Abstract
Neisseria gonorrhoeae strain MS11 is able to express 11 different opacity (Opa) proteins on its outer surface. A number of these Opa proteins have been shown to function as adhesins through binding of CD66 receptors present on human cells. CD66 antigens, or carcinoembryonic antigen family members, constitute a family of glycoproteins belonging to the immunoglobulin superfamily. Opa variants recognize this class of receptors in a differential manner such that certain Opa variants recognize up to four different CD66 receptors (CD66a, -c, -d, and -e), whereas others recognize only two (CD66a and -e) or none. We explored the basis for this receptor tropism in the present study. Our data show that glycoforms of CD66e and deglycosylated CD66e are recognized by gonococci in an Opa-specific manner. Binding by Opa variants of recombinant N-terminal domains of CD66 receptors expressed in Escherichia coli reflected the adherence specificities of Opa variants to HeLa cells expressing native CD66 molecules. These data indicate that recognition of CD66 receptors by Opa variants is mediated by the protein backbone of the CD66 N-domains. Furthermore, by using chimeric constructs between different CD66 N-domains we identified distinct binding regions on the CD66e N-domain for specific groups of Opa variants, suggesting that the differential recognition of CD66 receptors by Opa variants is dictated by the presence of specific binding regions on the N-domain of the receptor.
Neisseria gonorrhoeae, the causative agent of gonorrhea, is strictly adapted to its human host. Sexual transmission of the pathogen leads to colonization of the urogenital tract through bacterial interactions with mucosal epithelial cells. Colonization of the mucosa is facilitated by gonococcal pili, which have been reported to bind to CD46 or membrane cofactor protein (1). After the initial adherence, bacteria may bind more tightly to the cell surface by means of colony opacity-associated (Opa) proteins (2, 3). The expression of Opa proteins is phase-variable, and multiple proteins can be expressed by individual organisms. Individual strains of N. gonorrhoeae may have up to 11 genes encoding a family of Opa proteins with regions of variable amino acid sequences arrayed on the surface of the organism (4, 5).
Distinct Opa variants are capable of establishing contact with epithelial cells through cell surface heparan-sulfate proteoglycans (6, 7). Recently, another family of host cell receptors has been described that interact with a wide variety of gonococcal and meningococcal Opa variants (8, 9). These receptors belong to the CD66 antigen family, also designated as the carcinoembryonic antigen (CEA) gene family, and are found on epithelial cells and neutrophils (10, 11), two cell types that are targeted by gonococci during natural infection. CD66 antigens are recognized differentially by Opa variants of N. gonorrhoeae strain MS11 (12–14). OpaD, -E, -F, -H, -J, and -K variants recognize both CD66a [or BGPa (biliary glycoprotein a)] and CD66e (CEA), whereas OpaB, -C, -G, and -I variants bind CD66a, CD66c [or NCA (nonspecific cross-reacting antigen)], CD66d [or CGM1a (CEA gene family member 1a)] and CD66e. Bacteria expressing OpaA do not bind any of the tested CD66 receptors (CD66a–e). One member of this receptor family, CD66b (or CGM6), does not act as a receptor for any Opa variant.
CD66 antigens belong to the immunoglobulin (Ig) superfamily (15, 16). They contain a single Ig variable (IgV)-like N-terminal domain (N-domain), followed by a differing number of Ig constant-like, C2 type, internal domains. The molecules are anchored to the plasma membrane by a transmembrane domain (CD66a and -d) or by a glycosyl-phosphatidylinositol linkage (CD66b, -c, and -e) (10). CD66 antigens are N-glycosylated molecules, with up to 50% of their molecular mass attributed to oligosaccharide residues. The number and composition of CD66 sugar chains depend on the tissue status—i.e., development stage or malignant transformation. For example, the carbohydrate moieties of CD66e derived from healthy adult feces, fetal meconium, or human colon carcinoma are composed differently (17), indicating that colon epithelial cells are capable of expressing different glycoforms of CD66e. These findings suggest that the carbohydrate moieties of CD66 antigens will be dependent on the differentiation status of the tissue where they are present. Neutrophils express differently glycosylated forms of CD66 molecules in response to stimulation by the chemotactic peptide fMLP (18). Thus, if the carbohydrate moieties of CD66 receptors play any role in recognition by Opa proteins, then binding of gonococci to tissues through these receptors may depend not only on the presence of the receptor but also on the tissue status. Therefore, it is important to determine the nature of the Opa–CD66 interaction.
In the present study we have characterized the molecular basis for the recognition of CD66 receptors by gonococcal Opa variants. We utilized cells expressing CD66e glycoforms and chemically deglycosylated CD66e to show that the CD66 carbohydrate moieties are not important for binding of Opa variants. Furthermore, we found that recombinant N-domains of CD66 antigens, expressed by Escherichia coli, mirror binding patterns found for native receptor molecules expressed on cells. The use of chimeric N-domains of CD66e and CD66b has further defined the important binding domains within the CD66e molecule and has provided a molecular basis for the differential binding of Opa variants to CD66 receptors expressed on the cell surface.
MATERIALS AND METHODS
Bacterial Strains.
