Abstract
A 97-kD component of nuclear pore-targeting complex (the β-subunit of nuclear pore–targeting complex [PTAC]/importin/karyopherin) mediates the import of nuclear localization signal (NLS)-containing proteins by anchoring the NLS receptor protein (the α-subunit of PTAC/importin/karyopherin) to the nuclear pore complex (NPC). The import requires a small GTPase Ran, which interacts directly with the β-subunit. The present study describes an examination of the behavior of the β-subunit in living cells and in digitonin-permeabilized cells. In living cells, cytoplasmically injected β-subunit rapidly migrates into the nucleus. The use of deletion mutants reveals that nuclear migration of the β-subunit requires neither Ran- nor α-subunit–binding but only the NPC-binding domain of this molecule, which is also involved in NLS-mediated import. Furthermore, unlike NLS-mediated import, a dominant-negative Ran, defective in GTP-hydrolysis, did not inhibit nuclear migration of the β-subunit. In the digitonin-permeabilized cell-free import assay, the β-subunit transits rapidly through the NPC into the nucleus in a saturating manner in the absence of exogenous addition of soluble factors. These results show that the β-subunit undergoes translocation at the NPC in a Ran-unassisted manner when it does not carry α-subunit/NLS substrate. Therefore, a requirement for Ran arises only when the β-subunit undergoes a translocation reaction together with the α-subunit/NLS substrate. The results provide an insight to the yet unsolved question regarding the mechanism by which proteins are directionally transported through the NPC, and the role of Ran in this process.
Nuclear pore complexes (NPCs)1 mediate the bidirectional nucleocytoplasmic exchange of molecules via several different mechanisms. Ions, small metabolites, and proteins <20–40 kD in size can passively diffuse through 90-Å aqueous channels that span the NPC. Most macromolecules, which are too large to passively diffuse through the aqueous channels of the NPC, are thought to be transported through gated-channels of the NPC by active mechanisms (for reviews see Fabre and Hurt, 1994; Rout and Wente, 1994; Davis, 1995). These include the nuclear import of nuclear localization signal (NLS)-containing karyophiles and small nuclear RNPs (for reviews see Dingwall and Laskey, 1991; Garcia-Bustos et al., 1991; Goldfarb and Michaud, 1991; Görlich and Mattaj, 1996; Panté and Aebi, 1996), nuclear export of nuclear export signal–containing proteins (for reviews see Gerace, 1995; Görlich and Mattaj, 1996; Panté and Aebi, 1996; Yoneda, 1996) and RNAs (for reviews see Elliott et al., 1994; Izaurralde and Mattaj, 1995), and shuttling proteins, such as heat shock-related proteins (Mandell and Feldherr, 1990) and heterogeneous nuclear RNP A1 (Michael et al., 1995; for reviews see Laskey and Dingwall, 1993; Piñol-Roma and Dreyfuss, 1993). Based on sensitivity to chilling and WGA, the transport of calmodulin has been shown to be facilitated and to occur in an energy-independent manner (Pruschy et al., 1994).
The NLS-mediated import process involves a series of sequential steps, which are mediated by several soluble/ cytoplasmic factors. The import is initiated by NLS-dependent complex formation, termed the nuclear pore-targeting complex (PTAC), in the cytoplasm (Görlich et al., 1995a ; Imamoto et al., 1995c ). PTAC is a stable protein complex composed of a karyophilic protein and two cytosolic components, which we originally named PTAC58 and PTAC97, based on their molecular masses (Imamoto et al., 1995a ,b). These components have also been referred to as importin α and β (Görlich et al., 1994, 1995a ,b), karyopherin α and β (Moroianu et al., 1995a ; Radu et al., 1995a ), or hSRP1 α/ NLS receptor (Adam and Gerace, 1991; Weis et al., 1995) as well as nuclear transport factor p97 (Adam and Adam, 1994; Chi et al., 1995). In this report, we will refer to the 58-kD component of PTAC as the α-subunit, and the 97-kD component as the β-subunit. The α-subunit directly binds to the NLS of karyophile (Adam and Gerace, 1991; Imamoto et al., 1995b ; Moroianu et al., 1995a ; Weis et al., 1995) and the β-subunit to the NH2-terminal portion of the α-subunit (Görlich et al., 1996a ; Weis et al., 1996). The β-subunit provides NPC-binding sites for PTAC, since this protein directly interacts with several FXFG or GLFG repeat-containing nucleoporins (Iovine et al., 1995; Kraemer et al., 1995; Radu et al., 1995b ). NLS-containing karyophiles target the cytoplasmic face of the NPC with α- and β-subunits as a single entity in the energy-independent stages of NLS-mediated nuclear import.
Subsequent energy-dependent steps of nuclear import, in which karyophiles translocate through gated channels of the NPC to the nuclear interior, have been shown to be mediated by small GTPase Ran, along with a dimeric protein called p10/NTF2 (Melchior et al., 1993; Moore and Blobel, 1993,1994; Paschal and Gerace, 1995). Ran, in its GTP form, has been shown to bind directly to the β-subunit (Floer and Blobel, 1996; Paschal et al., 1996). Using an in vitro transport assay, it has been shown that the α- and β-subunits dissociate upon NPC translocation and that the α-subunit accumulates in the nucleus along with the NLS substrate, while the β-subunit remains bound to the nucleoplasmic face of the NPC (Görlich et al., 1995b ; Moroianu et al., 1995b ). Dissociation of the α-subunit from the β-subunit is induced by the binding of GTP-Ran to the β-subunit (Rexach and Blobel, 1995; Görlich et al., 1996b ). Moreover, GTP-Ran causes the dissociation of β-subunit from FXFG repeat-containing nucleoporins on binding to the β-subunit (Rexach and Blobel, 1995).
The issue of how proteins actually directionally move through the NPC is not well understood. Several laboratories have carried out experiments directed at explaining the detailed translocation mechanism, and, as a result of these, different translocation models have been proposed. One model proposes that a repeated docking and undocking of PTAC, promoted by GTP-Ran, generates the vectorial movement through the NPC (Rexach and Blobel, 1995), and another proposes that a multi-complex formation between α-subunit, β-subunit, GDP-Ran, and p10 at the central gate of NPC is required for vectorial movement (for reviews see Koepp and Silver, 1996; Nigg, 1997). More recently, Chi et al. (1996) proposed that multi-complex formation between α-subunit, β-subunit, GDP-Ran, and RanBP1 at the central gate of the NPC is required for vectorial movement. Görlich et al. (1996b) proposed that the asymmetric distribution of GDP- and GTP-Ran in the cytoplasm and nucleoplasm contributes to the vectorial movement. In all cases, Ran has been proposed to play a key role in translocating the NLS-containing karyophile through the NPC with directionality, although its exact action remains to be elucidated.
Meanwhile, distantly related β-subunit homologues have been identified from mammalian and yeast sources that mediate different transport pathways. One of these, termed transportin or karyopherin β2, has been shown to mediate the import of heterogeneous nuclear RNP A1, by binding directly to a novel NLS of this protein named the M9 sequence (Pollard et al., 1996; Bonifaci et al., 1997). Karyopherin β3 was shown to bind to ribosomal proteins, and it has been suggested that it mediates the nuclear transport of ribosomal proteins (Yaseen and Blobel, 1997). In yeast, three β-subunit homologues have been identified from the yeast genome database. One of these, termed Kap104, was shown to bind directly to yeast mRNA binding proteins, termed Nab2p and Nab4p, and has been proposed to mediate the nuclear transport of these proteins (Aitchison et al., 1996). Neither of these β-subunit homologues associates with the α-subunit. All of the β-subunit homologues were shown to interact directly with NPC components, but not all of them bind directly to Ran (Yaseen and Blobel, 1997). Therefore, the issue of which of the interactions among transport factors is sufficient to exert the movement of molecules through NPC remains unclear.
In the present study, we examined the behavior of the β-subunit and found that it can undergo NPC translocation in both a Ran-assisted and -unassisted manner. The β-subunit, which is distributed throughout the cytoplasm and nucleus, rapidly migrates into the nucleus when injected into the cytoplasm of living cells. The use of deletion mutants, which lack the ability to bind α-subunit, GTP-Ran, or both, reveals that this protein migrates into the nucleus without binding to either α-subunit or GTP-Ran but requires its own NPC-binding domain, which possesses necessary and sufficient information to directionally translocate NPC. Moreover, dominant-negative Ran, defective in GTP hydrolysis, failed to inhibit nuclear migration of β-subunit in living cells. In the digitonin-permeabilized cell-free transport assay, the β-subunit moves rapidly through the NPC into the nucleus in the absence of exogenous addition of soluble factors. The data collected herein show that, in marked contrast to NLS-mediated import, which occurs when the β-subunit carries α-subunit and bound NLS substrate, Ran is not required when the β-subunit undergoes translocation through the NPC without carrying α-subunit/NLS substrate. Sensitivity to WGA and saturability of import show that the β-subunit migrates into the nucleus through gated channels of the NPC via specific molecular interactions of the NPC-binding domain of the β-subunit with NPC components.
