Abstract
We examined the activity of human T cells engineered to express variants of a single TCR (1G4) specific for the cancer/testis Ag NY-ESO-1, generated by bacteriophage display with a wide range of affinities (from 4 μM to 26 pM). CD8+ T cells expressing intermediate- and high-affinity 1G4 TCR variants bound NY-ESO-1/HLA-A2 tetramers with high avidity and Ag specificity, but increased affinity was associated with a loss of target cell specificity of the TCR gene-modified cells. T cells expressing the highest affinity TCR (KD value of 26 pM) completely lost Ag specificity. The TCRs with affinities in the midrange, KD 5 and 85 nM, showed specificity only when CD8 was absent or blocked, while the variant TCRs with affinities in the intermediate range—with KD values of 450 nM and 4 μM— demonstrated Ag-specific recognition. Although the biological activity of these two relatively low-affinity TCRs was comparable to wild-type reactivity in CD8+ T cells, introduction of these TCR dramatically increased the reactivity of CD4+ T cells to tumor cell lines.
T cells expressing Ag-specific TCRs mediate the elimination of tumor and virally infected cells by recognition of Ag in the form of individual peptides bound to MHC molecules. This recognition is programmed during thymic development where T cells undergo stringent selection processes to ensure that only those T cells that are self-MHC-restricted (positive selection) but not self-reactive (negative selection) are matured and released to the periphery (1, 2). The selection process yields a population of T cells that, while not highly self-reactive, have the potential to recognize foreign peptides bound to an MHC product with high specificity and sensitivity. The TCRs on peripheral T cells, which have undergone avidity selection, produce a narrow window of monomeric TCR-Ag affinities of between 1 and 100 μM (3, 4). Although interactions between TCR and the restriction MHC element have been detected only in the presence of specific peptides, it is believed that this thymic-positive selection imposes (imprints) a low self-MHC affinity onto the peripheral T cell pool (5, 6). Evidence indicates that T cells require interaction with endogenous peptide-MHC (pMHC)3 complexes for long-term survival, suggesting that the continuous cross-reactivity of self Ags is needed for maintenance of immunological memory (7–9). Hence, an important aspect of TCR recognition and T cell biology is that positive selection generates a repertoire of T cells that can cross-react with self-peptide Ags to ensure T cell survival but this cross-reactivity to self-pMHC occurs at affinities too weak to trigger the T cell activation under normal physiological conditions.
The narrow window of TCR-Ag-binding affinities that separate positive and negative selection (10) limits the avidity of the T cell for its cellular target. It has been reported that T cell avidity appears to be correlated with the efficient in vivo elimination of tumor cells and virally infected cells (11, 12). Hence, studying the behavior of T cells expressing high-affinity TCRs that are directed at disease-specific Ags is of both theoretical and practical importance.
Recent approaches aimed at engineering higher affinity TCRs in vitro have been influenced by consensus crystallographic structural data that reveal a conserved diagonal docking orientation of the TCR α-/TCR β-chain heterodimer at the pMHC surface which positions the CDR2 loops of both chains over the MHC groove-flanking helices and the CDR3 loops of both chains over the bound peptide (13). Using a yeast display approach, Holler et al. (14) generated mutations within the α-chain CDR3 region of the murine 2C TCR to produce high-affinity TCRs outside of normal in vivo constraints. The mutant TCR generated in their study had >100-fold higher affinity (KD ~ 9 nM) for the pMHC ligand while retaining a high degree of peptide specificity with in vitro-binding assays. When the high-affinity mutant TCR was transfected into in both CD8+ and CD8+ hybridoma T cells, the transfected CD8+ cells reacted specifically to Ag but the CD8+ hybridoma was cross-reactive with self-pMHC complexes (15, 16). Using bacteriophage display and directed molecular evolution of the TCR CDR3 and CDR2 loops, a panel of TCRs were generated with Ag affinities of up to a million times higher than wild-type (wt) TCR (17). One of the panels of TCRs studied in this report was directed against the epitope of the cancer/testis Ag NY-ESO-1 (p157–165)/HLA-A2. The soluble form of these high-affinity TCRs bound only to NY-ESO-1/HLA-A2 and not to a panel of control peptide Ags, suggesting that the binding is highly Ag specific. Furthermore, high-affinity TCR monomers and tetramers showed no cross-reactivity to endogenous cellular pMHC Ags (17, 18).
To examine whether T cells expressing these high-affinity TCRs would exert enhanced reactivity toward tumor cells, we introduced a panel of these high-affinity TCRs with KD values of 4 μM, 450 nM, 84 nM, 5 nM, and 26 pM into stimulated human PBLs. Although CD8+ T cells expressing intermediate-and high-affinity 1G4 TCR variants showed greatly increased binding to NY-ESO-1/HLA-A2 tetramers with high Ag specificity, increasing affinity was associated with a loss of specificity of the gene-modified cells. In contrast to CD8+ T cells, engineered CD4+ T cells expressing intermediate-affinity TCRs demonstrated a pronounced increase in sensitivity for recognition of target cells pulsed with the cognate peptide, and greatly enhanced recognition of appropriate tumor targets. These results suggest that genetically modified CD4+ T cells that express TCRs with intermediate affinity may be useful for TCR-based adoptive cancer therapies.