N. gonorrhoeae MS11 variants were propagated on gonococcal clear typing agar (19). Wild-type variants MS11 mk expressing chromosomally encoded Opa proteins were kindly provided by J. Swanson and are designated according to Swanson et al. (20) by capital letters—e.g., OpaA–OpaK. Recombinant MS11 Opa variants (21) were a generous gift of T. F. Meyer (Max-Planck-Institut für Biologie, Tübingen, Germany). Nomenclature of recombinant strains is numerical—e.g., Opa50-Opa60 (21). Opa protein expression was verified by SDS/PAGE and immunoblotting of bacterial lysates followed by detection with anti-Opa antibody 4B12 (20). Only nonpiliated bacteria were used. For experiments, bacteria were grown for 3 hr in 10 ml of Hepes medium [10 mM Hepes/145 mM NaCl/5 mM KCl/0.5 mM MgCl2/1 mM CaCl2/5 mM glucose/1.5% proteose peptone no. 3 (Difco), pH 7.4] in a Gyrotory shaker at 37°C. Bacterial suspensions were spun for 5 min at 2,000 × g and resuspended in 1 ml of Hepes buffer (Hepes medium without proteose peptone).
Cells and Cell Culture.
Stably transfected HeLa cell lines expressing defined CD66 proteins (22, 23) were a gift from F. Grunert (Institut für Immunbiologie, Freiburg, Germany) and were cultured as described (12). Parent Chinese hamster ovary (CHO) cells, Pro5, and glycosylation-defective mutants, Lec2 and Lec8, were obtained from the American Type Culture Collection (Manassas, VA). CHO cells were cultured in alpha-minimal essential medium supplemented with 10% fetal calf serum, 2 mM glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. The cell cultures were maintained at 37°C in a humidified atmosphere at 5% CO2 in air. Media and sera were purchased from GIBCO/BRL.
Transfection Procedure.
Plasmid pdKCR-neo containing a full-length CEA (CD66e) cDNA was kindly provided by S. Oikawa (Institute for Biomedical Research, Suntory Ltd., Osaka, Japan) (24). For construction of stably transfected Pro5, Lec2, and Lec8 cell lines expressing CD66e, cells were cotransfected with 20 μg of pSG5CEA and 2 μg of pSV2neo plasmids by using the calcium phosphate precipitation method according to the procedure recommended in the Cell Phect Transfection Kit (Pharmacia). Neomycin-resistant cells were selected by growth in the presence of 0.5 mg/ml geneticin G418 (GIBCO/BRL) for 14 days. Surviving cells were cloned by limiting dilution, and G418-resistant colonies were isolated, expanded, and screened for CD66e expression by fluorescence-activated cell sorter analysis and Western blotting.
Western Blot Analysis.
Lysates were prepared by boiling bacteria in SDS/PAGE sample buffer. Samples were electrophoresed on either 8% (for intact CD66e) or 13.5% (for N-domains) polyacrylamide gels containing SDS and were immunoblotted onto nitrocellulose (25). After blocking with 3% BSA in PBS plus 0.1% Tween-20 (PBS/T), CD66e was detected by horseradish peroxidase (HRP)-conjugated anti-CEA antiserum (1:1000, Dako) and N-domain fusion proteins by anti-His antibody (1:15,000, Pharmacia) followed by HRP-conjugated staphylococcal protein A (1:20,000, Sigma). Blots were developed with the enhanced chemiluminescence protocol (Amersham).
Infection Assay.
Gonococci (1.5 × 107) were added to 2 × 105 cells, cultured on 12 mm diameter glass coverslips, in 1 ml of Hepes buffer for 2 hr at 37°C. Nonadherent bacteria were removed by three washes. After fixation, intra- and extracellular bacteria were visualized by a differential staining procedure (12) and counted for at least 50 cells per coverslip.
Deglycosylation of CEA.
CEA was extracted from human tumor tissue by phosphatidylinositol-specific phospholipase C (PI-PLC) treatment and purified exactly as described (26). CEA was 99% pure as judged by Coomassie brilliant blue staining of the material after SDS/PAGE (data not shown). Chemical deglycosylation was carried out according to Edge et al. (27). Five-hundred μg of CEA was dissolved in a mixture of 33 μl of anisole and 67 μl of trifluoromethanesulfonic acid (TFMSA) at 4°C. The mixture was stirred for 4 hr at 4°C, diluted with 200 μl of diethyl ether, and neutralized by adding 300 μl of 50% (vol/vol) aqueous pyridine at −40°C. The solution was dialyzed against 10% pyridine/PBS and then against 0.1% SDS/PBS.
Binding of Bacteria to Immobilized CEA.
CEA was dried onto the wells of ELISA plates (Greiner, Frickenhausen, Germany) at 1 μg per well. The wells were blocked with 3% BSA for 2 hr at 37°C. Bacteria were radiolabeled by adding 11 nM [35S]methionine/[35S]cysteine (1,000 Ci/mmol; Express Protein Labelling Mix, NEN; 1 Ci = 37 GBq) to the Hepes medium during growth. After three washes to remove unincorporated radiolabel, 108 bacteria in Hepes buffer were added per well and incubated for 1 hr at 37°C. After five washes with Hepes buffer, adherent bacteria were detached with 0.1% SDS and counted in a liquid scintillation counter.
Isolation of CD66e Glycoforms from Transfected Cells.
Cells were detached from tissue culture flasks with 0.5 mM EDTA in PBS, spun at 400 × g, resuspended in Hanks’ balanced salt solution (HBSS), and incubated for 2 hr at 37°C with 0.29 unit of PI-PLC (ICN) plus protease inhibitor mixture (Boehringer Mannheim). Cells were spun for 5 min at 400 × g and the supernatant, containing soluble CD66e, was concentrated by using a 30,000 Mr cut-off Centricon device (Amicon).