Materials and Methods
Cell Culture
Madin-Darby bovine kidney (MDBK) cells and human embryonic lung (HEL) cells were incubated in DME containing 10% bovine serum. MDBK cells were plated on 8-well multi-test slides 12–24 h before use for cell-free import assays.
Microinjection
HEL cells were plated on marked coverslips, and microinjection experiments were performed as described previously (Yoneda et al., 1987). After injection and incubation, cells were fixed with 3.7% formaldehyde in PBS (137 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4, pH 7.2). The injected fluorescently labeled proteins were detected by fluorescent microscopy (Axiophot; Carl Zeiss, Inc., Thornwood, NY) or confocal laser scanning microscopy (Carl Zeiss, Inc.). Confocal images were collected using ×63 objective, and the final figures were produced using Adobe Photoshop (Adobe Systems, Inc., Mountain View, CA).
Expression and Purification of Recombinant Proteins
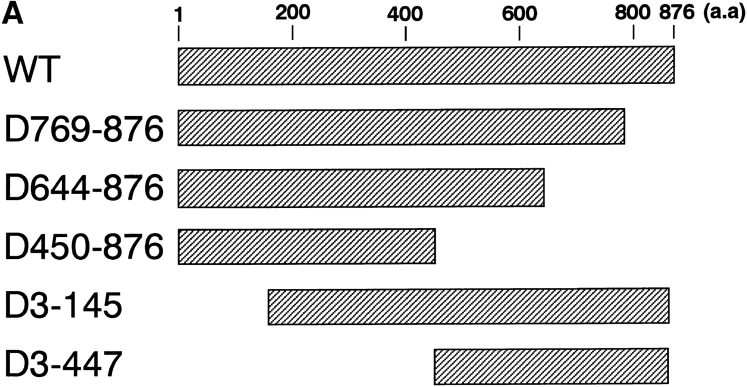
An epitope-tagged β-subunit was constructed in a glutathione-S-transferase (GST) gene fusion vector pGEX-2T (Pharmacia Biotech, Inc., Piscataway, NJ) as described previously (Imamoto et al., 1995a ). This vector is referred to as the pGEX HA-PTAC97. The expression vectors of the D769-876 and D644-876 mutants were constructed from the pGEX HA-PTAC97 by cutting out the HindIII–KpnI and EcoRI–KpnI fragments, respectively, followed by blunting and ligation. The expression vectors of the D3-145 and D3-447 mutants were constructed from pGEX HA-PTAC97 by cutting out the SacI–NdeI and SacI–AccI fragments, respectively, followed by blunting and ligation with the NotI linker (AGCGGCCGCT). To construct the expression vector of the D450-876 mutant, an NdeI–AccI (KpnI) fragment, which is 145–449 amino acids of β-subunit, was amplified from the pGEX HA-PTAC97 by PCR using the synthetic oligonucleotides (5′-TTGGGGAGCTCGAGCATATGAAAGAGTCCACATTGG-3′ and 5′-GAAAAGGTACCAGGTAGACATCGTTGATGGCGGCTTC-3′) which incorporate a KpnI site after an AccI site. After digesting with NdeI and KpnI, this PCR product was inserted into NdeI and KpnI sites of pGEX HA-PTAC97. These resultant plasmids were sequenced to confirm the fidelity of the region of amplification by PCR and in-frame ligation of the fused region.
The green fluorescent protein (GFP) chimeras of recombinant β-subunit or deletion mutants were generated as follows. The fragment encoding the entire amino acids of GFP and amino acids 1–3 of β-subunit was amplified from pS65T-C1 (#6089-1; Clonetech Laboratories, Palo Alto, CA) by PCR using the synthetic oligonucleotides (5′-CCGTAAGATCTATGAGTAAAGGAGAAGAACTTTTC-3′ and 5′-CCGTTGAGCTCCATGGATCCTTTGTATAGTTCATCCATGCC-3′) which incorporate a BglII site in frame followed by a start codon of GFP. The amplified PCR product was digested with BglII and SacI, followed by incorporating into the BamHI and SacI sites of the pGEX HA-PTAC97 or the expression vector of GST-fused deletion mutants (D450-876 and D3-447) of β-subunit described above.
The resultant expression vectors were transformed into Escherichia coli strain BL21(DE3). The E. coli cells were grown in LB medium containing 50 μg/ml ampicillin at 37°C to a density of 1.2 (OD550). Expression was induced by addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and incubation for 14 h at 20°C, except that the D450-876 chimeras were induced by addition of 0.1 mM IPTG. Lysis of bacteria and purification of the fusion protein with glutathione-Sepharose were performed as described previously (Imamoto et al., 1995a ). The GST portion of chimeras was cleaved off by a 2 h incubation at room temperature with 1 NIH unit of thrombin (Sigma Chemical Co., St. Louis, MO) per 100 μg of chimeras. GST and thrombin were separated from recombinant proteins on a MonoQ column (Pharmacia Biotech, Inc.) at flow rate of 0.5 ml/min with a linear gradient from 0.05 to 1.0 M NaCl in 20 mM Hepes, pH 7.3, 2 mM DTT, 1 μg/ml each of aprotinin, leupeptin, and pepstatin. The recombinant proteins were dialyzed against 20 mM Hepes, pH 7.3, 100 mM potassium acetate, 2 mM DTT, and 1 μg/ml each of aprotinin, leupeptin, and pepstatin.
E. coli strain BL21 (DE3) expressing human Ran was kindly provided by Dr. T. Nishimoto (Graduate School of Medical Science, Kyushu University, Fukuoka, Japan). The recombinant wild-type Ran and G19V Ran were expressed, purified, and charged with GTP or GDP based on methods described previously (Dasso et al., 1994; Sekimoto et al., 1996). The GST– α-subunit was expressed and purified as described previously (Imamoto et al., 1995b ). Aliquots of each recombinant protein were frozen in liquid nitrogen and stored at −80°C.
Antibodies
The GST–β-subunit, purified on a glutathione-Sepharose column, was used to immunize two rabbits. Immunizations were performed as described previously (Imamoto et al., 1995b ). Anti–β-subunit antibodies were performed independently from anti-serum of each rabbit, by first depleting the antiserum of anti-GST antibodies by passage over GST affinity columns and then on recombinant β-subunit–conjugated Sepharose. Antibodies bound to β-subunit–Sepharose were eluted with 0.1 M glycine-HCl (pH 2.5) and neutralized. After dialysis against phosphate buffer (pH 7.2) containing 0.3 M NaCl, the antibodies were further incubated with GST-Sepharose to absorb residual traces of anti-GST antibodies. Preimmune IgG fraction was obtained by two cycle precipitation with 40% saturated ammonium sulfate.
Indirect Immunofluorescence
HEL cells and MDBK cells, plated on coverslips, were washed twice in PBS and fixed with 3.7% formaldehyde in PBS (room temperature for 20 min). After permeabilization with 0.5% Triton X-100 in PBS (room temperature for 5 min), cells were incubated with 10 μg/ml of affinity-purified anti–β-subunit antibodies for 1.5 h at room temperature. Rabbit antibodies were detected with FITC-conjugated goat antibodies to rabbit IgG (Tago Inc., Burlingame, CA). The samples were examined using an Axiophot microscope (Carl Zeiss, Inc.). For the confocal experiments, cells were treated identically. Images were collected using ×63 objective on the Zeiss confocal laser scanning microscope (Carl Zeiss, Inc.), and the final figure was produced using Adobe Photoshop (Adobe Systems, Inc., Mountain View, CA).
Preparation of NLS-containing Transport Substrates
Allophycocyanin (APC; Calbiochem-Novabiochem Corp., La Jolla, CA) and biotinylated bovine serum albumin (bBSA) were chemically conjugated to synthetic peptide containing the amino acid sequence of SV40 large T-antigen NLS (CYGGPKKKRKVEDP), as described previously (Imamoto et al., 1995a ).