Materials and Methods
PBL, cell lines, and RNA electroporation
All of the PBLs used in this study were from metastatic melanoma patients treated at the Surgery Branch (National Cancer Institute (NCI), National Institutes of Health, Bethesda, MD). Melanoma cell lines 624.38mel, A375mel, 1390mel, 1363mel, SK32mel, 526mel, and 888mel were generated at the Surgery Branch as previously described (19). HLA-A2+ and HLA-A2− EBV-B and renal cell carcinoma cell lines (RCCs) were also generated at the Surgery Branch as described (20). All cell lines were cultured in medium consisting of RPMI 1640 supplemented with 10% heat-inactivated FBS (Biofluids), 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen Life Technologies). Lymphocytes were cultured in AIM-V medium (Invitrogen Life Technologies) supplemented with 5% human AB serum (Valley Biomedical) and 300 IU/ml IL-2 (Chiron) at 37°C and 5% CO2. The MART-1 and gp100-specific T cell clones JKF6 and L2D8 were isolated from patient tumor-infiltrating lymphocyte (TIL) cultures (Surgery Branch, NCI). In vitro-transcribed (IVT) RNA electroporation of anti-CD3 Ab (OKT3) stimulated human PBLs and cell lines was conducted as described in our previous report (21).
Peptide synthesis
Synthetic peptides used in this study were made using a solid phase method on a peptide synthesizer (Gilson) at the Surgery Branch (NCI). The quality of each peptide was evaluated by mass spectrometry (Biosynthesis). The sequences of the peptide used in this study are as follow: NY-ESO-1, 157–165 (165V) (SLLMWITQV); MART-1, 26–35 (27L) (ELAGIGILTV); gp100, 209–217 (210M) (IMDQVPFSV); Flu-MP, 58–66 (GILGFVFTL).
High-affinity TCR α- and β-chain genes
High-affinity TCR genes were generated by bacterial phage display (17). The 1G4 c58/c61 α TCR/β TCR in this publication is identical with “1G4c113” or “clone 113”; c10/c1 corresponds to “clone10” and c12/c2 corresponds to “clone 12” (17). The mutated TCR α-chain c5 and β-chain genes c59 and c100 were not previously reported (Y. Li and P. E. Molloy, unpublished observations).
1G4 wt and high-affinity mutant NY-ESO-1/HLA-A2-specific TCR genes were reconstructed as full-length TCR α- and β-chains (using the TCR β-chain 2 constant region) and were designed with native interchain disulfide bonds. The open reading frames and 5′ untranslated regions of these genes were optimized for maximal expression in human cells (22) by GENEART. Additional, nonoptimized, TCR genes for NY-ESO-1 (ET-8F) and gp100 were described previously (23, 24).
Single-chain trimer and retroviral vector construction
PCR primers were designed to amplify plasmid-encoded genes and introduce a T7 promoter at the 5′ end and a poly(A) tract at the 3′ the genes for the α and β TCR chains, α and β CD8, HLA-A2 and both NY-ESO-1 (165V)- and MART-1(27L)-single chain (peptide-β2-microglobulin (β2m)-HLA-A2) trimers (SCT), respectively. By using these purified PCR products as templates, RNA was generated via IVT as described previously (21). SCT PCR templates for both NY-ESO-1 and MART-1 were generated by PCR as previously described (25). The pIRES.neo. OVA.hβ2m.A201 plasmid was obtained from T. Hansen (Washington University School of Medicine, St. Louis, MO). This plasmid encodes the following elements starting from the N terminus: the mouse β2m signal peptide, SIINFEKL (OVA-derived H-2Kb-restricted epitope), the first flexible linker (G3ASG4SG4S), the mature portion of human β2m, the second flexible linker (G4S)4, the mature portion of HLA-A201 and its 3′ untranslated region. The entire insert was then subcloned into the NotI and HindIII sites of pCR2.1 to create pCR2.1.OVA.h β2m.A201. The region encoding the SIINFEKL epitope is flanked by unique AgeI site and NheI sites within the β2m signal peptide and the first linker, respectively. For the final constructs, the SIINFEKL epitope was exchanged for the MART-1 or NY-ESO-1 HLA-A2-restricted epitope (details available upon request). Positive recombinants were identified by restriction mapping and confirmed by DNA sequencing.
The retroviral vector backbone used in this study, pMSGV, is a derivative of the murine stem cell virus (MSCV)-based splice-gag vector (pMSGV), which uses a MSCV long terminal repeat and has been previously described (26). The TCR α-polio internal ribosomal entry site (IRES)-TCR β DNA fragment for each pair of wt or high-affinity 1G4 TCR chains were generated and assembled by PCR and cloned into pMSGV (XhoI/EcoRI) to generate MSGV-TCR α-IRES-TCR β retroviral vectors. Generation of PG13 packaging cell lines and retroviral transduction of stimulated PBLs were conducted as previously described (23).
FACS analysis, CD4/CD8 cell separation, and Ab-blocking assays
Cell surface expression of human CD3, CD4, and CD8 molecules on PBL was determined by specific Ab conjugate staining (FITC- or PE-conjugated Abs; BD Biosciences). Human Vβ8-, Vβ13.1-, and HLA-A2-specific mAb-PE conjugates were supplied by Immunotech. PE-labeled NY-ESO-1 (165V), MART-1 (27L), and gp100 (210M) peptide/HLA-A2 streptavidin tetramers were purchased from Beckman Coulter. Immunofluorescence, analyzed as the relative log fluorescence of ~1 × 105 live cells, was measured using a FACSCalibur flow cytometer (BD Biosciences).