Construction and Isolation of Recombinant N-Domains.
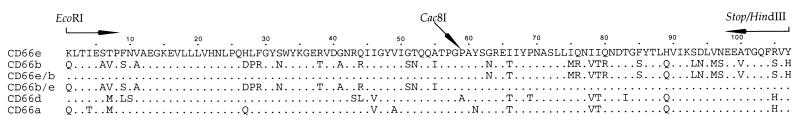
Clones of N-domains of CD66 antigens were constructed by PCR amplifying the appropriate sequence from DNA isolated from HeLa transfectants. The 5′ primers contained N-domain sequence plus an EcoRI site and the 3′ primers contained N-domain sequence plus a HindIII site and a stop codon replacing the last N-domain codon (Fig. 1). Primers were designed using known CD66 N-domain sequences (22, 24, 28, 29). The PCR products were inserted into the pCR2.1 vector by using the TA cloning protocol (Invitrogen). The EcoRI–HindIII-flanked N-domain fragments were then subcloned into the pRSET-A vector (Invitrogen) and electroporated into E. coli strain BL21 (DE3) (Novagen). This strategy resulted in expression of 107 N-domain residues fused to a 48-residue spacer containing a six-His stretch at the N-terminal part of the protein. Chimeras between CD66b and CD66e N-domains were made by digesting the EcoRI–HindIII-flanked N-domains with Cac8I (New England Biolabs). The resulting fragments were purified and the EcoRI-CD66b fragment was ligated onto the HindIII-CD66e fragment and vice versa. Ligation products were digested with EcoRI and HindIII, ligated into pRSET-A, and electroporated into E. coli BL21 cells. Sequences of all clones were verified by sequencing a PCR fragment containing the entire insert by using the Dye terminator cycle sequencing kit with a 373 Stretch DNA Sequencer (Applied Biosystems). For isolation of recombinant N-domains E. coli cells were grown in 10 ml of Luria broth containing 100 μg/ml carbenicillin to an OD at 600 nm of 0.6. Then 1 mM isopropyl β-d-thiogalactopyranoside was added for 90 min, whereafter cells were collected (10 min at 2,000 × g) and frozen at −70°C. Cleared cell lysates were prepared by sonicating cell pellets in 0.7 ml of lysis buffer [50 mM Tris⋅HCl, pH 8.5/1 mM phenylmethylsulfonyl fluoride (Sigma)] twice for 1 min each, followed by centrifugation for 15 min at 13,000 × g. His-tagged proteins were purified in the presence of 8 M urea through Ni-nitrilotriacetate (NTA) spin columns as recommended by the manufacturer (Qiagen). Purified samples were dialyzed against PBS by ultrafiltration in a 3,000 Mr cut-off Centricon device.
Figure 1.
Amino acid alignment of CD66 N-domains used in this study. Protein sequences were deduced from DNA sequencing of the clones. Dots indicate residues identical to those in CD66e. Arrows indicate the regions corresponding to the primer binding sites for PCR amplification of N-domains from HeLa-CD66 DNA. The location of the Cac8I site present in the DNA used to construct CD66b/e chimeric N-domains is indicated at the corresponding residue 59.
Binding of Soluble Receptors by Gonococci.
Gonococci (108) were incubated with the appropriate ligand in 200 μl of Hepes buffer for 20 min at 37°C. To assay binding of N-domains, 7.5 μl of cleared E. coli lysate was added. To measure binding of CHO cell-derived CD66e, 10% of the material derived from a semiconfluent 75-cm2 tissue culture flask was used per sample. Bacteria were collected by centrifugation (5 min at 2,000 × g), washed twice with 1 ml of Hepes buffer, solubilized in 30 μl of SDS/PAGE sample buffer, and processed for immunoblotting. Binding under low-stringency conditions was performed in Hepes buffer with 280 mM sucrose replacing NaCl and KCl.
RESULTS
Opa+ Gonococci Recognize CD66e Deficient in Sialic Acid or Galactose Residues.
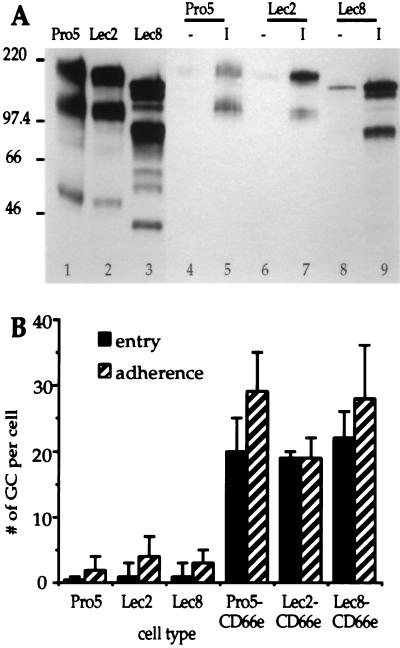
The interaction of Opa-expressing gonococci with carbohydrate-modified CD66e was investigated by using parental (Pro5) and mutant (Lec2 and Lec8) CHO cell lines transfected with recombinant CD66e cDNA. Lec2 cells are deficient in CMP-sialic acid transport (30) and Lec8 cells in UDP-galactose transport into the Golgi compartment (31), resulting in a drastic reduction in sialylation or galactosylation of macromolecules. CD66e isolated by PI-PLC treatment from the mutant transfectants migrated much faster in SDS/PAGE than did the parental Pro5-CD66e, consistent with a decreased glycosylation of the molecules (Fig. 2A, lanes 1–3). The two major bands visible in each preparation are often observed in CEA preparations from different sources (data not shown); they most likely represent different CEA glycoforms, the lower one being CEA containing high-mannose type oligosaccharides instead of mature complex-type chains. The bands running lower than 70 kDa are probably proteolytic degradation products. Binding experiments showed that OpaI-expressing bacteria bound all CD66e glycoforms irrespective of their carbohydrate composition, whereas Opa− bacteria did not bind significant amounts of any CD66e type (Fig. 2A, lanes 4–9). Furthermore, infection assays performed with the different cell types revealed no differences between the mutant and the parent cell lines expressing CD66e; i.e., they bound and internalized OpaI variants to similar levels (Fig. 2B). Thus, sialic acid or galactose residues from CD66e are not required for recognition of CD66e by OpaI-expressing gonococci.