Solution Binding Assay
The ability of the deletion mutant proteins to form a complex with the α-subunit and biotin-labeled SV40 T-antigen NLS conjugate (T-bBSA; Imamoto et al., 1995a ) was examined in the solution binding assay. The amounts of mutant proteins incorporated into the complex correspond to those that directly bind to the α-subunit, since β-subunit does not directly associate with the NLS substrate (Imamoto et al., 1995a ). Binding assays were performed in transport buffer (TB: 20 mM Hepes, pH 7.3, 110 mM potassium acetate, 2 mM magnesium acetate, 5 mM sodium acetate, 1 mM glycoletherdiaminetetraacetic acid [EGTA], 2 mM DTT, and 1 μg/ml each of aprotinin, leupeptin, and pepstatin) in a final volume of 100 μl. Recombinant GST chimeras of β-subunit or deletion mutants were incubated with α-subunit and T-bBSA (0.25 mg/ml) for 1 h on ice. The solution was then incubated for 1 h at 4°C with 30 μl of immobilized avidin (Pierce, Rockford, IL), which was washed with TB and resuspended in a 50% slurry. The immobilized avidin was collected by centrifugation and washed five times by resuspension in 0.5 ml of TB. The bound proteins were eluted from immobilized avidin by a 30-min incubation at 37°C with 30 μl of the elution buffer (0.2% SDS, TE, pH 8.0). The eluted proteins were resolved by SDS-PAGE. The intensity of protein bands, stained with coomassie brilliant blue, was quantified by densitometric scanning (CS-930; Shimazu, Kyoto, Japan). The molar ratio of β-subunit or deletion mutant proteins bound to α-subunit was estimated by dividing the intensity of the protein bands by the molecular mass of each protein.
Ran Overlay Assay
Direct binding of β-subunit to GTP-Ran was performed using an overlay assay, based on methodology described by Lounsbury et al. (1994). Recombinant wild-type β-subunit and its deletions were separated by 12.5% SDS-PAGE and transferred to nitrocellulose and then incubated, first in buffer containing 20 mM Hepes, pH 7.3, 100 mM sodium acetate, 5 mM magnesium acetate, 0.25% Tween 20, 0.5% BSA, and 5 mM DTT, and then preincubated for 30 min at room temperature in binding buffer (20 mM Hepes, pH 7.3, 100 mM potassium acetate, 5 mM magnesium acetate, 0.05% tween 20, 0.5% BSA, and 5 mM DTT) in the presence of 100 μM GTP. Blots were rinsed with binding buffer and then overlaid with 0.126 pmol/μl [γ-32P]GTP-Ran (sp act, 15,400 cpm/pmol) in binding buffer for 30 min at room temperature. Binding ability to Ran was also examined in the solution binding assay using GST chimeras of wild-type β-subunit and its deletions, which gave the same results that were obtained in the overlay assay.
Cell-free Import Assay
Digitonin-permeabilized MDBK cells were prepared as described previously (Adam et al., 1990; Imamoto et al., 1995a ). Digitonin-permeabilized cells were incubated with TB containing testing proteins, in 10 μl, under each condition indicated in the respective figure legends. After incubation, cells were fixed with 3.7% formaldehyde in TB. T-APC and GFP fusion proteins were detected by Axiophot microscopy (Carl Zeiss, Inc.).
Results
β-Subunit Is Localized Both in the Cytoplasm and within the Nucleus
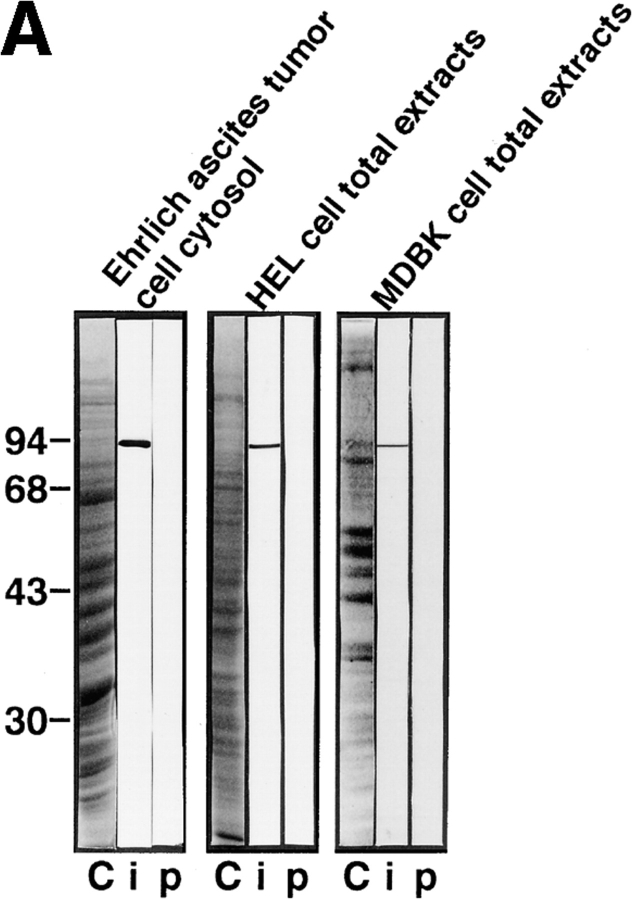
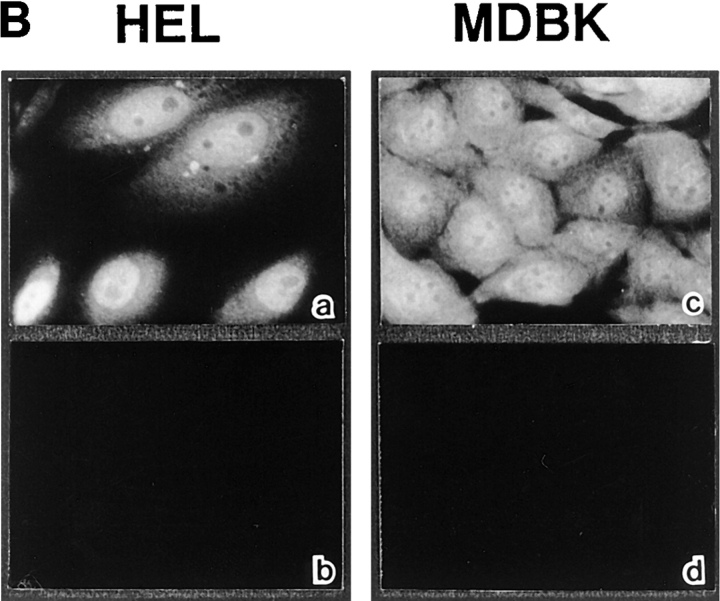
We initially examined the intracellular localization of a 97-kD component of the PTAC (β-subunit) by using affinity-purified antibodies raised against recombinant β-subunit. The affinity-purified anti–β-subunit antibodies recognized a single protein band that corresponds to endogenous β-subunit from cytosol prepared from Ehrlich ascites tumor cells, as well as total extracts prepared from human embryonic lung (HEL) cells, and MDBK cells (Fig. 1 A). Staining with these antibodies showed that the β-subunit is located not only in the cytoplasm but also within the nucleus in these tissue-cultured mammalian cells (Fig. 1 B, a and c). Intranuclear staining was further confirmed by confocal microscopy (Fig. 2, a and b). These staining patterns were abolished on incubation of the antibodies with recombinant β-subunit (Fig. 1 B, b and d), confirming that they represent a subcellular localization of endogenous β-subunit.
Figure 1.
Subcellular localization of β-subunit in tissue-cultured mammalian cells. (A) SDS-PAGE profiles of proteins stained by coomassie brilliant blue staining (indicated as C) of Ehrlich ascites tumor cells cytosol, or total extracts of HEL cells, and MDBK cells, and Western blots of the corresponding proteins incubated with affinity-purified anti–β-subunit antibodies (indicated as i) or preimmune IgG (indicated as p). (B) HEL cells (a and b) and MDBK cells (c and d) were stained with affinity-purified anti– β-subunit antibodies preincubated with recombinant GST (a and c) or GST–β-subunit (b and d) trapped in glutathione-Sepharose.
Figure 2.
Confocal microscopy reveals that β-subunit is localized inside the nucleus. HEL cells (a) and MDBK cells (b) stained with affinity-purified anti–β-subunit antibodies, as in Fig. 1, or subcellular localization of GFP–wild-type β-subunit that was microinjected into the cytoplasm of HEL cells (c) as in Fig. 4, was examined with a confocal laser scanning microscope (Carl Zeiss, Inc.). 0.5-μm sections, corresponding to the equator of the nucleus, are shown.
β-Subunit Rapidly Migrates into the Nucleus in Living Cells
We examined whether the β-subunit migrates into the nucleus by using tissue-cultured cell microinjection. To visualize the injected recombinant β-subunit, we modified this protein in three different ways: tagging with influenza virus hemagglutinin (HA) epitope, tagging with GFP to the NH2 terminus of β-subunit, or labeling with FITC. None of these modifications affected the activity of the β-subunit in terms of supporting nuclear import in the digitonin-permeabilized cell-free transport assay (data not shown). We found that all these modified recombinant β-subunits rapidly migrated into the nucleus within 30 min after injection into the cytoplasm. Intranuclear localization of cytoplasmically injected GFP–β-subunit, observed by confocal microscopy, is shown in Fig. 2 c.