CD4+ and CD8+ cells were separated using a magnetic bead-based approach for both negative and positive selection of those population subsets (Dynal Biotech and Miltenyi Biotec).
In Ab-blocking experiments, peptide-pulsed T2 cells (5 × 104 cells/100 μl) were incubated with each mAb at a concentration of 10 μg/ml for 30 min at 37°C in a flat-bottom 96-well plate. T cells (5 × 104 cells/well) were then added and incubated with target cells overnight at 37°C. The supernatants were harvested and assayed for IFN-γ production by ELISA.
Cytokine release assays
PBL cultures were tested for reactivity in cytokine-release assays using commercially available ELISA kits (IFN-γ; Endogen). T2 cells were pulsed with peptide at indicated concentrations in R/10 medium for 2 h at 37°C, followed by washing (three times) before initiation of cocultures. Stimulator APCs and responder T cells were cocultured for 24 h. Cytokine secretion was measured in culture supernatants diluted to be in the linear range of the assay.
51Cr-release assays
The ability of the TCR-transduced T cells to lyse Ag-specific peptide-pulsed target cells was measured using a 51Cr-release assay as described previously (19). Briefly, 1 × 106 target cells were labeled for 1 h at 37°C with 200 μCi 51Cr sodium chromate (GE Healthcare). Labeled target cells (5 × 103) were pulsed with peptide for 1 h and incubated with effector cells at the ratios indicated in the text for 4 h at 37°C in 0.2 ml of R/10 medium. Harvested supernatants were counted using a Wallac 1470 Wizard gamma counter (PerkinElmer). Total and spontaneous 51Cr release was determined by incubating 5 × 103-labeled target cells in either 2% SDS or R/10 medium for 4 h at 37°C. Each data point was determined as an average of quadruplicate wells. The percent-specific lysis was calculated as follows: percent-specific lysis = ((specific 51Cr release − spontaneous 51Cr release)/(total 51Cr release − spontaneous 51Cr release)) × 100.
Results
High-affinity TCRs bind pMHC tetramers with high Ag specificity
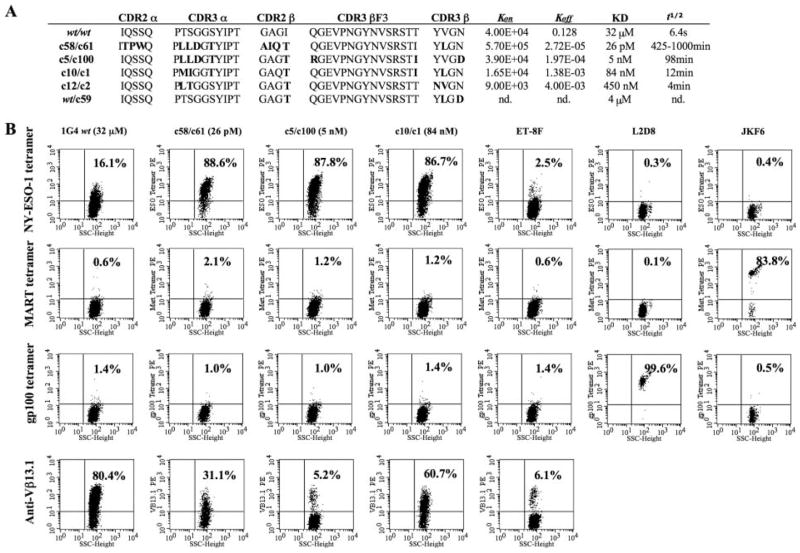
Previous studies demonstrated that soluble versions of high-affinity TCRs selected against NY-ESO-1 p157–165 Ag bound specifically to NY-ESO-1 peptide-HLA-A2 when assayed by Biacore surface plasmon resonance and flow cytometry (17, 18). A selection was made of genes encoding TCR α/TCR β pairs with a wide range of monovalent in vitro Ag-binding kinetics to determine whether differing biological activities were observed in the multivalent cellular context (KD 4 μM to 26 pM, Fig. 1A). To test whether human primary T lymphocytes expressing these TCRs retained specificity, IVT RNA for both α- and β-chains from the wt 1G4 or mutant TCR pairs were coelectroporated into CD8+ T cells and the transfected T cells were subjected to flow cytometry analysis by pMHC-tetramer staining. Both wt and high-affinity TCR-transfected CD8 T cells specifically bound to the NY-ESO-1 tetramer (they did not bind to control MART-1 and gp100 tetra-mers, Fig. 1B). The previously reported NY-ESO-1 TCR, ET-8F (23) bound less tetramer than the wt 1G4 (2.5 and 16%, respectively, background 0.3–0.4%) suggesting that 1G4 is a higher affinity TCR than ET-8F.
FIGURE 1.
High-affinity TCRs and tetramer staining. A, Summary of high-affinity TCRs used in this study. Amino acid sequences are indicated in single-letter code with changes in sequence relative to the wt 1G4 TCR shown in bold. KD were obtained using Biacore SPR with soluble version of the selected TCRs (nd, not determined). B, Specific HLA-tetramer binding of high-affinity TCR-transfected CD8+ T cells. Flow cytometry analysis of HLA-tetramer and anti-Vβ13.1 Ab staining for high-affinity NY-ESO-1/HLA-A2-restricted TCR genes electroporated as IVT RNA into CD8+ T cells. Controls were NY-ESO-1 TCR ET-8F and TIL clones L2D8 and JKF6 that are specific for gp100 and MART-1, respectively. The numbers in each quadrant indicates the percentage (%) of stained cells, living cells gated for by propidium iodide (PI) exclusion and CD8 gated, data representative of five experiments.
