Abstract
Formylglutamate amidohydrolase (FGase) catalyzes the terminal reaction in the five-step pathway for histidine utilization in Pseudomonas putida. By this action, N-formyl-L-glutamate (FG) is hydrolyzed to produce L-glutamate plus formate. Urocanate, the first product in the pathway, induced all five enzymes, but FG was able to induce FGase alone, although less efficiently than urocanate did. This induction by FG resulted in the formation of an FGase with electrophoretic mobility identical to that of the FGase induced by urocanate. A 9.6-kilobase-pair HindIII DNA fragment containing the P. putida FGase gene was cloned into the corresponding site on plasmid pBEU1 maintained in Escherichia coli. Insertion of the fragment in either orientation on the vector resulted in expression, but a higher level was noted in one direction, suggesting that the FGase gene can be expressed from either of two vector promoters with different efficiencies or from a single vector promoter in addition to a less efficient Pseudomonas promoter. FGase was purified 1,110-fold from the higher-expression clone in a yield of 10% through six steps. Divalent metal ions stimulated activity, and among those tested (Co, Fe, Zn, Ca, Ni, Cd, Mn, and Mg), Co(II) was the best activator, followed by Fe(II). FGase exhibited a Km of 14 mM for FG and a specific activity of 100 mumol/min per mg of protein in the presence of 5 mM substrate and 0.8 mM CoCl2 at 30 degrees C. The enzyme was maximally active in the range of pH 7 to 8. FGase was found to be a monomer of molecular weight 50,000. N-Acetyl-L-glutamate was not a substrate for the enzyme, but both it and N-formyl-L-aspartate were competitive inhibitors of formylglutamate hydrolysis, exhibiting Ki values of 6 and 9 mM, respectively. The absence of FGase activity as an integral part of histidine breakdown in most other organisms and the somewhat uncoordinated regulation of FGase synthesis with that of the other hut enzymes in Pseudomonas suggest that the gene encoding its synthesis may have evolved separately from the remaining hut genes.