In the digitonin-permeabilized cell-free transport assay, it was shown that the β-subunit does not accumulate in the nucleus, while α-subunit does so with NLS substrate during NLS-mediated nuclear import. In the following studies, we attempted to analyze whether nuclear import of the β-subunit is related to NLS-mediated nuclear import.
Nuclear Migration of β-Subunit Occurs without Binding to α-Subunit and Ran in Living Cells
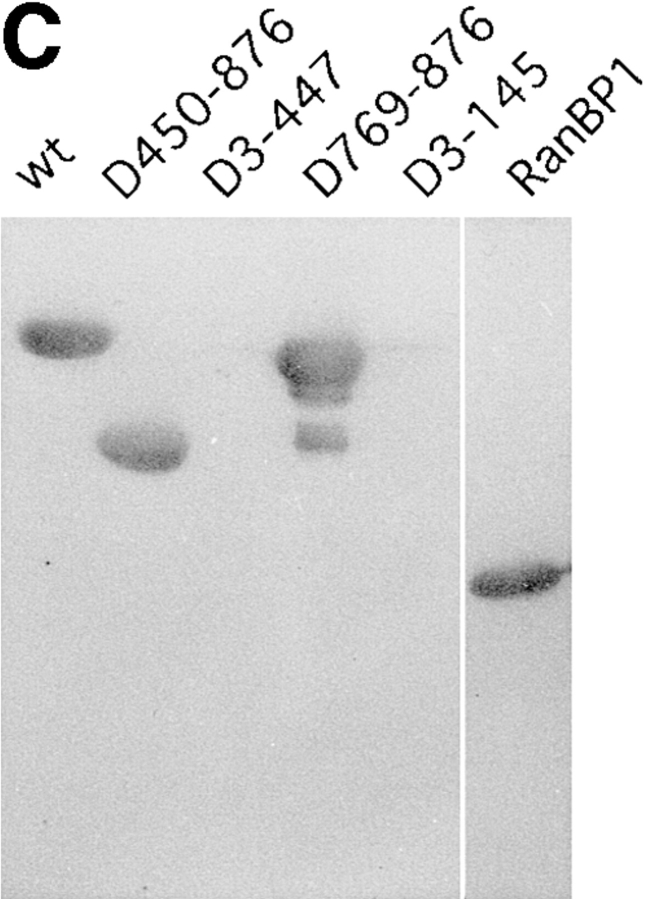
To determine the functional domain of the β-subunit, which is required for its nuclear locating ability, deletion mutants were generated, permitting an initial mapping of the α-subunit-, Ran-, and NPC-binding domains of the β-subunit. Deletion mutants that were generated and used in this study are shown in Fig. 3 A. In the solution binding assay, COOH-terminal deletions, such as D769-876 and D644-876, showed a decreased binding activity for the α-subunit, and D450-876 completely lost binding affinity (Fig. 3 B). In contrast, NH2-terminal deletions, such as D3-145 and D3-447, showed binding activity to the α-subunit that was equivalent to the wild-type β-subunit. These results show that the α-subunit binding domain is located in the COOH-terminal portion of the molecule.
Figure 3.
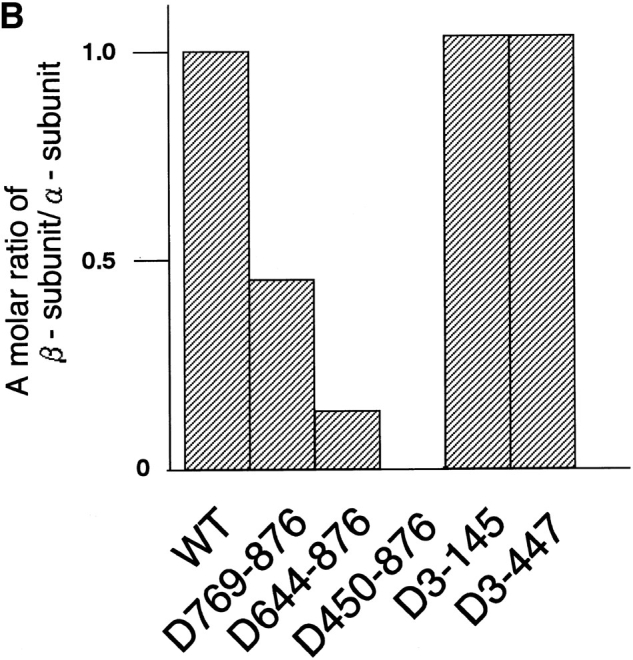
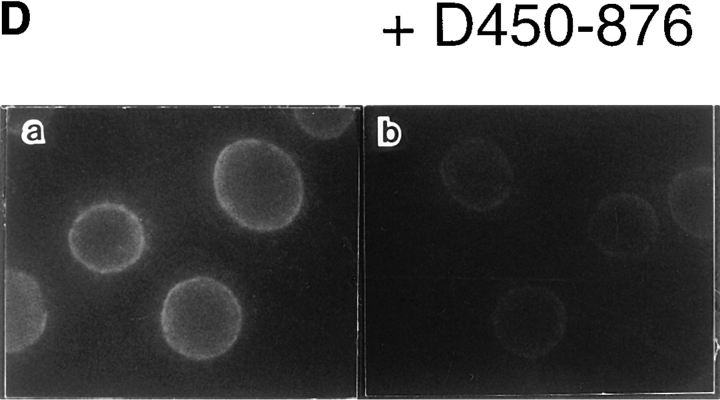
Mapping of the α-subunit-, Ran-, and NPC-binding domains of β-subunit. (A) The constructions of the deletion mutants of β-subunit. (B) α-Subunit–binding activity of deletion mutants of β-subunit. GST–β-subunit or its deletion mutants were incubated with 30 pmol GST–α-subunit and T-bBSA (0.25 mg/ ml) for 30 min at 37°C and then trapped in immobilized avidin. The bound proteins were resolved by SDS-PAGE and quantified by densitometric scanning, and molar ratios of β-subunit or its deletion mutants bound to α-subunit were determined. (C) Ran-binding activity of deletion mutants of β-subunit was determined by overlay assay. 100 pmol of wild-type β-subunit and its deletions, or RanBP1, were subjected to SDS-PAGE, transferred to nitrocellulose sheet, and incubated with [γ-32P]GTP-Ran as described under experimental procedures. [γ-32P]GTP-Ran bound to the recombinant proteins were detected by autoradiography. (D) NH2-terminal portion of β-subunit competitively inhibits NPC docking of PTAC. Digitonin-permeabilized cells were incubated with 10 μl testing solution containing 12 pmol GST–β-subunit, 12 pmol GST–α-subunit, and T-APC (0.1 mg/ml) in the presence (b) or absence (a) of 144 pmol D450-876 mutant for 20 min on ice. After incubation, the cells were fixed with 3.7% formaldehyde in TB, and T-APC was detected by Axiophot microscopy (Carl Zeiss Inc.).
It is noteworthy that, D450-876 specifically binds to GTP-Ran (Fig. 3 C). This portion of the β-subunit also contains a domain that interacts directly with the NPC, since D450-876 competitively inhibited the NPC docking of PTAC, when examined in the digitonin-permeabilized cells (Fig. 3 D). D3-447, which lacks the entire NH2-terminal portion of the β-subunit, neither binds to Ran (Fig. 3 C) nor NPC (see Figs. 4 and 7). A deletion mutant lacking the NH2-terminal 143 amino acids (D3-145) did not bind to Ran (Fig. 3 C). Since neither amino acids 1–300 and 145– 449 β-subunit fragments bound to GTP-Ran in overlay assay, two stretches of NH2- and COOH-terminal amino acids of D450-876 appear to be necessary for efficient Ran binding (data not shown). These mapping results are consistent with recently published reports by other groups (Chi et al., 1997; Kutay et al., 1997).
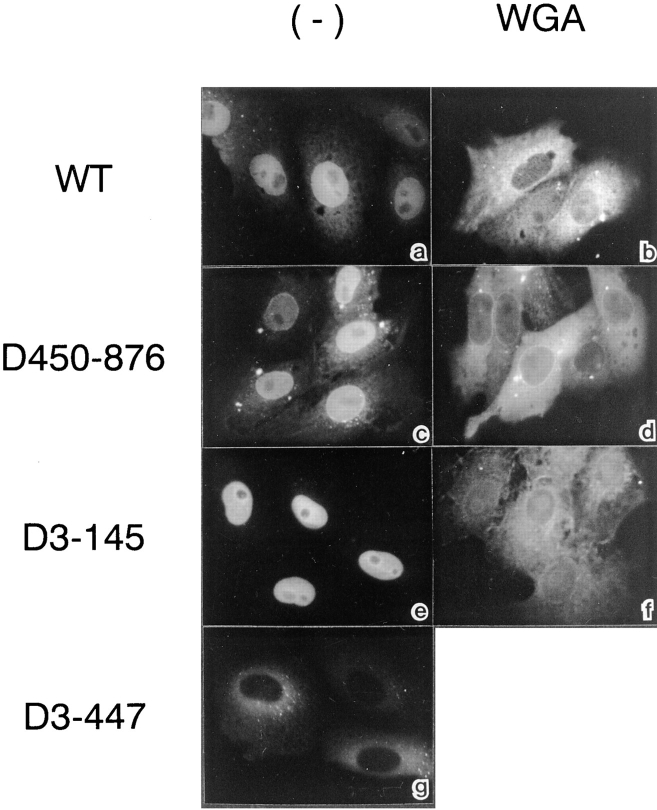
Figure 4.