The percentage of specific tetramer binding of the mutant high-affinity TCR-transfected T cells were ~5-fold higher than that of wt TCR-transfected T cells. The levels of staining observed when using an anti-Vβ13.1 Ab (IMMU222) were consistently lower in T cells transfected with the mutant TCR than those observed with cells that had been transfected with the wt TCR, while tetramer binding was significantly higher in the cells transfected with the mutant TCRs. These findings indicated that these mutations of the CDR3 region change the conformation of the TCR protein and therefore, its ability to be recognized by the anti-Vβ13.1 Ab, and suggest that the differences in transfection efficiency were not responsible for the enhanced tetramer binding observed with the high-affinity TCRs.
Cellular Ag recognition by high-affinity TCR expressed on CD8+ T cells is affinity dependent
To test whether T cells redirected with high-affinity TCRs would convey better function than wt TCR-expressing T cells, we compared the function of CD8+ T cells expressing the highest affinity TCRs (monomer KD values ranging from 84 nM to 26 pM) with two wt TCRs by coculture of TCR-transfected cells with peptide-pulsed T2 cells (Table I, Expt. 1). Transfer of wt 1G4 TCR demonstrated specific cytokine production to 0.1 nM NY-ESO-1 peptide. As seen in tetramer-binding assays (Fig. 1B), CD8 cells electroporated with wt ET-8F were less reactive (no reactivity observed below 10 nM peptide) than 1G4-engineered cells. In contrast to the wild-type TCRs, CD8 T cells expressing high-affinity TCRs exhibited cross-reactivity as demonstrated by recognition of MART-1 peptide-pulsed cells (Table I, Expt. 1). A lack of specificity was also observed in a second experiment conducted using CD8+ PBL cells from a different donor. In this experiment, T cells transfected with the highest affinity TCR, c58/c61 (26 pM), as well as the next highest affinity TCR, c5c/100 (5 nM) released equivalent levels of cytokine IFN-γ in response to culture with unpulsed T2 cells (Table I, Expt. 2). The CD8+ T cells transfected with the TCR c12/c2 (450 nM), retained Ag specificity, but demonstrated reduced Ag reactivity relative to T cells transfected with the 1G4 wt TCR (Table I, Expt. 2). This lack of specific T cell recognition observed in cells transfected with the high-affinity TCR was not due to novel hybrid α TCRs/β TCRs formed between the endogenously expressed and the transfected TCR chains, as no reactivity was seen when either exogenous TCR α (c58) or β (c61) chain was separately transfected into CD8+ T cells (data not shown).
Table I.
Nonspecific production of IFN-γ by TCR-transferred CD8 T cellsa
| NY-ESO-1 Peptide (nM)
|
||||||
|---|---|---|---|---|---|---|
| 10 | 10.0 | 0.1 | 0.01 | 0 | MART-1 | |
| Expt. 1 | ||||||
| 1G4 wt (32 μM) | 17,144 (1,575) | 7,447 (818) | 2,195 (338) | 0 | ND | 0 |
| c58/c61 (26 pM) | 23,745 (861) | 25,569 (1,255) | 22,226 (114) | 23,847 (490) | ND | 31,148 (11,244) |
| c5/c100 (5 nM) | 5,107 (288) | 3,775 (192) | 3,235 (86) | 3,514 (177) | ND | 4,722 (155) |
| c10/c1 (84 nM) | 5,961 (521) | 5,611 (677) | 3,011 (136) | 3,553 (195) | ND | 5,992 (219) |
| ET-8F wt | 2,683 (771) | 0 | 0 | 0 | 0 | ND |
| Expt. 2 | ||||||
| 1G4 wt (32 μM) | 3,763 (67) | 1,131 (28) | 0 | 0 | ND | 0 |
| c58/c61 (26 pM) | 4,356 (200) | 3,212 (33) | 4,189 (145) | 4,205 (62) | 3,808 (117) | ND |
| c5/c100 (5 nM) | 2,603 (38) | 1,893 (41) | 2,441 (45) | 2,410 (180) | 2,182 (91) | ND |
| c12/c2 (450 nM) | 1,516 (13) | 0 | 0 | 0 | 0 | ND |
| ET-8F wt | 381 (5) | 0 | 0 | 0 | 0 | ND |
| gp100 TCR | 0 | ND | ND | ND | 0 | ND |
IFN-γ production by purified CD8 T cells electroporated with IVT RNA for wt 1G4 or mutant high-affinity TCRs. Lymphocytes were cocultured overnight with T2 cells pulsed with serially diluted NY-ESO-1 peptide 157–165 (165V) at the indicated concentrations. Control peptide was 1000 nM MART-1 27–35. Control TCRs were anti-NY-ESO-1 ET-8F wt and a gp100-specific TCR. Two independent electroporations were performed using different PBL donors (Expts. 1 and 2). Values in bold denote specific reactivity. Data are the mean values (picograms per milliliter) of triplicate samples, with SD in parentheses (representative of four experiments).