Full text
PDF

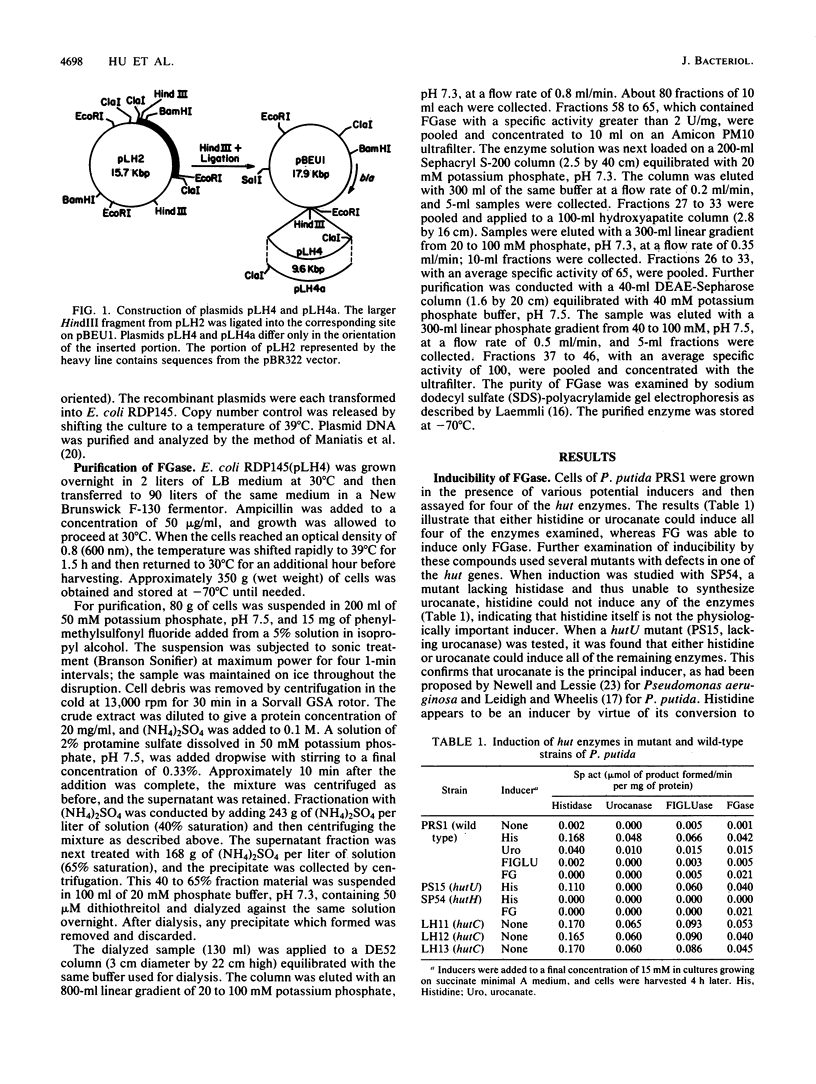
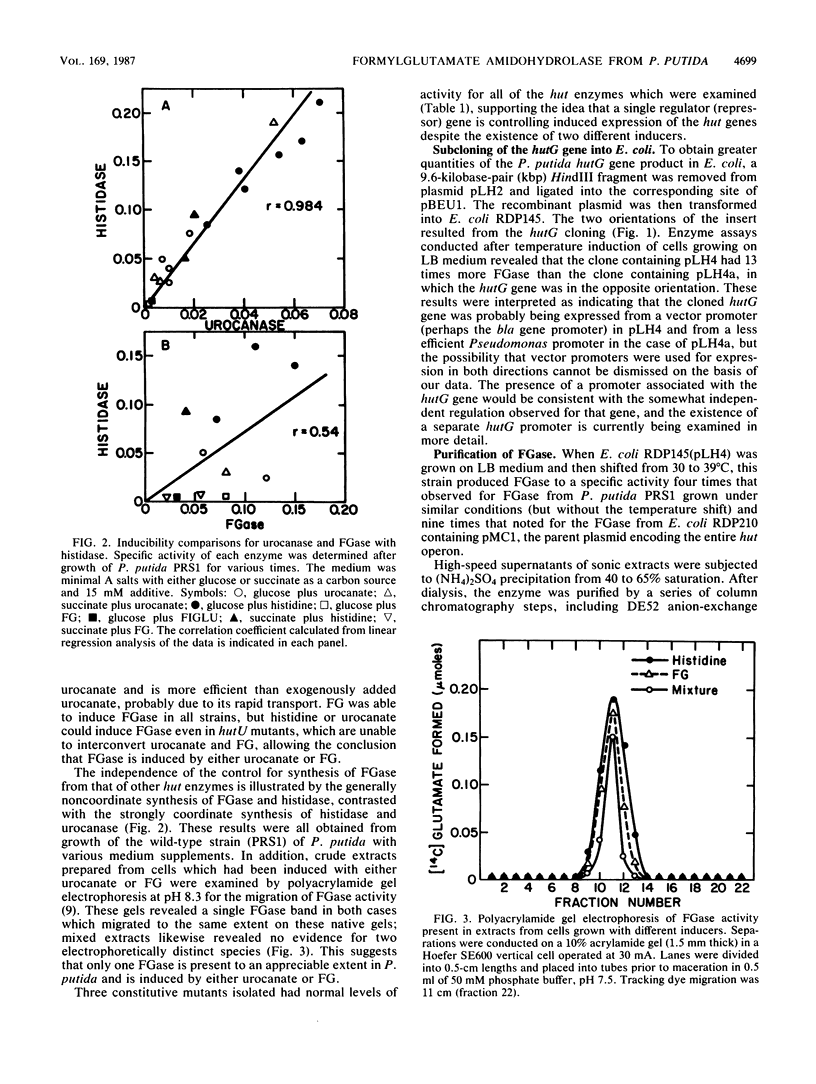
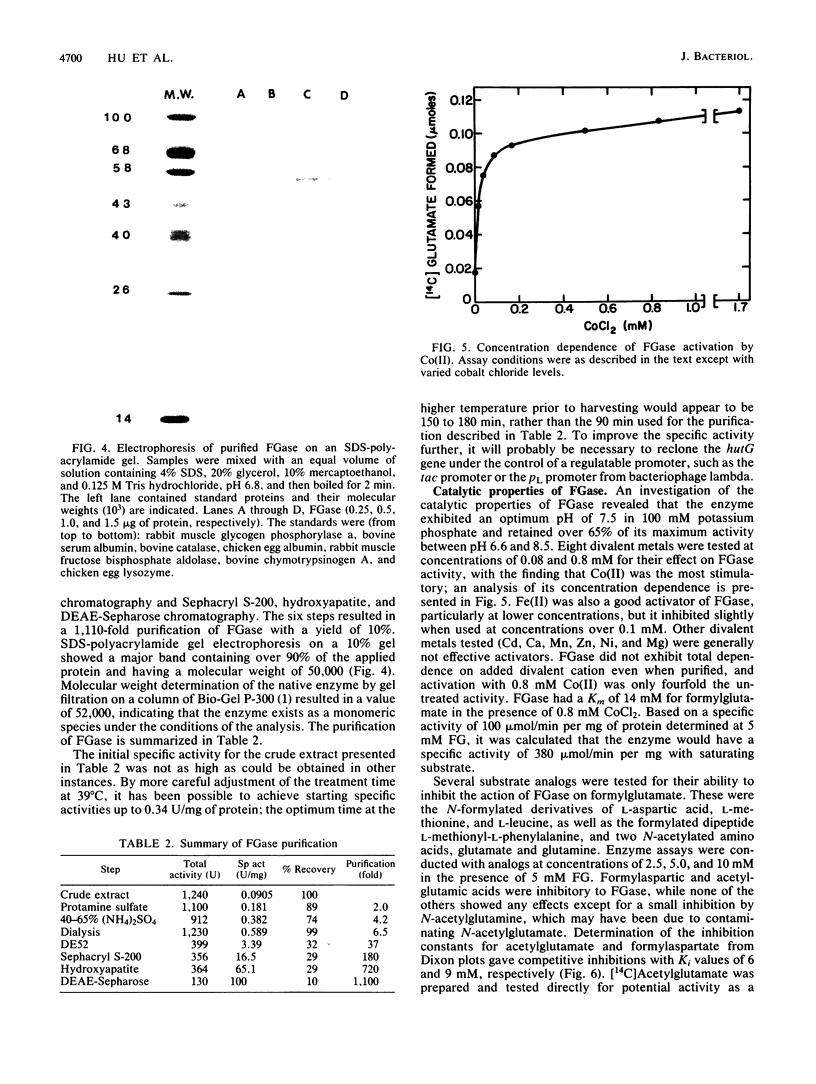


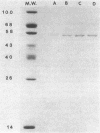
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews P. Estimation of the molecular weights of proteins by Sephadex gel-filtration. Biochem J. 1964 May;91(2):222–233. doi: 10.1042/bj0910222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOREK B. A., WAELSCH H. The enzymatic degradation of histidine. J Biol Chem. 1953 Nov;205(1):459–474. [PubMed] [Google Scholar]
- Boylan S. A., Bender R. A. Genetic and physical maps of Klebsiella aerogenes genes for histidine utilization (hut). Mol Gen Genet. 1984;193(1):99–103. doi: 10.1007/BF00327421. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Buzby J. S., Porter R. D., Stevens S. E., Jr Plasmid transformation in Agmenellum quadruplicatum PR-6: construction of biphasic plasmids and characterization of their transformation properties. J Bacteriol. 1983 Jun;154(3):1446–1450. doi: 10.1128/jb.154.3.1446-1450.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasin L. A., Magasanik B. Induction and repression of the histidine-degrading enzymes of Bacillus subtilis. J Biol Chem. 1968 Oct 10;243(19):5165–5178. [PubMed] [Google Scholar]
- Consevage M. W., Porter R. D., Phillips A. T. Cloning and expression in Escherichia coli of histidine utilization genes from Pseudomonas putida. J Bacteriol. 1985 Apr;162(1):138–146. doi: 10.1128/jb.162.1.138-146.