The NPC-binding domain of β-subunit is necessary and sufficient for its nuclear migration. HEL cells injected with 1.5 mg/ml GFP–wild-type β-subunit (a and b), GFP-D450-876 (c and d), FITC-D3-145 (e and f), or GFP-D3-447 (g) were incubated for 30 min at 37°C. b, d, and f show cells co-injected with 1 mg/ml WGA. After incubation, cells were fixed with 3.7% formaldehyde in PBS, and the localization of proteins was examined by Axiophot microscopy (Carl Zeiss Inc.).
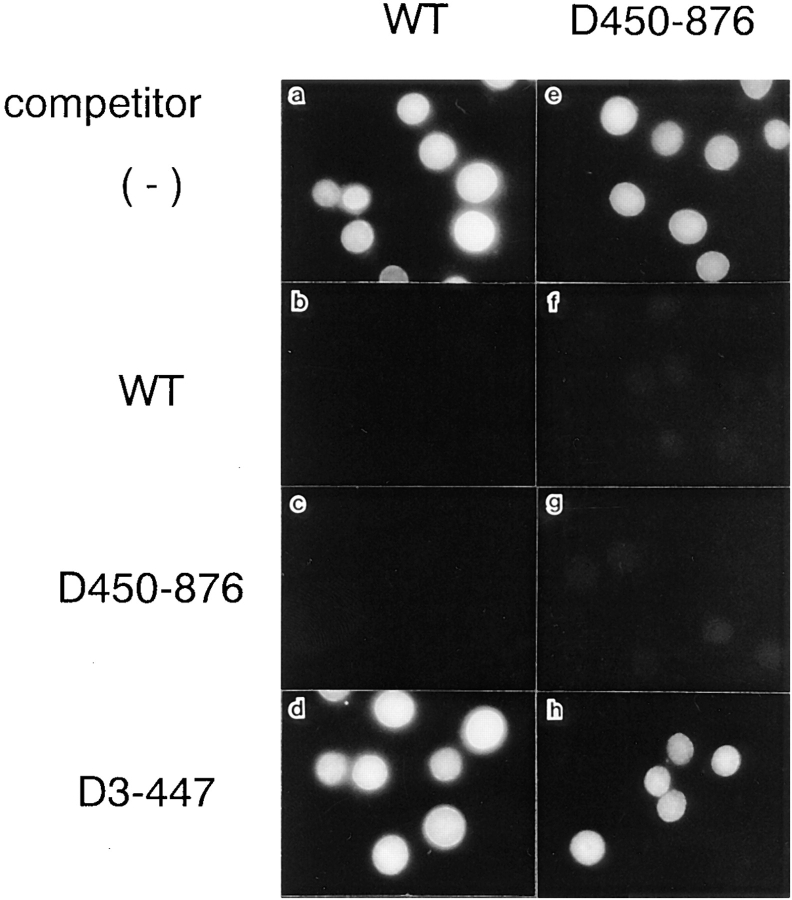
Figure 7.
Nuclear migration of β-subunit is saturable. Digitonin-permeabilized cells were incubated with 10 μl of testing solution containing 3 pmol GFP–β-subunit (a–d) or GFP-D450-876 (e–h) in the absence (a and e) or presence of either 24 pmol GST– β-subunit (b and f), GST-D450-876 (c and g), or GST-D3-447 (d and h) for 15 min on ice. Localization of GFP-chimeras was examined after fixation of the permeabilized cells as in Fig. 3 D.
Having established which of the β-subunit mutant proteins cannot bind to the α-subunit (D450-876), Ran (D3-145), or to both Ran and NPC (D3-447), we examined which of these proteins are capable of migrating into the nucleus in living cells. As shown in Fig. 4, cytoplasmically injected D450-876 and D3-145 rapidly migrated into the nucleus, while D3-447 did not. As with the wild-type β-subunit, nuclear migration of D450-876 and D3-145 was inhibited by WGA, indicating that all of these proteins migrate through the gated channels of the NPC. These results show that the nuclear migration of β-subunit does not require α-subunit and Ran binding, but only the NPC-binding domain, which is involved in PTAC docking.
It has been shown that in the digitonin-permeabilized transport assay, both α-subunit and Ran binding to the β-subunit is essential for NLS-mediated import (Chi et al., 1996; Görlich et al., 1996b ; Kutay et al., 1997). In contrast, the above microinjection results show that the β-subunit is capable of migrating into the nucleus of living cells in the absence of these interactions. These findings indicate that the β-subunit migrates into the nucleus alone, independent of NLS-mediated nuclear import. We refer to such behavior as the “unassisted” nuclear migration of the β-subunit.
β-Subunit Migrates into the Nucleus of Digitonin-permeabilized Cells in the Absence of Exogenous Addition of Soluble Factors
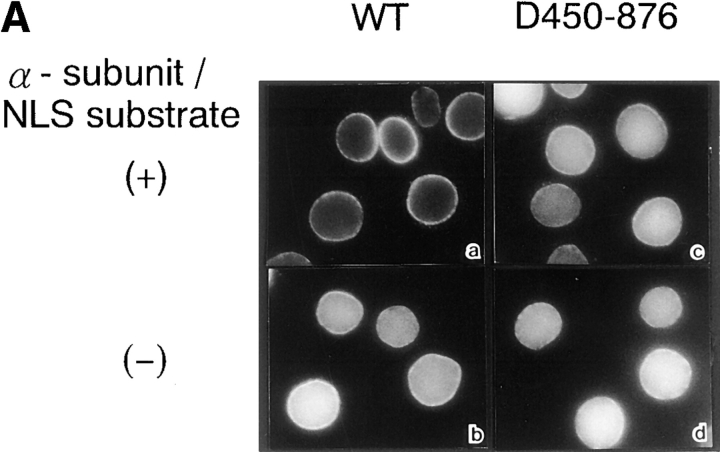
In further investigations, we examined the behavior of the β-subunit using the digitonin-permeabilized cell-free transport assay. In the presence of α-subunit/NLS substrate, the β-subunit accumulated at the nuclear rim, as reported previously (Imamoto et al., 1995a ; Görlich et al., 1995b ; Moroianu et al., 1995b ). However, wild-type β-subunit was found to efficiently accumulate in the nucleus in the absence of added soluble factors. Consistently, D450-876, lacking the α-subunit–binding domain, efficiently migrated into the nucleus, irrespective of the presence or absence of α-subunit/NLS substrate (Fig. 5 A).
Figure 5.
Unassisted nuclear migration of β-subunit occurs in the absence of exogenous addition of soluble factors in digitonin-permeabilized cell-free transport assay. (A) Digitonin-permeabilized MDBK cells were incubated with 10 μl of testing solution containing 3 pmol GFP–β-subunit (a and b) or GFP-D450-876 (c and d) in the presence (a and c) or absence (b and d) of 3 pmol GST– α-subunit and T-APC (0.1 mg/ml) for 20 min. (B) Digitonin-permeabilized MDBK cells were incubated with 10 μl testing solution containing 4 pmol of GFP–β-subunit (a–c), GFP-D450-876 (d–f), or FITC-D3-145 (g–i) in the absence (a, d, and g) or presence of either 20 pmol GTP-Ran (b, e, and h) or GDP-Ran (c, f, and i) for 15 min. After incubation, localization of β-subunit and its mutant proteins was examined after fixation of the permeabilized cells as in Fig. 3 D.
We next examined the effect of small GTPase Ran on the nuclear accumulation of β-subunit. As shown in Fig. 5 B, wild-type β-subunit and D450-876 significantly accumulated at the nuclear rim, but did not enter the nucleus in the presence of GTP-Ran. GDP-Ran had a much less inhibitory effect on the nuclear accumulation of both proteins. The effect of Ran on the nuclear migration of β-subunit is likely to be the result of its direct binding to the β-subunit, since the nuclear accumulation of D3-145, which lacks Ran-binding ability, was not affected by GTP-Ran.