Figure 2.
Interaction of MS11 Opa variants with CD66e glycoforms. (A) Immunoblot showing binding of CD66e glycoforms by gonococcal Opa variants. CD66e glycoforms were isolated from CD66e-transfected CHO cell lines by using PI-PLC (lanes 1–3). Binding of CD66e glycoforms to Opa− (-) or OpaI (I) expressing gonococci is shown in lanes 4–9. Bound CD66e was detected with anti-CEA rabbit serum. Mr markers (× 10−3) are indicated on the left. The cell types from which CD66e was derived are indicated above the blot. (B) Adherence and entry of OpaI variants to Pro5, Lec2, and Lec8 cells and their CD66e-expressing counterparts. GC, gonococci. Data are means ± SE of three independent experiments.
OpaI Gonococci Recognize Deglycosylated CD66e.
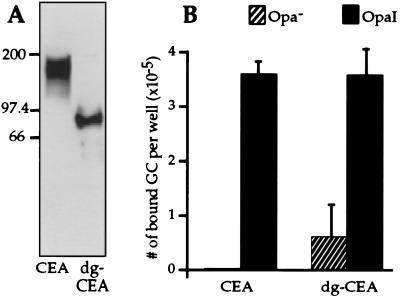
To determine whether oligosaccharide residues in the inner core of the CD66e carbohydrate moiety were involved in Opa-CD66e recognition, we utilized an assay previously used to measure heterophilic interactions between CD66 family members (32). Native and deglycosylated human tumor-derived CEA (CD66e) was immobilized onto the wells of ELISA plates, and interaction with gonococci was evaluated by measuring binding of radiolabeled Opa variants to these plates. Deglycosylation by TFMSA of human tumor-derived CEA resulted in a decrease of its apparent Mr from 180,000 to 85,000 (Fig. 3A). Because TFMSA treatment leaves the innermost GlcNAc attached to Asn residues, the calculated Mr of PI-PLC- and TFMSA-treated CEA containing potentially 28 GlcNAc residues would be 80,000 (33). The appearance of an 85,000 band after TFMSA treatment of CEA has been observed by others (15, 33). Although it is not known whether the difference between calculated and apparent Mr is significant and may represent some residual carbohydrate, the more than 50% reduction in apparent Mr of CEA after deglycosylation indicates that the CEA is virtually devoid of carbohydrate.
Figure 3.
Effect of deglycosylation of CEA on recognition by MS11 Opa variants. (A) Immunoblot showing native CEA (CEA) and CEA treated with TFMSA (dg-CEA). Each lane contains 50 ng of material, which was detected with anti-CEA rabbit serum. Mr markers (× 10−3) are indicated on the left. (B) Binding of MS11 Opa variants to native and deglycosylated CEA. Radiolabeled bacteria were added to the wells of ELISA plates containing native (CEA) or deglycosylated (dg-CEA) human tumor CEA for 1 hr at 37°C. Nonadherent bacteria were removed by washing and the number of bound bacteria was determined by liquid scintillation counting of detached bacteria. GC, gonococci. Data are means ± SE of three independent experiments.
As shown in Fig. 3B, OpaI variants bound specifically to immobilized native CEA. After deglycosylation of CEA, OpaI variants were still capable of binding CEA. Thus, our results indicate that carbohydrate moieties of CEA are not required for binding of OpaI-expressing gonococci.
OpaI Gonococci Recognize the N-Domain of CD66e, Expressed in E. coli.
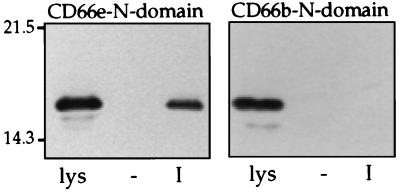
Because the carbohydrate moiety of CD66e did not appear to contribute to Opa recognition, we tested whether recombinant CD66e produced by E. coli is recognized by Opa-producing gonococci. For this purpose, we constructed a recombinant plasmid encoding the N-domain of CD66e as a His-tag fusion protein. In a similar manner, the N-domain of CD66b was cloned and expressed. CD66b is not recognized by any Opa variant (12–14) and therefore served as a negative control in our assay. Binding of CD66b and CD66e N-domains by Opa variants was measured with cleared lysates of induced recombinant E. coli clones with the results shown in Fig. 4. These data show that OpaI but not Opa− bacteria recognized the N-domain of CD66e. The N-domain of CD66b was not bound by any of the Opa variants, indicating the specificity of this assay for evaluating only the CD66-related protein present in the lysate. CD66e N-domain purified from bacterial lysates by using Ni-NTA columns had the same Opa-specific binding characteristics as the material in crude lysates (data not shown). Thus, this experiment demonstrated that the CD66e N-domain protein backbone is sufficient for recognition by OpaI variants.