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coote J. G., Hassall H. The control of the enzymes degrading histidine and related imidazolyl derivates in Pseudomonas testosteroni. Biochem J. 1973 Mar;132(3):423–433. doi: 10.1042/bj1320423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVIS B. J. DISC ELECTROPHORESIS. II. METHOD AND APPLICATION TO HUMAN SERUM PROTEINS. Ann N Y Acad Sci. 1964 Dec 28;121:404–427. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- Früh H., Leisinger T. Properties and localization of N-acetylglutamate deacetylase from Pseudomonas aeruginosa. J Gen Microbiol. 1981 Jul;125(1):1–10. doi: 10.1099/00221287-125-1-1. [DOI] [PubMed] [Google Scholar]
- George D. J., Phillips A. T. Identification of alpha-ketobutyrate as the prosthetic group of urocanase from Pseudomonas putida. J Biol Chem. 1970 Feb 10;245(3):528–537. [PubMed] [Google Scholar]
- Kaminskas E., Kimhi Y., Magasanik B. Urocanase and N-formimino-L-glutamate formiminohydrolase of Bacillus subtilis, two enzymes of the histidine degradation pathway. J Biol Chem. 1970 Jul 25;245(14):3536–3544. [PubMed] [Google Scholar]
- Kendrick K. E., Wheelis M. L. Histidine dissimilation in Streptomyces coelicolor. J Gen Microbiol. 1982 Sep;128(9):2029–2040. doi: 10.1099/00221287-128-9-2029. [DOI] [PubMed] [Google Scholar]
- Kimhi Y., Magasanik B. Genetic basis of histidine degradation in Bacillus subtilis. J Biol Chem. 1970 Jul 25;245(14):3545–3548. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leidigh B. J., Wheelis M. L. Genetic control of the histidine dissimilatory pathway in Pseudomonas putida. Mol Gen Genet. 1973 Feb 2;120(3):201–210. doi: 10.1007/BF00267152. [DOI] [PubMed] [Google Scholar]
- Lessie T. G., Neidhardt F. C. Formation and operation of the histidine-degrading pathway in Pseudomonas aeruginosa. J Bacteriol. 1967 Jun;93(6):1800–1810. doi: 10.1128/jb.93.6.1800-1810.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund P., Magasanik B. N-formimino-L-glutamate formiminohydrolase of Aerobacter aerogenes. J Biol Chem. 1965 Nov;240(11):4316–4319. [PubMed] [Google Scholar]