These results show that the nuclear migration of β-subunit occurs in the absence of added soluble factors, when it is not bound to either α-subunit or GTP-Ran, and that the β-subunit itself possesses the necessary and sufficient information to translocate the NPC.
The Unassisted Nuclear Migration of β-Subunit Occurs without the Support of GTP-Hydrolysis of Ran
The above results indicate that the β-subunit, when it does not carry α-subunit with NLS substrate, is capable of migrating into the nucleus in the absence of Ran. To further verify that the movement of the β-subunit through the NPC is not supported by Ran, we examined the effect of GTP-G19V Ran, a point-mutated Ran, defective in GTP hydrolysis, on the unassisted nuclear migration of β-subunit. This dominant-negative Ran, defective in GTP hydrolysis, has been demonstrated to strongly inhibit the Ran-dependent import of known substrates in living cells (Dickmanns et al., 1996; Sekimoto et al., 1996). In contrast, in living cells, it was found that co-injection of GTP-G19V Ran did not affect the nuclear migration of either wild-type β-subunit or D3-145, while it strongly inhibited the nuclear migration of SV40 T-antigen NLS-containing karyophile (Fig. 6 A). These in vivo results further confirm that GTP hydrolysis of Ran is not required for movement of the β-subunit through the NPC.
Figure 6.
The import of β-subunit occurs without the support of GTP hydrolysis of Ran. (A) 1 mg/ml SV40 T-antigen NLS substrate (a and b), 1 mg/ml GFP–wild-type β-subunit (c and d), or 0.7 mg/ml FITC-D3-145 (e and f) were injected with (b, d, and f) or without (a, c, and e) 3 mg/ml GTP-G19V Ran in the cytoplasm of HEL cells. After incubation for 30 min at 37°C, localization of β-subunit or D3-145 was examined after fixation of the cells as in Fig. 4. (B) Digitonin-permeabilized MDBK cells were incubated with 10 μl of testing solution containing 3 pmol GFP–β-subunit (a–c) or FITC-D3-145 (d–f) in the absence (a and d) or presence of 40 pmol G19V GTP-Ran (b and e) for 15 min. c and f show the import assay performed with permeabilized cells pretreated with 0.2 mg/ml WGA for 15 min. Localization of β-subunit or its mutant proteins was examined after fixation of the permeabilized cells as in Fig. 3 D.
It should be noted, however, that in the digitonin-permeabilized cell-free transport assay, GTP-G19V Ran showed an inhibitory effect on the nuclear import of wild-type β-subunit, but failed to inhibit the import of D3-145 (Fig. 6 B). These in vitro results are consistent with those obtained with GTP-Ran, described above, and indicate that the inhibitory effect of GTP-G19V Ran appears to be due to the direct binding of this molecule to the β-subunit but not the inhibition of GTP hydrolysis. Moreover, different results, obtained in in vitro and in vivo studies, indicate that GTP-G19V Ran cannot stably bind to the β-subunit in the cytoplasm of living cells (discussed below).
Nuclear Migration of β-Subunit Is Saturable
The unassisted β-subunit import was promoted by amino acid residues 1–447 of this molecule. To confirm that the import is promoted through specific interaction(s), we examined the saturability of the import. In the digitonin-permeabilized cell-free system, the nuclear migration of GFP-tagged β-subunit and D450-876 decreased on the addition of an eightfold excess of nonlabeled β-subunit or D450-876 (Fig. 7). Addition of the NH2-terminal deletion, D3-447, which lacks the NPC-binding domain, had no effect on nuclear migration of the β-subunit. Since nuclear migration of the β-subunit occurs in the absence of added soluble factors, it is likely that the β-subunit transits through the NPC, from the cytoplasmic face toward the nuclear interior, by interacting directly with saturable site(s) within the NPC.
Discussion
Our present study shows that a 97-kD component of the nuclear pore–targeting complex (β-subunit) can behave in two different ways in cells. One is to support NLS-mediated import and the other is to migrate into the nucleus in an unassisted manner. The former reaction, in which the β-subunit carries the α-subunit with NLS substrate, is absolutely dependent on the presence of Ran and requires the Ran-binding domain of the β-subunit to exert a translocation reaction. The latter reaction, when β-subunit is alone, does not require Ran for the translocation through the NPC. Therefore, the β-subunit can exert protein translocation reaction through NPC via two contrasting mechanisms, namely, “Ran-assisted” versus “Ran-unassisted” import.
The Ran-unassisted nuclear migration of the β-subunit was strongly inhibited by WGA, both in living cells and in the digitonin-permeabilized cells (Figs. 4 and 6 B). Therefore, it is most likely that this protein migrates into the nucleus through gated channels of the NPC, similar to NLS-mediated import. Nuclear migration of the β-subunit is not the result of protein degradation, which was confirmed by judging the integrity of the GFP–β-subunit, which migrated into the nucleus of permeabilized cells by SDS-PAGE, followed by immunoblotting (data not shown).
The unassisted nuclear migration of the β-subunit was promoted by its NPC-interacting domain, which is located within its NH2-terminal portion and which is also involved in the NPC docking of PTAC. A competition study, showing that the unassisted nuclear migration of β-subunit occurs in a saturating manner (Fig. 7), indicates that the β-subunit is translocated through the NPC via specific interaction(s) with saturable site(s) of the NPC. Since D3-145 migrated into the nucleus in a manner similar to the wild-type β-subunit and D450-876, a structure necessary for the nuclear migration of the β-subunit through interaction with NPC components, would be predicted to be located within amino acids 146–447 of this protein.
Unlike NLS-mediated import, the β-subunit import was observed without the exogenous addition of ATP/GTP. Also, nuclear accumulation of β-subunit occurred even on ice (see Fig. 7), although the rate of import was reduced compared to the reaction performed at 30°C. Pretreatment of permeabilized cells with apyrase or hexokinase/glucose reduced but did not abolish the import (Imamoto, N., S. Kose, and Y. Yoneda, manuscript in preparation). Meanwhile, sensitivity to WGA and saturability of import provide strong support for the view that the unassisted nuclear migration of β-subunit through NPC occurs via a mechanism that involves specific molecular interactions of the NPC-binding domain of the β-subunit with NPC components. Differences in the level of requirement of ATP/ GTP may be due to differences in the requirement of Ran, although this must be addressed in further studies.
It was shown in previous studies that the β-subunit binds directly with several yeast nucleoporins, containing FXFG repeats (NUP1, NUP2, NUP36, NUP156, and NUP96) and GLFG repeat (NUP116) (Iovine et al., 1995; Kraemer et al., 1995; Radu et al., 1995b ). However, in a solution-binding assay, it was shown that the β-subunit, when bound to GTP-Ran, cannot access the repeat-containing nucleoporins. Our present data show that the β-subunit is stably complexed at the nuclear rim when it is bound to α-subunit/NLS substrate but transits rapidly through the NPC when it is not bound to these factors (Fig. 5 A). NPC docking of PTAC was significantly reduced, but was not completely inhibited in the presence of 12-fold excess of D450-876 β-subunit fragment, which may indicate that the mode of interaction of the β-subunit with NPC is different when it is alone or complexed with the α-subunit/NLS substrate (see Fig. 3 D). The rim binding of the β-subunit that occurs in the presence of α-subunit/NLS substrate can be thought of as an intermediate state of NLS-mediated import, probably through repeat-containing nucleoporins. It is possible, however, that the β-subunit could be complexed with different site(s) of the NPC in the presence of GTP-Ran, in contrast to those in the presence of α-subunit/NLS substrate. By analogy with the finding that the β-subunit forms a stable complex with Ran and its binding protein RanBP1 (Chi et al., 1996; Imamoto, N., S. Kose, T. Tachibana, and Y. Yoneda, unpublished observation), one such candidate might be a 358-kD Ran-binding NPC protein (RanBP2/NUP358), which contains four Ran-binding domains similar to RanBP1 (Yokoyama et al., 1995; Wu et al., 1995). The rim binding of the β-subunit that occurs in the presence of GTP-Ran will require further studies to understand the physiological significance.
Previous published results did not detect the nuclear accumulation of β-subunit in the digitonin-permeabilized cell-free import assay (Görlich et al., 1995; Moroianu et al., 1995b ; Kutay et al., 1997). However, our present data show that the β-subunit transits rapidly through NPC in the permeabilized cell-free import assay. Although we cannot clearly explain reasons for such contradictions, the nuclear migration of β-subunit would not be detected in the import assay when carried out in the presence of either α-subunit, GTP-Ran, or crude cytosol. In our experiments, nuclear migration of the β-subunit in the in vitro system can be observed only in the absence of exogenous addition of soluble factors. α-Subunit and GTP-Ran prevent the nuclear migration of β-subunit, and crude cytosol, in all probability, contains these and other soluble factor(s) that prevent the nuclear migration of the β-subunit in vitro. Also, nuclear migration of β-subunit might be affected by factor(s) that could be differently retained in the permeabilized cells upon preparation.