To determine whether the cross-reactivity observed for T2 cells was HLA-A2 restricted, a panel of HLA-A2+ or HLA-A2− target cell lines were tested (Table II). Five HLA-A2+ cell lines (C1R-A2, 1558EBV, 697EBV, RCC1, and RCC3) were pulsed with NY-ESO-1 peptide and used as stimulators for TCR-electroporated CD8 cells. As was observed with T2 cells, non-peptide-pulsed stimulator cells induced cross-reactive cytokine production from CD8 cells electroporated with TCRs c58/c61 (26 pM) and c5/c100 (5 nM); however, T cells transfected with intermediate-affinity TCR c10/c1 (84 nM) recognized unpulsed HLA-A2+ cell lines to a varying degree. Cross-reactivity of the c10/c1 TCR (84 nM) was observed with C1R-A2 and the two HLA-A2+ EBV cell lines but not the RCC lines. The RCC lines had lower amounts of HLA-A2 expression as determined by FACS analysis (mean fluorescence intensity 94–220, Table II). Peptide-pulsed, as well as unpulsed, HLA-A2− cell lines (C1R, 1350EBV, RCC2, and RCC4) failed to stimulate significant cytokine release from any of the transfectants, indicating that the cross-reactivity observed with the high-affinity TCRs was HLA-A2 restricted. Cells that were transfected with a combination of the wt α-chain and the c59 β-chain TCR (4 μM) appeared to maintain their specificity, and had comparable or slightly higher reactivity than those cells transfected with the wt α-and β-chain constructs.
Table II.
HLA-A2 restriction of TCR transfected CD8 T Cellsa
| Cell Line | HLA-A2 (MFI) | Peptide conc. | 1G4wt (32 μM) | c58/c61 (26 pM) | c5/c100 (5 nM) | c10/c1 (84 nM) | wtα/c59 (4 μM) | No RNA |
|---|---|---|---|---|---|---|---|---|
| C1R-A2 | + (783) | 100 nM | 342 (26) | 217 (15) | 126 (35) | 129 (2) | 272 (78) | 33 (25) |
| C1R-A2 | 10 nM | 310 (20) | 269 (2) | 92 (12) | 171 (42) | 309 (118) | 30 (7) | |
| C1R-A2 | 1 nM | 172 (38) | 279 (5) | 106 (31) | 218 (62) | 161 (9) | 26 (7) | |
| C1R-A2 | 0.1 nM | 30 (7) | 267 (22) | 90 (20) | 187 (3) | 102 (8) | 29 (3) | |
| C1R-A2 | 8 (3) | 231 (21) | 68 (1) | 192 (42) | 73 (7) | 39 (13) | ||
| C1R | − (5) | 100 nM | 7 (2) | 37 (3) | 4 (2) | 7 (5) | 9 (4) | 19 (6) |
| C1R | 27 (6) | 38 (4) | 6 (2) | 12 (9) | 12 (8) | 20 (2) | ||
| 1558EBV | + (450) | 100 nM | 1288 (195) | 880 (180) | 1113 (181) | 793 (174) | 1437 (12) | 0 |
| 1558EBV | 10 nM | 1112 (169) | 958 (192) | 1049 (89) | 563 (69) | 1738 (119) | 0 | |
| 1558EBV | 1 nM | 0 | 1160 (230) | 951 (306) | 557 (33) | 691 (63) | 0 | |
| 1558EBV | 0.1 nM | 0 | 1184 (192) | 966 (169) | 434 (238) | 214 (82) | 0 | |
| 1558EBV | 0 | 1042 (35) | 679 (4) | 306 (121) | 69 (32) | 0 | ||
| 1350EBV | − (5) | 100 nM | 0 | 0 | 0 | 0 | 0 | 0 |
| 1350EBV | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 697EBV | + (390) | 100 nM | 2357 (691) | 2216 (431) | 3659 (219) | 1326 (69) | 2970 (742) | 0 |
| 697EBV | 10 nM | 1510 (225) | 2055 (121) | 1770 (466) | 945 (66) | 1913 (492) | 0 | |
| 697EBV | 1 nM | 386 (48) | 1777 (129) | 972 (177) | 604 (58) | 1155 (72) | 0 | |
| 697EBV | 0.1 nM | 0 | 1069 (42) | 292 (6) | 342 (136) | 0 | 0 | |
| 697EBV | 0 | 1699 (335) | 956 (51) | 796 (67) | 14 (2) | 0 | ||
| RCC1 | + (94) | 100 nM | 610 (91) | 509 (66) | 39 (6) | 183 (18) | 993 (141) | 0 |
| RCC1 | 10 nM | 175 (39) | 574 (187) | 0 | 134 (10) | 305 (76) | 0 | |
| RCC1 | 1 nM | 0 | 526 (93) | 0 | 64 (30) | 0 | 0 | |
| RCC1 | 0.1 nM | 0 | 541 (118) | 0 | 53 (15) | 0 | 0 | |
| RCC1 | 0 | 538 (25) | 0 | 41 (4) | 0 | 0 | ||
| RCC2 | − (7) | 100 nM | 0 | 0 | 0 | 0 | 0 | 0 |
| RCC2 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| RCC3 | + (200) | 100 nM | 524 (100) | 385 (55) | 89 (51) | 328 (45) | 637 (40) | 75 (43) |
| RCC3 | 10 nM | 108 (40) | 222 (80) | 33 (13) | 79 (42) | 320 (77) | 36 (14) | |
| RCC3 | 1 nM | 42 (40) | 269 (30) | 41 (11) | 59 (3) | 161 (6) | 50 (5) | |
| RCC3 | 0.1 nM | 38 (15) | 345 (87) | 65 (1) | 125 (33) | 331 (52) | 17 (8) | |
| RCC3 | 50 (43) | 466 (24) | 71 (37) | 103 (12) | 159 (146) | 19 (7) | ||
| RCC4 | − (4) | 100 nM | 42 (40) | 34 (24) | 46 (10) | 58 (23) | 231 (162) | 14 (9) |
| RCC4 | 37 (22) | 38 (18) | 30 (19) | 42 (22) | 149 (94) | 30 (5) |
IFN-γ production by purified CD8 T cells electroporated with IVT RNA for wt 1G4 or mutant high affinity TCRs following co-culture with different HLA-A2+ or HLA-A2− cell lines. Stimulator cells were pulsed with serially diluted NY-ESO-1 peptide 157–165 (165V) at the indicated concentrations, and IFN-γ production was measured by ELISA (values are pg/ml). Data are the mean values of duplicate samples, with SD in parentheses, where values in bold denote specific reactivity. HLA-A2 expression of each cell line was shown with mean fluorescent intensity (MFI) measured by flow cytometry given in parenthesis.
