.L. (1999) MENT, a heterochromatin protein that mediates higher order chromatin folding, is a new serpin family member. J. Biol. Chem., 274, 5626–5636. [DOI] [PubMed] [Google Scholar]
- Hall I.M., Shankaranarayana,G.D., Noma,K., Ayoub,N., Cohen,A. and Grewal,S.I. (2002) Establishment and maintenance of a heterochromatin domain. Science, 297, 2232–2237. [DOI] [PubMed] [Google Scholar]
- Hoyer-Fender S., Costanzi,C. and Pehrson,J.R. (2000) Histone macroH2A1.2 is concentrated in the XY-body by the early pachytene stage of spermatogenesis. Exp. Cell Res., 258, 254–260. [DOI] [PubMed] [Google Scholar]
- Jacobs S.A. and Khorasanizadeh,S. (2002) Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science, 295, 2080–2083. [DOI] [PubMed] [Google Scholar]
- Jeppesen P. and Turner,B.M. (1993) The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell, 74, 281–289. [DOI] [PubMed] [Google Scholar]
- Kellum R., Raff,J.W. and Alberts,B.M. (1995) Heterochromatin protein 1 distribution during development and during the cell cycle in Drosophila embryos. J. Cell Sci., 108, 1407–1418. [DOI] [PubMed] [Google Scholar]
- Koutzamani E., Loborg,H., Sarg,B., Lindner,H.H. and Rundquist,I. (2002) Linker histone subtype composition and affinity for chromatin in situ in nucleated mature erythrocytes. J. Biol. Chem., 277, 44688–44694. [DOI] [PubMed] [Google Scholar]
- Lachner M., O’Carroll,D., Rea,S., Mechtler,K. and Jenuwein,T. (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature, 410, 116–120. [DOI] [PubMed] [Google Scholar]
- Litt M.D., Simpson,M., Gaszner,M., Allis,C.D. and Felsenfeld,G. (2001). Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science, 293, 2453–2455. [DOI] [PubMed] [Google Scholar]
- McQueen H.A., McBride,D., Miele,G., Bird,A.P. and Clinton.,M. (2001) Dosage compensation in birds. Curr. Biol., 11, 253–257. [DOI] [PubMed] [Google Scholar]
- Meehan R.R., Kao, C-F. and Pennings,S. (2003) HP1 binding to native chromatin in vitro is determined by the hinge region and not by the chromodomain EMBO J., 22, 3164–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzler-Guillemain C., Luciani,J., Depetris,D., Guichaoua,M.R. and Mattei,M.G. (2003) HP1β and HP1γ, but not HP1α, decorate the entire XY body during human male meiosis. Chromosome Res., 11, 73–81. [DOI] [PubMed] [Google Scholar]
- Minc E., Courvalin,J.C. and Buendia,B. (2000) HP1γ associates with euchromatin and heterochromatin in mammalian nuclei and chromosomes. Cytogenet. Cell Genet., 90, 279–284. [DOI] [PubMed] [Google Scholar]
- Muchardt C., Guilleme,M., Seeler,J.S., Trouche,D., Dejean,A. and Yaniv,M. (2002) Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1alpha. EMBO rep., 3, 975–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen A.L., Ortiz,J.A., You,J., Oulad-Abdelghani,M., Khechumian,R., Gansmuller,A., Chambon,P. and Losson,R. (1999) Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J., 18, 6385–6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen A.L., Oulad-Abdelghani,M., Ortiz,J.A., Remboutsika,E., Chambon,P. and Losson,R. (2001) Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol. Cell, 7, 729–739. [DOI] [PubMed] [Google Scholar]
- Nielsen P.R., Nietlispach,D., Mott,H.R., Callaghan,J., Bannister,A., Kouzarides,T., Murzin,A.G., Murzina,N.V. and Laue,E.D. (2002) Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature, 416, 103–107. [DOI] [PubMed] [Google Scholar]