- Meiss H. K., Brill W. J., Magasanik B. Genetic control of histidine degradation in Salmonella typhimurium, strain LT-2. J Biol Chem. 1969 Oct 10;244(19):5382–5391. [PubMed] [Google Scholar]
- Newell C. P., Lessie T. G. Induction of histidine-degrading enzymes in Pseudomonas aeruginosa. J Bacteriol. 1970 Oct;104(1):596–598. doi: 10.1128/jb.104.1.596-598.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. R., Magasanik B. The two operons of the histidine utilization system in Salmonella typhimurium. J Biol Chem. 1971 May 25;246(10):3330–3341. [PubMed] [Google Scholar]
- TABOR H., MEHLER A. H. Isolation of N-formyl-L-glutamic acid as an intermediate in the enzymatic degradation of L-histidine. J Biol Chem. 1954 Oct;210(2):559–568. [PubMed] [Google Scholar]
- Tyler B. M., Goldberg R. B. Transduction of chromosomal genes between enteric bacteria by bacteriophage P1. J Bacteriol. 1976 Mar;125(3):1105–1111. doi: 10.1128/jb.125.3.1105-1111.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlin B. E., Clark A. J. Overproduction of the Escherichia coli recA protein without stimulation of its proteolytic activity. J Bacteriol. 1981 Oct;148(1):386–390. doi: 10.1128/jb.148.1.386-390.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheelis M. L., Stanier R. Y. The genetic control of dissimilatory pathways in Pseudomonas putida. Genetics. 1970 Oct;66(2):245–266. doi: 10.1093/genetics/66.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh W. K., Ornston L. N. Origins of metabolic diversity: substitution of homologous sequences into genes for enzymes with different catalytic activities. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5365–5369. doi: 10.1073/pnas.77.9.5365. [DOI] [PMC free article] [PubMed] [Google Scholar]