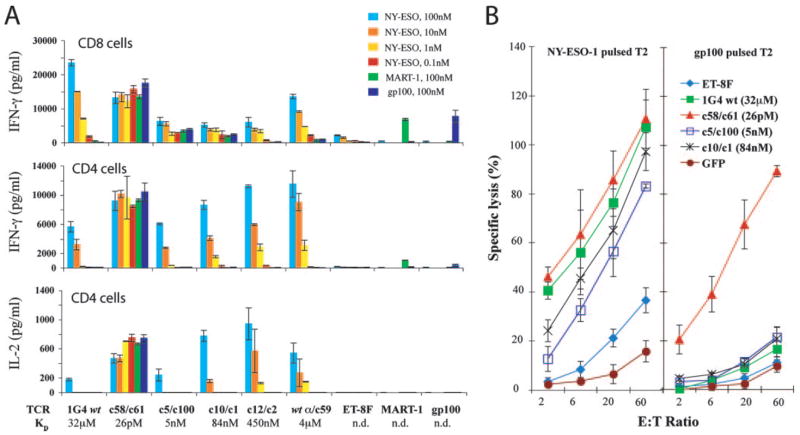
The effects of TCR affinity on target cell lysis were next investigated. The results demonstrated that CD8+ T cells transfected with the wt 1G4 as well as the wt ET-8F TCR lyse target cells pulsed with the NY-ESO-1 peptide but failed to lyse targets pulsed with the control influenza peptide (Fig. 2). Nonspecific killing of peptide-pulsed T2 was observed for CD8+ T cells transfected with mutant TCRs at affinities ranging from 84 nM to 26 pM, while those transfected with the wt α/c59 (4 μM) TCR lysed target cells pulsed with the cognate peptide but did not lyse control targets. The cell lysis assay results were thus consistent with data obtained from cytokine production assays and further demonstrated that the specificity of target cell recognition was dependent upon TCR affinity.
FIGURE 2.
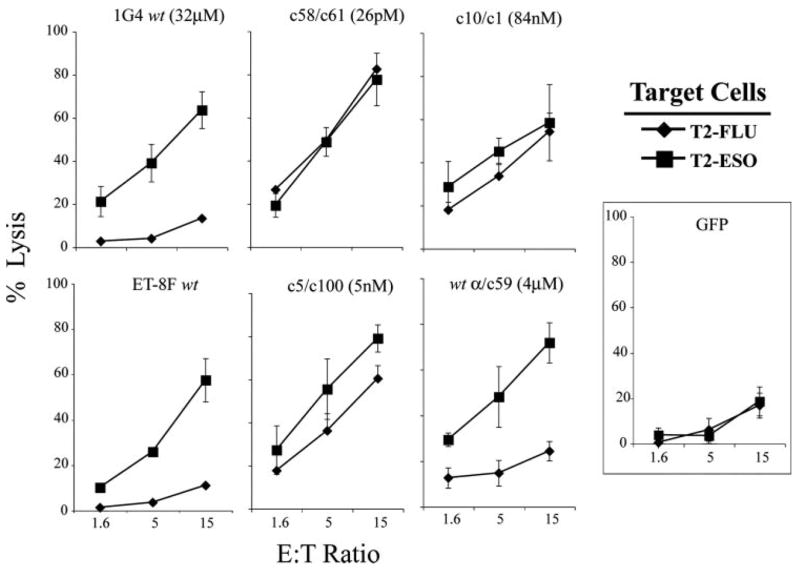
Nonspecific lysis of high-affinity TCR-transfected CD8+ T cells. Purified CD8+ T cells were electroporated with IVT TCR RNA for wt 1G4, wt ET-8F, and four mutated TCRs; c58/c61 (26 pM), c5/c100 (5 nM), c10/c1 (84 nM), or wt α/c59 (4 μM) TCR. T cells were cocultured with 51Cr-labeled peptide-pulsed T2 cells; 1 μM NY-ESO-1 or 1 μM Flu-MP and the amount of lysis was determined after 4 h. GFP RNA-transfected cells served as control. E:T was as shown; data are representative of two experiments.