In living cells, cytoplasmically injected β-subunit rapidly migrated into the nuclei, irrespective of the protein concentration in the needle (from 0.5 mg/ml; corresponding, approximately, to 0.2-fold of endogenous β-subunit to 10 mg/ml protein; corresponding, approximately, to fourfold of the endogenous β-subunit). Further, GTP-G19V Ran did not prevent the nuclear migration of β-subunit in living cells, while it prevented nuclear migration of the β-subunit in vitro. These observations suggest that there may be factor(s) in the cytoplasm of living cells that actively dissociate β-subunit from GTP-Ran. We speculate that a cellular mechanism, which remains to be elucidated, exists by which the unassisted nuclear migration of β-subunit occurs, even in the presence of endogenous α-subunit and Ran.
Indirect immunofluorescence studies using affinity-purified antibodies against recombinant β-subunit demonstrated that a significant amount of endogenous β-subunit is located within the nucleus in tissue-cultured mammalian cells (Figs. 1 and 2), while previous reports indicated that it is located in the cytoplasm and on the nuclear envelope, but not within the nucleus (Chi et al., 1995). In yeast, Koepp et al. (1996), using affinity-purified anti-Kap95p antibodies, detected Kap95p (yeast β-subunit) on the nuclear envelope but not within the nucleus. However, Iovine and Wente (1997) very recently demonstrated that Kap95p recycles between the cytoplasm and nucleus by showing that nuclear export signal–mutated Kap95p accumulates in the nucleus. Our preliminary results indicate that the β-subunit shuttles between the nucleus and cytoplasm, since nucleoplasmically injected β-subunit rapidly exits the nucleus. The tagging of GFP to the NH2 terminus of the β-subunit did not affect its nuclear export, indicating that the nuclear accumulation of the β-subunit observed in this study is not due to its inability to exit the nucleus caused by the tagging (data not shown). We presume that discrepancies between previous and present studies regarding the detection of nuclear β-subunit are due to differences in antibodies used, or, alternatively, differences in cellular and/ or fixation conditions that may affect the equilibrium of β-subunit between the cytoplasm and nucleus.
The present data, which show that the β-subunit migrates into the nuclear interior independently from the NLS-containing substrate, provide an intriguing possibility that this protein possesses another role in cells, in addition to supporting NLS-mediated import. The extent of nuclear migration of injected β-subunit, as well as the distribution of endogenous β-subunit, differed among cells, indicating that its unassisted import could be regulated by cellular physical conditions. The intracellular distribution of endogenous β-subunit should also be affected by its export. Whether the export of the β-subunit occurs in a Ran- assisted or -unassisted manner is now under investigation. The intranuclear amount of β-subunit may affect the ratio of α- to β-subunit in the cytoplasm or the GTPase cycle of Ran, both in the nucleus and cytoplasm, which, in turn, could lead to the regulation of NLS-mediated nuclear import. Further studies are required to explore these and other possibilities.
Acknowledgments
We thank Drs. T. Ohba and T. Nishimoto for E. coli strain expressing human Ran, and Dr. S. Yuba for helpful discussions.
This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas (No. 07282103), Grant-in-Aid for Scientific Research (B) (No. 08458229), Grant-in-Aid for Scientific Research (C) (No. 08680764), and Grant-in-Aid for COE Research (No. 07CE2006) from the Japanese Ministry of Education, Science, Sports and Culture, the Nissan Science Foundation, and the Naito Foundation.
Abbreviations used in this paper
- GFP
green fluorescent protein: GST, glutathione-S-transferase
- MDBK
Madin-Darby bovine kidney
- NLS
nuclear localization signal
- NPC
nuclear pore complex
- PTAC
nuclear pore–targeting complex
Footnotes
S. Kose and N. Imamoto contributed equally to this work.
Address all correspondence to Yoshihiro Yoneda, Department of Anatomy and Cell Biology, Osaka University Medical School, 2-2 Yamada-oka, Suita, Osaka 565, Japan. Tel.: (81) 6 879 3211. Fax: (81) 6 879 3219.
Takuya Shimamoto's present address is Department of Microbiology, Osaka University Medical School, Suita, Osaka 565, Japan.
References
- Adam EJH, Adam SA. Identification of cytosolic factors required for nuclear location sequence-mediated binding to the nuclear envelope. J Cell Biol. 1994;125:547–555. doi: 10.1083/jcb.125.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam SA, Gerace L. Cytosolic proteins that specifically bind nuclear location signals are receptors for nuclear import. Cell. 1991;66:837–847. doi: 10.1016/0092-8674(91)90431-w. [DOI] [PubMed] [Google Scholar]
- Adam SA, Sterne-Marr R, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison JD, Blobel G, Rout MP. Kap 104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- Bonifaci N, Moroianu J, Radu A, Blobel G. Karyopherin β2 mediates nuclear import of a mRNA binding protein. Proc Natl Acad Sci USA. 1997;94:5055–5060. doi: 10.1073/pnas.94.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi NC, Adam EJH, Adam SA. Sequence and characterization of cytoplasmic nuclear import factor p97. J Cell Biol. 1995;130:265–274. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi NC, Adam EJH, Visser GD, Adam SA. RanBP1 stabilizes the interaction of Ran with p97 in nuclear protein import. J Cell Biol. 1996;135:559–569. doi: 10.1083/jcb.135.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi NC, Adam EJH, Adam SA. Different binding domains for Ran-GTP and Ran-GDP/RanBP1 on nuclear import factor p97. J Biol Chem. 1997;272:6818–6822. doi: 10.1074/jbc.272.10.6818. [DOI] [PubMed] [Google Scholar]
- Dasso M, Seki T, Ohba T, Nishimoto T. A mutant form of the Ran/TC4 protein disrupts nuclear function in Xenopus laevisegg extracts by inhibiting the RCC1 protein, a regulator of chromosome condensation. EMBO (Eur Mol Biol Organ) J. 1994;13:5732–5744. doi: 10.1002/j.1460-2075.1994.tb06911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L. The nuclear pore complex. Annu Rev Biochem. 1995;64:865–896. doi: 10.1146/annurev.bi.64.070195.004245. [DOI] [PubMed] [Google Scholar]
- Dickmanns A, Bischoff FR, Marshallsay C, Lührmann R, Ponstingl H, Fanning E. The thermolability of nuclear protein import in tsBN2 cells is suppressed by microinjected Ran-GTP or Ran-GDP, but not by RanQ69L or RanT24N. J Cell Sci. 1996;109:1449–1457. doi: 10.1242/jcs.109.6.1449. [DOI] [PubMed] [Google Scholar]
- Dingwall C, Laskey RA. Nuclear targeting sequences—a consensus? . Trends Cell Biol. 1991;4:357–365. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- Elliott DJ, Stutz F, Lescure A, Rosbash M. mRNA nuclear export. Curr Opin Genet Dev. 1994;4:305–309. doi: 10.1016/s0959-437x(05)80058-9. [DOI] [PubMed] [Google Scholar]
- Fabre E, Hurt EC. Nuclear transport. Curr Opin Cell Biol. 1994;6:335–342. doi: 10.1016/0955-0674(94)90023-x. [DOI] [PubMed] [Google Scholar]
- Floer M, Blobel G. The nuclear transport factor karyopherin β binds stoichiometrically to Ran-GTP and inhibits the Ran GTPase activating protein. J Biol Chem. 1996;271:5313–5316. doi: 10.1074/jbc.271.10.5313. [DOI] [PubMed] [Google Scholar]
- Garcia-Bustos J, Heitman J, Hall MN. Nuclear protein localization. Biochem Biophys Acta. 1991;1071:83–101. doi: 10.1016/0304-4157(91)90013-m. [DOI] [PubMed] [Google Scholar]
- Gerace L. Nuclear export signals and the fast track to the cytoplasm. Cell. 1995;82:341–344. doi: 10.1016/0092-8674(95)90420-4. [DOI] [PubMed] [Google Scholar]
- Goldfarb D, Michaud N. Pathways for the nuclear transport of proteins and RNAs. Trends Cell Biol. 1991;1:20–40. doi: 10.1016/0962-8924(91)90065-h. [DOI] [PubMed] [Google Scholar]