Figure 4.
Binding of E. coli-expressed N-domains of CD66e and CD66b by MS11 Opa variants. Opa variants were incubated with cleared lysates of E. coli containing the appropriate N-domains and processed for immunoblotting. The N-domains were detected with anti-His antibody. Lanes labeled lys show the cleared lysate, in an amount representing 100% binding; lanes - and I show the amount of N-domain bound by Opa− and OpaI variants, respectively. The figure is representative of three independent experiments.
Differential Recognition of CD66e N-Domain by MS11 Opa Variants.
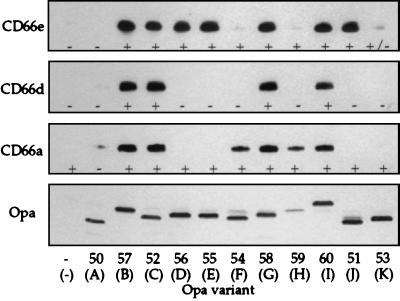
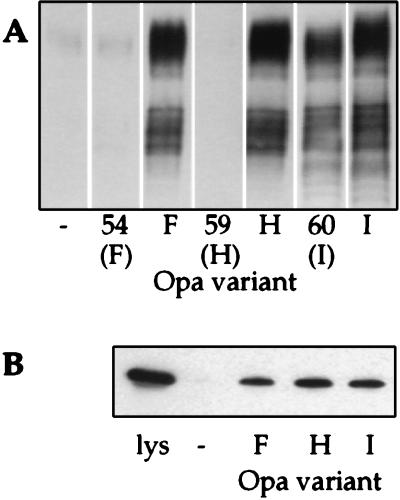
Binding of the recombinant N-domain of CD66e by all Opa variants of strain MS11 was assessed by using a series of recombinant Opa variants in MS11 (Opa50–60) (21). Wild-type opa genes exhibit differential rates of phase-variation, and therefore certain wild-type Opa variants are difficult to maintain stably in vitro. This difficulty is avoided by using recombinant Opa variants whose recombinant opa genes are engineered such that they are unable to undergo phase-variation (21). As shown in Fig. 5 Top, 7 of 11 Opa variants recognized the protein backbone of the CD66e N-domain. These results are consistent with the specificity observed for binding of gonococci to HeLa-CD66e cells, with the exception of Opa54 and Opa59. To examine whether binding of CD66e by Opa54 and Opa59 variants required other components of CD66e, we measured binding of intact CD66e, released from Pro5-CD66e cells, to these variants. As shown in Fig. 6A, they did not bind to soluble, intact CD66e either, although Opa54 and Opa59 variants recognized Pro5-CD66e cells in an Opa-dependent manner (data not shown). However, when we repeated the binding assay with wild-type Opa variants, which express up to three times as much Opa protein per bacterium (21), we did find binding of the wild-type counterparts of Opa54 and Opa59 (OpaF and OpaH, respectively) to intact CD66e and to the recombinant CD66e N-domain (Fig. 6). Moreover, binding experiments performed at a lower ionic strength showed Opa-specific binding of the CD66e N-domain by both Opa54 and Opa59 (data not shown), suggesting that these Opa variants bind to CD66e with a lower avidity than other Opa variants.
Figure 5.
Binding of E. coli expressed N-domains of different CD66 receptors by recombinant MS11 Opa variants. Gonococci expressing different Opa proteins were incubated with cleared lysates of E. coli containing the N-domains of CD66e, -d, or -a and processed for immunoblotting. Bound N-domain was detected with anti-His antibody. The + and - designations refer to the results of our previous study, where interaction of Opa variants with CD66e, -d, and -a expressed on HeLa cells was studied: + indicates recognition; - indicates no recognition (12). Opa protein expression of the variants was evaluated by immunoblotting with 4B12 antibody and is shown in Bottom. Below the designation of the recombinant Opa variants used the nomenclature of wild-type Opa homologs is indicated in parentheses. The figure is representative of three independent experiments.
Figure 6.
Interaction of Opa54/F and Opa59/H variants with CD66e and recombinant CD66e-N-domain. (A) Binding of CD66e, derived from Pro5-CD66e cells through PI-PLC treatment, by recombinant Opa variants and their wild-type counterparts. Bound CD66e was detected with anti-CEA rabbit serum. Below the designation of the recombinant Opa variants the nomenclature of the wild-type Opa homologs is indicated in parentheses. (B) Binding of recombinant CD66e-N-domain by wild-type Opa variants. Opa−, F, H, and I variants were incubated with cleared lysates of E. coli containing the CD66e N-domain and processed for immunoblotting. Bound N-domain was detected with anti-His antibody. Results with Opa60/I are shown as a positive control. The figure is representative of three independent experiments.
Binding of Opa Variants to E. coli-Expressed CD66a- and CD66d-N-Domains.