- Orlando V. (2003) Polycomb, epigenomes and control of cell identity. Cell, 112, 599–606. [DOI] [PubMed] [Google Scholar]
- Partridge J.F., Scott,K.S., Bannister,A.J., Kouzarides,T. and Allshire,R.C. (2002) cis-acting DNA from fission yeast centromeres mediates histone H3 methylation and recruitment of silencing factors and cohesin to an ectopic site. Curr. Biol., 12, 1652–1660. [DOI] [PubMed] [Google Scholar]
- Peters A.H. et al. (2001) Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell, 107, 323–337. [DOI] [PubMed] [Google Scholar]
- Peters A.H., Mermoud,J.E., O’Carroll,D., Pagani,M., Schweizer,D., Brockdorff,N. and Jenuwein,T. (2002) Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat. Genet., 30, 77–80. [DOI] [PubMed] [Google Scholar]
- Plath K. et al. (2003) Role of histone H3 lysine 27 methylation in X inactivation. Science, 300, 131–135. [DOI] [PubMed] [Google Scholar]
- Remboutsika E., Lutz,Y., Gansmuller,A., Vonesch,J.L., Losson,R. and Chambon,P. (1999) The putative nuclear receptor mediator TIF1alpha is tightly associated with euchromatin. J. Cell Sci., 112, 1671–1683. [DOI] [PubMed] [Google Scholar]
- Richards E.J. and Elgin,S.C. (2002) Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell, 108, 489–500. [DOI] [PubMed] [Google Scholar]
- Rowley A.F. and Radcliffe,N.A. (eds) (1988) Vertebrate Blood Cells. Cambridge University Press, Cambridge, UK. [Google Scholar]
- Ruiz-Carrillo A., Wangh,L.J., Littau,V.C. and Allfrey,V.G. (1974) Changes in histone acetyl content and in nuclear non-histone protein composition of avian erythroid cells at different stages of maturation. J. Biol. Chem., 249, 7358–7368. [PubMed] [Google Scholar]
- Schmid M., Enderle,E., Schindler,D. and Schempp,W. (1989) Chromosome banding and DNA replication patterns in bird karyotypes. Cytogenet. Cell Genet., 52, 139–146. [DOI] [PubMed] [Google Scholar]
- Silva J. et al. (2003) Establishment of histone H3 methylation on the inactive X chromosome requires transient recruitment of Eed–Enx1 polycomb group complexes. Dev. Cell, 4, 481–495. [DOI] [PubMed] [Google Scholar]
- Smothers J.F. and Henikoff,S. (2000) The HP1 chromo shadow domain binds a consensus peptide pentamer. Curr. Biol., 10, 27–30. [DOI] [PubMed] [Google Scholar]
- Smothers J.F. and Henikoff,S. (2001) The hinge and chromo shadow domain impart distinct targeting of HP1-like proteins. Mol. Cell. Biol., 21, 2555–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verreault A., Kaufman,P.D., Kobayashi,R. and Stillman,B. (1996) Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell, 87, 95–104. [DOI] [PubMed] [Google Scholar]
- Wang J., Mager,J., Chen,Y., Schneider,E., Cross,J.C., Nagy,A. and Magnuson,T. (2001) Imprinted X inactivation maintained by a mouse Polycomb group gene. Nat. Genet., 28, 371–375. [DOI] [PubMed] [Google Scholar]
- Weintraub H. (1984) Histone-H1-dependent chromatin superstructures and the suppression of gene activity. Cell, 38, 17–27. [DOI] [PubMed] [Google Scholar]
- Widom J. (1998) Chromatin structure: linking structure to function with histone H1. Curr. Biol., 8, R788–R791. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K., Hidema,S. and Mizuno,S. (1998) Chicken chromobox proteins: cDNA cloning of CHCB1, -2, -3 and their relation to W-heterochromatin. Exp. Cell Res., 242, 303–314. [DOI] [PubMed] [Google Scholar]