CD8 molecules affect the cross-reactivity of high-affinity TCR-expressing T cells
The CD8 coreceptor protein enhances the formation of stable TCR-pMHC complex as an active participant in the T cell recognition complex (27, 28) To examine the influence of CD8 molecules on the specificity of T cell activation observed with the 1G4 TCR variants, studies were conducted using purified populations of CD8+ and CD4+ T cells. In contrast to results in CD8+ T cells, CD4+ T cells engineered with c5/c100 (5 nM) and c10/c1 (84 nM) TCRs were Ag specific as evidenced by both IFN-γ and IL-2 production upon stimulation with NY-ESO-1-pulsed but not with MART-1 or gp100 peptide-pulsed T2 cells (Fig. 3A). As observed with the transfected CD8+ T cells, CD4+ T cells that expressed the highest affinity c58/c61 (26 pM) TCR demonstrated cross-reactivity as evidenced by recognition of target cells pulsed with control peptides (Fig. 3A). As previously noted, the secretion of IFN-γ by CD8+ T cells expressing intermediate-affinity c10/c1 (84 nM) and c12/c2 (450 nM) TCRs were significantly less reactive than the CD8+ T cells expressing 1G4 wt (Fig. 3A), but these same TCRs transfected into CD4+ T cells secreted both IFN-γ and IL-2 at higher amounts than when 1G4 wt TCR was transfected into CD4+ T cells (Fig. 3A, lower panels).
FIGURE 3.
Specificity of high-affinity TCR-transfected CD4+ T cells. A, CD8+ or CD4+ purified T cells were electroporated with IVT RNA from the TCRs as indicated and cocultured with T2 pulsed with serially diluted NY-ESO-1 peptide, or 100 nM MART-1 or gp100 peptide. IFN-γ production for both CD8+ and CD4+ cells, and IL-2 production by CD4+ cells is shown (nd, not determined; data are representative of three experiments). B, TCR-transfected CD4+ T cells were cocultured with 51Cr-labeled peptide-pulsed T2 cells; 1 μM NY-ESO-1 or 1 μM gp100 and the amount of lysis was determined after 4 h. E:T was as shown. GFP IVT RNA-transfected cells served as control; data are representative of two experiments.
51Cr-release cell lysis assays were conducted to test whether CD4+ T cells expressing these high-affinity TCRs were capable of specifically killing target cells. CD4+ T cells transfected with the 1G4 wt TCR, as well as those transfected with the intermediate-affinity TCRs, specifically lysed T2 cells pulsed with the NY-ESO-1 peptide, whereas those transfected with the high-affinity c58/c61 TCR lysed target cells pulsed with both the NY-ESO-1 and control gp100 peptide (Fig. 3B).
To test whether partial inactivation of CD8 function could reduce this TCR cross-reactivity, TCRs were transfected into CD8+ T cells and cocultured with targets in the presence of CD8-specific Abs (Fig. 4A). In the presence of anti-CD8 Ab the TCR-transfected T cells were still capable of responding to NY-ESO-1 peptide-pulsed T2 cells but to a reduced level compared with the control Ab (or no Ab) groups. To further examine the role of CD8 molecules in the nonspecific activation observed in cells transfected with intermediate- and high-affinity TCRs, CD4+ T cells that had been retrovirally transduced with constructs encoding the 1G4 TCR variants were transfected with IVT RNAs encoding either the CD8 α-chain alone or were cotransfected with RNAs encoding both the CD8 α- and β-chains. The results indicated that CD4+ T cells expressing TCRs c5/c100 (5 nM) and c1/c10 (84 nM), that were then transfected with the CD8 α- and β-chain constructs, released significant levels of IFN-γ in response to target cells pulsed with both the specific NY-ESO-1 and the control gp100 peptides (Fig. 4B).
FIGURE 4.
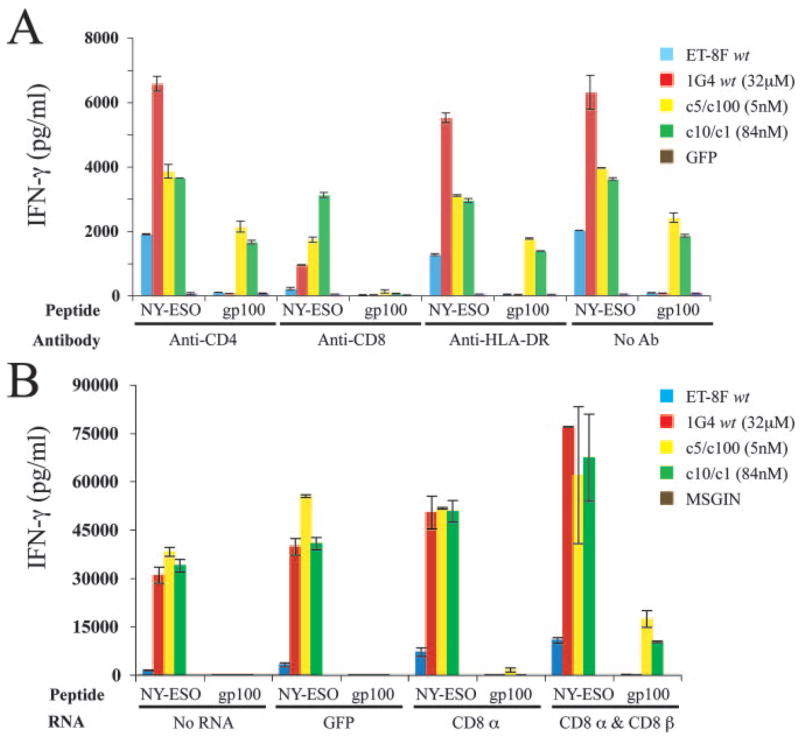
Effect of CD8 coreceptor on T cell activation of high-affinity TCR-transfected T cells. A, IFN-γ production by TCR, or GFP control, transfected CD8+ T cells were incubated with Abs as indicated and then cocultured with 100 nM NY-ESO-1 or gp100 peptide-pulsed T2 cells. Cytokine levels determined following overnight coculture; data are representative of two experiments. B, IFN-γ production by CD4+ T cells retrovirally transduced with TCR genes then electroporated with IVT RNA for CD8α, CD8α/CD8β or GFP. Engineered CD4+ T cells were cocultured overnight with peptide-pulsed T2 cells (100 nM NY-ESO-1 or gp100) and cytokine levels were determined. GFP-expressing retroviral vector MSGIN served as control.