To determine whether the N-domain protein backbone of other CD66 receptors would also provide the binding site for Opa proteins, we constructed CD66a and CD66d N-domain His-tag fusion proteins and analyzed the recognition of these molecules by Opa variants in the same manner as described above. The N-domain of CD66d was bound by Opa52, Opa57, Opa58, and Opa60 variants, which reflects exactly the pattern found for bacterial binding to HeLa-CD66d cells (Fig. 5). The N-domain of CD66a was bound by Opa52, Opa57, Opa58, Opa60, plus Opa54 and Opa59 variants (Fig. 5). Thus, the Opa54 and Opa59 variants can readily bind the protein backbone of a CD66 N-domain: apparently binding avidity for the CD66a N-domain is higher than that for the CD66e-N-domain. Opa51, Opa53, Opa55, and Opa56 variants did not bind to soluble CD66a N-domain, in contrast to their adherence to CD66a-transfected cells (Fig. 5). This difference was maintained when experiments were performed at lower ionic strength or when the respective wild-type variants (OpaD and OpaE) were used (data not shown).
Identification of two Opa Binding Regions in the N-Domain of CD66e.
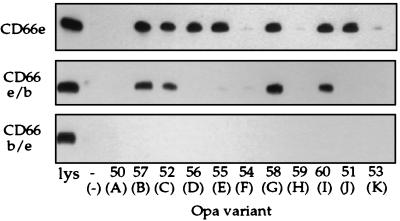
To further localize CD66e N-domain sites involved in binding of Opa proteins, we constructed chimeras of CD66e and CD66b N-domains, using a unique, allelic, restriction site within the DNA sequence encoding residue 59 in both molecules (Fig. 1). Interestingly, only the Opa52, Opa57, Opa58, and Opa60 variants recognized the chimera containing the first half of the CD66e N-domain. The other Opa variants that recognized the intact CD66e N-domain, including OpaF and OpaH variants, were unable to bind this chimera. None of the Opa variants bound the chimera containing the second half of the CD66e-N-domain (Fig. 7). These data suggest that CD66e contains at least two different regions required for Opa protein binding: one in the first 59 CD66e-N-domain residues involved in binding of Opa52, Opa57, Opa58, and Opa60, and one that is composed of residues in both the first and second half of the CD66e-N-domain for Opa51, Opa54, Opa55, Opa56, and Opa59 variants.
Figure 7.
Binding of E. coli-expressed CD66e, CD66e/b, and CD66b/e N-domain chimeras by recombinant MS11 Opa variants. Opa variants were incubated with cleared lysates of E. coli expressing the indicated N-domains and processed for immunoblotting. Bound N-domain was detected with anti-His antibody. The lane designated lys contains only the E. coli lysate. Below the designation of the recombinant Opa variants used the nomenclature for wild-type Opa homologs is indicated in parentheses. The figure is representative of three independent experiments.
DISCUSSION
The outer-membrane Opa protein families of N. gonorrhoeae and Neisseria meningitidis function as bacterial adhesins, binding to several members of the CD66 family of receptors on epithelial and polymorphonuclear cells. CD66 antigens are heavily glycosylated molecules belonging to the Ig superfamily. They consist of an N-terminal IgV-like domain (N-domain) and a variable number of IgC-like domains. Gonococcal Opa variants recognize these receptors in a differential manner. Our results demonstrate that the protein backbones of the N-domains of CD66 receptors contain recognition sites for gonococcal Opa adhesins and that distinct Opa variants recognize different binding regions, providing a molecular basis for the Opa-related CD66 receptor tropism displayed by gonococci.
The complex carbohydrate moieties typically present on CD66 molecules are not required for the adherence of Opa+ gonococci, as demonstrated by the observation that gonococci bound several glycoforms of CD66e as well as chemically deglycosylated CD66e in an Opa-dependent fashion. Definite proof of the involvement of peptide sequences was provided by the Opa-dependent binding of recombinant, nonglycosylated CD66 N-domains produced by E. coli. The focus on the N-domain was prompted by the fact that Opa+ bacteria recognize CD66d, which contains an IgV N-domain but lacks any of the internal IgC domains, and by previous data that neisserial Opa proteins bind to the glycosylated N-domain of CD66a (34). The finding that Opa+ gonococci recognize peptide determinants was unexpected, as other bacterial pathogens, such as Salmonella and E. coli expressing type I fimbriae, bind to CD66 molecules through N-domain-linked oligosaccharides in a mannose-sensitive manner (35, 36). The ability of Opa variants to interact with peptide determinants in the N-domain of CD66e is consistent with structural analyses which predict that only the N-domain of CD66e contains regions not covered by carbohydrate that would be accessible for protein–protein interactions (37).
The pattern of binding of Opa variants to the recombinant CD66e N-domain reflected the differential recognition of the native receptor expressed on transfected eukaryotic cells. These data imply that the specificity of the interaction is maintained in the isolated nonglycosylated N-domain. Immunochemical analysis previously demonstrated that E. coli-derived CD66e N-domain retained reactivity with six different antibodies raised to intact CD66e, suggesting conformational similarity between recombinant product and native protein. This reactivity was not found for other CD66e domains: approximately one-third of peptide epitopes were lost in expressing recombinant internal CD66e domains in E. coli (38). The better preservation of conformation in the recombinant IgV N-domain may be related to the absence of disulfide bonds, necessary for the accurate folding of the internal IgC domains.