- Görlich D, Mattaj W. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- Görlich D, Prehn S, Laskey RA, Hartmann E. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Görlich D, Kostka S, Kraft R, Dingwall C, Laskey RA, Hartmann E, Prehn S. Two different subunits of importin cooperate to recognize nuclear location signals and bind them to the nuclear envelope. Curr Biol. 1995a;5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- Görlich D, Vogel F, Mills A, Hartmann E, Laskey RA. Distinct functions for the two importin subunits in nuclear protein import. Nature. 1995b;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- Görlich D, Henklein P, Laskey RA, Hartmann E. A 41 amino acid motif in importin-α confers binding to importin-β and hence transit into the nucleus. EMBO (Eur Mol Biol Organ) J. 1996a;15:1810–1817. [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Panté N, Kutay U, Aebi U, Bischoff FR. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO (Eur Mol Biol Organ) J. 1996b;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Imamoto N, Shimamoto T, Kose S, Takao T, Tachibana T, Matsubae M, Sekimoto T, Shimonishi Y, Yoneda Y. The nuclear pore-targeting complex binds to nuclear pores after association with a karyophile. FEBS Lett. 1995a;368:415–419. doi: 10.1016/0014-5793(95)00699-a. [DOI] [PubMed] [Google Scholar]
- Imamoto N, Shimamoto T, Takao T, Tachibana T, Kose S, Matsubae M, Sekimoto T, Shimonishi Y, Yoneda Y. In vivo evidence for involvement of a 58 kDa component of nuclear pore-targeting complex in nuclear protein import. EMBO (Eur Mol Biol Organ) J. 1995b;14:3617–3626. doi: 10.1002/j.1460-2075.1995.tb00031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamoto N, Tachibana T, Matsubae M, Yoneda Y. A karyophilic protein forms a stable complex with cytoplasmic components prior to nuclear pore binding. J Biol Chem. 1995c;270:8559–8565. doi: 10.1074/jbc.270.15.8559. [DOI] [PubMed] [Google Scholar]
- Iovine MK, Wente SR. A nuclear export signal in Kap95p is required for both recycling the import factor and interaction with the nucleoporin GLFG repeat regions of Nup116p and Nup100p. J Cell Biol. 1997;137:797–811. doi: 10.1083/jcb.137.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovine MK, Watkins JL, Wente SR. The GLFG repetitive region of the nucleoporin Nup116p interacts with Kap95p, an essential yeast nuclear import factor. J Cell Biol. 1995;131:1699–1713. doi: 10.1083/jcb.131.6.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Mattaj IW. RNA export. Cell. 1995;81:153–159. doi: 10.1016/0092-8674(95)90323-2. [DOI] [PubMed] [Google Scholar]
- Koepp DM, Silver PA. A GTPase controlling nuclear trafficking: running the right way or walking RANdomly? . Cell. 1996;87:1–4. doi: 10.1016/s0092-8674(00)81315-x. [DOI] [PubMed] [Google Scholar]
- Koepp DM, Wang DH, Corbett AH, Silver PA. Dynamic localization of the nuclear import receptor and its interactions with transport factors. J Cell Biol. 1996;133:1163–1176. doi: 10.1083/jcb.133.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer DM, Strambio-de-Castillia C, Blobel G, Rout MP. The essential yeast nucleoporin NUP159 is located on the cytoplasmic side of the nuclear pore complex and serves in karyopherin-mediated binding of transport substrate. J Biol Chem. 1995;270:19017–19021. doi: 10.1074/jbc.270.32.19017. [DOI] [PubMed] [Google Scholar]
- Kutay U, Izaurralde E, Bischoff FR, Mattaj IW, Görlich D. Dominant-negative mutants of importin-β block multiple pathways of import and export through the nuclear pore complex. EMBO (Eur Mol Biol Organ) J. 1997;16:1153–1163. doi: 10.1093/emboj/16.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskey RA, Dingwall C. Nuclear shuttling: the default pathway for nuclear proteins. Cell. 1993;74:585–586. doi: 10.1016/0092-8674(93)90505-k. [DOI] [PubMed] [Google Scholar]
- Lounsbury KM, Beddow AL, Macara IG. A family of proteins that stabilizes the Ran/TC4 GTPase in its GTP-bound conformation. J Biol Chem. 1994;269:11285–11290. [PubMed] [Google Scholar]
- Mandell RB, Feldherr CM. Identification of two hsp70-related Xenopusoocyte proteins that are capable of recycling across the nuclear envelope. J Cell Biol. 1990;111:1775–1783. doi: 10.1083/jcb.111.5.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior FB, Paschal J, Evans J, Gerace L. Inhibition of nuclear import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J Cell Biol. 1993;123:1649–1659. doi: 10.1083/jcb.123.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael WM, Choi M, Dreyfuss G. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- Moore MS, Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- Moore MS, Blobel G. Purification of Ran-interacting protein that is required for protein import into the nucleus. Proc Natl Acad Sci USA. 1994;91:10212–10216. doi: 10.1073/pnas.91.21.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroianu J, Blobel G, Radu A. Previously identified protein of uncertain function is karyopherin α and together with karyopherin β docks import substrate at the nuclear pore complex. Proc Natl Acad Sci USA. 1995a;92:2008–2011. doi: 10.1073/pnas.92.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroianu J, Hijikata M, Blobel G, Radu A. Mammalian karyopherin α1β and α2β heterodimers: α1 or α2bind nuclear localization signal and β interacts with peptide repeat-containing nucleoporins. Proc Natl Acad Sci USA. 1995b;92:6532–6536. doi: 10.1073/pnas.92.14.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- Panté N, Aebi U. Toward the molecular dissection of protein import into nuclei. Curr Opin Cell Biol. 1996;8:397–406. doi: 10.1016/s0955-0674(96)80016-0. [DOI] [PubMed] [Google Scholar]
- Paschal BM, Gerace L. Identification of NTF2, a cytosolic factor for nuclear protein import that interacts with nuclear pore complex protein p62. J Cell Biol. 1995;129:925–937. doi: 10.1083/jcb.129.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschal BM, Delphin C, Gerace L. Nucleotide-specific interaction of Ran/TC4 with nuclear transport factors NTF2 and p97. Proc Natl Acad Sci USA. 1996;93:7679–7683. doi: 10.1073/pnas.93.15.7679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñol-Roma S, Dreyfuss G. hnRNP proteins: localization and transport between the nucleus and the cytoplasm. Trends Cell Biol. 1993;3:151–155. doi: 10.1016/0962-8924(93)90135-n. [DOI] [PubMed] [Google Scholar]
- Pollard V, Michael WM, Nakielny S, Siomi MC, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–993. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- Pruschy M, Ju Y, Spitz L, Carafoli E, Goldfarb DS. Facilitated nuclear transport of calmodulin in tissue culture cells. J Cell Biol. 1994;127:1527–1536. doi: 10.1083/jcb.127.6.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu A, Blobel G, Moore MS. Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc Natl Acad Sci USA. 1995a;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu A, Moore MS, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995b;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- Rout MP, Wente SR. Pores for thought: nuclear pore complex proteins. Trends Cell Biol. 1994;4:357–365. doi: 10.1016/0962-8924(94)90085-x. [DOI] [PubMed] [Google Scholar]
- Sekimoto T, Nakajima K, Tachibana T, Hirano T, Yoneda Y. Interferon-γ-dependent nuclear import of Stat1 is mediated by the GTPase activity of Ran/TC4. J Biol Chem. 1996;271:31017–31020. doi: 10.1074/jbc.271.49.31017. [DOI] [PubMed] [Google Scholar]
- Weis K, Mattaj I, Lamond A. Identification of hSRP1α as a functional receptor for nuclear localization sequences. Science. 1995;268:1049–1051. doi: 10.1126/science.7754385. [DOI] [PubMed] [Google Scholar]
- Weis K, Ryder U, Lamond AI. The conserved amino terminal domain of hSRP1 α is essential for nuclear protein import. EMBO (Eur Mol Biol Organ) J. 1996;15:1818–1825. [PMC free article] [PubMed] [Google Scholar]
- Wu J, Matunis MJ, Kraemer D, Blobel G, Coutavas E. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J Biol Chem. 1995;270:14209–14213. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- Yaseen NR, Blobel G. Cloning and characterization of human karyopherin β3. Proc Natl Acad Sci USA. 1997;94:4451–4456. doi: 10.1073/pnas.94.9.4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama N, Hayashi N, Seki T, Panté N, Ohba T, Nishii K, Kuma K, Hayashida T, Miyata T, Aebi U, et al. A giant nucleopore protein that binds Ran/TC4. Nature. 1995;376:184–188. doi: 10.1038/376184a0. [DOI] [PubMed] [Google Scholar]
- Yoneda Y. Nuclear export and its significance in retroviral infection. Trends Microbiol. 1996;4:1–2. doi: 10.1016/0966-842x(96)81494-6. [DOI] [PubMed] [Google Scholar]
- Yoneda Y, Imamoto-Sonobe N, Yamaizumi M, Uchida T. Reversible inhibition of protein import into the nucleus by wheat germ agglutinin injected into cultured cells. Exp Cell Res. 1987;173:586–595. doi: 10.1016/0014-4827(87)90297-7. [DOI] [PubMed] [Google Scholar]