Binding of recombinant N-domains of various CD66 receptors by Opa variants demonstrated that recognition sites for Opa proteins were located not only on the CD66e N-domain but also in the protein backbone of the homologous N-domains of other members of the CD66 family. Furthermore, the typical tropism of Opa variants for the various CD66 receptors was mirrored in the binding of the recombinant N-domains. This correlation is best illustrated by the observation that both native CD66d and recombinant CD66d N-domain are recognized by the same 4 of 11 MS11 Opa variants (Fig. 5). Thus, different N-domains may carry different binding sites for distinct Opa proteins. Further support for this concept was obtained from the results with the chimeras of CD66e and CD66b. The finding that the first 59 amino acid residues of the CD66e N-domain were sufficient for binding of four Opa variants (Opa52, Opa57, Opa58, Opa60), whereas the remaining Opa variants that bind CD66e required the entire 107 residues of the CD66e N-domain strongly suggests that these two groups of Opa variants utilize different binding regions within the same molecule. The observation that only the Opa variants that bind within the first 59 residues of CD66e recognize the CD66c and CD66d receptors (12–14) may indicate that the second binding region is absent from these molecules, supporting the idea that the presence of distinct Opa recognition regions controls receptor tropism.
The only exception to the correlation observed between the recognition of recombinant CD66 N-domain versus native CD66 receptors by Opa variants was the interaction of Opa51, Opa53, Opa55, and Opa56 variants with CD66a. These Opa variants did not bind the CD66a N-domain, whereas they do bind and enter HeLa-CD66a cells (12, 14). The reason for this discrepancy is unclear. Interestingly, however, the Opa-specific binding of recombinant CD66a N-domain corresponds exactly with the recognition of HeLa-CD66a cells by Opa-expressing E. coli (14), suggesting that perhaps other gonococcal determinants play a role during infection of HeLa-CD66a cells.
The Opa protein family contains regions of extensive homology interspersed with a semivariable (SV) and two hypervariable (HV1 and HV2) domains (4, 21). The variable domains are contained within surface-exposed loops and are predicted to account for the differences in biological activities among Opa variants. The differential binding of CD66 receptors by Opa variants supports the contention that variation in surface-exposed loops may serve to attune to heterogeneity of CD66 receptors on various cell types encountered during an infection. Finer mapping of the binding sites in Opa and CD66 proteins may further resolve the role of Opa protein variation in relation to CD66 receptor specificity.
Acknowledgments
We thank Dr. Masahide Kuroki (Fukuoka University, Fukuoka, Japan) for supplying human tumor CEA. We also thank Jos van Putten, John Swanson, and Ted Hackstadt for helpful suggestions on the manuscript. This work was supported in part by Grant 6 PO4 A 01808 from the State Committee for Scientific Research (KBN), Warsaw, Poland.
ABBREVIATIONS
- CEA
carcinoembryonic antigen
- PI-PLC
phosphatidylinositol-specific phospholipase C
- TFMSA
trifluoromethanesulfonic acid
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Kallstrom H, Liszewski M K, Atkinson J P, Jonsson A B. Mol Microbiol. 1997;25:639–647. doi: 10.1046/j.1365-2958.1997.4841857.x. [DOI] [PubMed] [Google Scholar]
- 2.Meyer T F, Pohlner J, van Putten J P M. Curr Top Microbiol Immunol. 1994;192:283–317. doi: 10.1007/978-3-642-78624-2_13. [DOI] [PubMed] [Google Scholar]
- 3.van Putten J P M, Duensing T D. Rev Med Microbiol. 1997;8:51–59. [Google Scholar]
- 4.Bhat K S, Gibbs C P, Barrera O, Morrison S G, Jähnig F, Stern A, Kupsch E-M, Meyer T F, Swanson J. Mol Microbiol. 1991;5:1889–1901. doi: 10.1111/j.1365-2958.1991.tb00813.x. [DOI] [PubMed] [Google Scholar]
- 5.Stern A, Brown M, Nickel P, Meyer T F. Cell. 1986;47:61–71. doi: 10.1016/0092-8674(86)90366-1. [DOI] [PubMed] [Google Scholar]
- 6.van Putten J P M, Paul S M. EMBO J. 1995;14:2144–2154. doi: 10.1002/j.1460-2075.1995.tb07208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen T, Belland R J, Wilson J, Swanson J. J Exp Med. 1995;182:511–517. doi: 10.1084/jem.182.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Virji M, Makepeace K, Ferguson D J P, Watt S M. Mol Microbiol. 1996;22:941–950. doi: 10.1046/j.1365-2958.1996.01551.x. [DOI] [PubMed] [Google Scholar]
- 9.Chen T, Gotschlich E C. Proc Natl Acad Sci USA. 1996;93:14851–14856. doi: 10.1073/pnas.93.25.14851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson J A, Grunert F, Zimmermann W. J Clin Lab Anal. 1991;5:344–366. doi: 10.1002/jcla.1860050510. [DOI] [PubMed] [Google Scholar]
- 11.Obrink B. Curr Opin Cell Biol. 1997;9:616–626. doi: 10.1016/S0955-0674(97)80114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bos M P, Grunert F, Belland R J. Infect Immun. 1997;65:2353–2361. doi: 10.1128/iai.65.6.2353-2361.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen T, Grunert F, Medina-Marino A, Gotschlich E C. J Exp Med. 1997;185:1557–1564. doi: 10.1084/jem.185.9.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray-Owen S D, Lorenzen D R, Haude A, Meyer T F, Dehio C. Mol Microbiol. 1997;26:971–980. doi: 10.1046/j.1365-2958.1997.6342006.x. [DOI] [PubMed] [Google Scholar]
- 15.Paxton R, Mooser G, Pande H, Lee T D, Shively J E. Proc Natl Acad Sci USA. 1987;84:920–924. doi: 10.1073/pnas.84.4.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams A F, Barclay A N. Annu Rev Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- 17.Fukushima K, Ohkura T, Kanai M, Kuroki M, Matsuoka Y, Kobata A, Yamashita K. Glycobiology. 1995;5:105–115. doi: 10.1093/glycob/5.1.105. [DOI] [PubMed] [Google Scholar]
- 18.Stocks S C, Kerr M A. Biochem Biophys Res Commun. 1993;195:478–483. doi: 10.1006/bbrc.1993.2068. [DOI] [PubMed] [Google Scholar]
- 19.Swanson J. Infect Immun. 1978;19:320–331. doi: 10.1128/iai.19.1.320-331.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swanson J, Barrera O, Sola J, Boslego J. J Exp Med. 1988;168:2121–2129. doi: 10.1084/jem.168.6.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kupsch E-M, Knepper B, Kuroki T, Heuer I, Meyer T F. EMBO J. 1993;12:641–650. doi: 10.1002/j.1460-2075.1993.tb05697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berling B, Kolbinger F, Grunert F, Thompson J A, Brombacher F, Buchegger F, von Kleist S, Zimmerman W. Cancer Res. 1990;50:6534–6539. [PubMed] [Google Scholar]
- 23.Daniel S, Nagel G, Johnson J P, Lobo F M, Hirn M, Jantscheff P, Kuroki M, von Kleist S, Grunert F. Int J Cancer. 1993;55:303–310. doi: 10.1002/ijc.2910550222. [DOI] [PubMed] [Google Scholar]
- 24.Oikawa S, Nakazato H, Kosaki G. Biochem Biophys Res Commun. 1987;142:511–518. doi: 10.1016/0006-291x(87)90304-4. [DOI] [PubMed] [Google Scholar]
- 25.Towbin H, Staehelin T, Gordon J. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuoka Y, Matsuo Y, Okamoto N, Kuroki M, Kuroki M, Ikehara Y. Tumor Biol. 1991;12:91–98. doi: 10.1159/000217693. [DOI] [PubMed] [Google Scholar]
- 27.Edge A S, Faltynek C R, Hof L, Reichert L E, Jr, Weber P. Anal Biochem. 1981;118:131–137. doi: 10.1016/0003-2697(81)90168-8. [DOI] [PubMed] [Google Scholar]
- 28.Nagel G, Grunert F, Kuijpers T W, Watt S M, Thompson J, Zimmerman W. Eur J Biochem. 1993;214:27–35. doi: 10.1111/j.1432-1033.1993.tb17892.x. [DOI] [PubMed] [Google Scholar]
- 29.Barnett T R, Kretschmer A, Austen D A, Goebel S J, Hart J T, Elting J J, Kamarck M E. J Cell Biol. 1989;108:267–276. doi: 10.1083/jcb.108.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deutscher S L, Nuwayhid N, Stanley P, Briles E I, Hirschberg C B. Cell. 1984;39:295–299. doi: 10.1016/0092-8674(84)90007-2. [DOI] [PubMed] [Google Scholar]
- 31.Deutscher S L, Hirschberg C B. J Biol Chem. 1986;261:96–100. [PubMed] [Google Scholar]
- 32.Yamanaka T, Kuroki M, Matsuo Y, Matsuoka Y. Biochem Biophys Res Commun. 1996;219:842–847. doi: 10.1006/bbrc.1996.0320. [DOI] [PubMed] [Google Scholar]
- 33.Hernando J J, von Kleist S, Grunert F. Int J Cancer. 1994;56:655–661. doi: 10.1002/ijc.2910560509. [DOI] [PubMed] [Google Scholar]
- 34.Virji M, Watt S M, Barker S, Makepeace K, Doyonnas R. Mol Microbiol. 1996;22:929–939. doi: 10.1046/j.1365-2958.1996.01548.x. [DOI] [PubMed] [Google Scholar]
- 35.Leusch H G, Drzeniek Z, Hefta S A, Markos-Pusztai Z, Wagener C. Int J Med Microbiol. 1993;275:118–122. doi: 10.1016/s0934-8840(11)80775-9. [DOI] [PubMed] [Google Scholar]
- 36.Sauter S L, Rutherfurd S M, Wagener C, Shively J E, Hefta S A. J Biol Chem. 1993;268:15510–15516. [PubMed] [Google Scholar]
- 37.Bates P A, Luo J, Sternberg M J E. FEBS Lett. 1992;301:207–214. doi: 10.1016/0014-5793(92)81249-l. [DOI] [PubMed] [Google Scholar]
- 38.Kuroki M, Murakami M, Wakisaka M, Ikeda S, Oikawa S, Oshima T, Nakazato H, Kosaki G, Matsuoka Y. Immunol Invest. 1992;21:241–257. doi: 10.3109/08820139209072262. [DOI] [PubMed] [Google Scholar]