Enhanced recognition of tumor lines by CD4+ T cells expressing high-affinity TCR variants
One of the motivations for generating these high-affinity TCRs was their potential application in the T cell adoptive immunotherapy of cancer patients. To this end, we tested the recognition of a panel of melanoma cell lines by the variant TCRs transfected into CD8+ or CD4+ T cells. CD8+ T cells transfected with 1G4 wt (32 μM) and c12/c2 (450 nM) TCRs recognized only the melanoma cell lines that are positive for both HLA-A2 and NY-ESO-1 protein, while the other three variant TCRs tested; c58/c61 (26 pM), c5/c100 (5 nM), and c10/c1 (84 nM) recognized both NY-ESO-1 protein positive and negative tumor lines (Fig. 5A).
FIGURE 5.
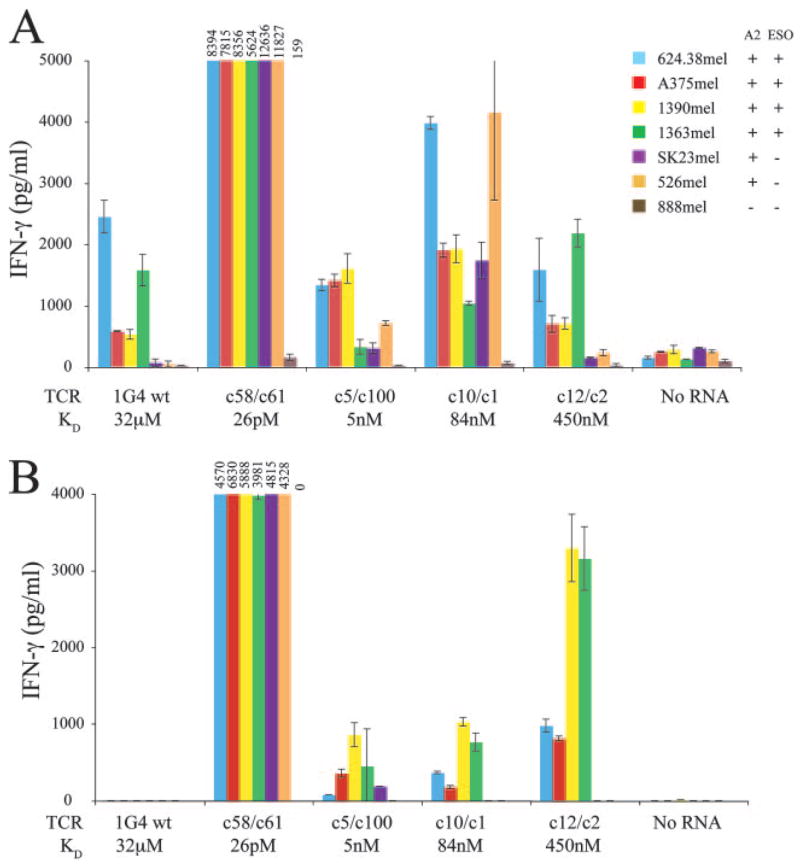
Increased tumor cell line recognition specificity by high-affinity TCR-transfected CD4+ T cells. Purified CD8+ (A) and CD4+ (B) T cells were electroporated with IVT TCR RNA for 1G4 wt, three mutant high-affinity TCRs; c58/c61 (26 pM), c5/c100 (5 nM), and c10/c1 (84 nM), or the medium-affinity c12/c2 (450 nM) TCR. T cells were cocultured with tumor cell lines overnight and production of IFN-γ determined. HLA-A2 and NY-ESO-1 expression by the melanoma cell lines were as indicated. Graphs truncated as shown with absolute values of cytokine production for TCR c58/c61 shown above each bar; data are representative of four experiments.
The CD4+ T cells transfected with the 1G4 wt TCR did not recognize any of the melanoma cell lines, presumably as a result of the relatively low levels of endogenously peptide presented on the surface of these target cells (Fig. 5B). In contrast, CD4+ T cells expressing the intermediate-affinity c12/c2 (450 nM) and c10/c1 (84 nM) TCRs recognized the four tumor targets expressing both HLA-A2 and NY-ESO-1, but failed to recognize HLA-A2 targets that did not express NY-ESO-1, indicating that they recognized tumor targets in an HLA-restricted, Ag-specific manner. The CD4+ T cells expressing the c5/c100 (5 nM) and c58/c61 (26 pM) TCRs both recognized the HLA-A2+/NY-ESO-1-negative melanoma line SK23 mel, while c58/c61 additionally recognized a second NY-ESO-1-negative line, 526 mel (both TCRs failed to recognize the HLA-A2−/NY-ESO-1-negative melanoma line 888mel).
Investigation into the basis of cellular cross-reactivity
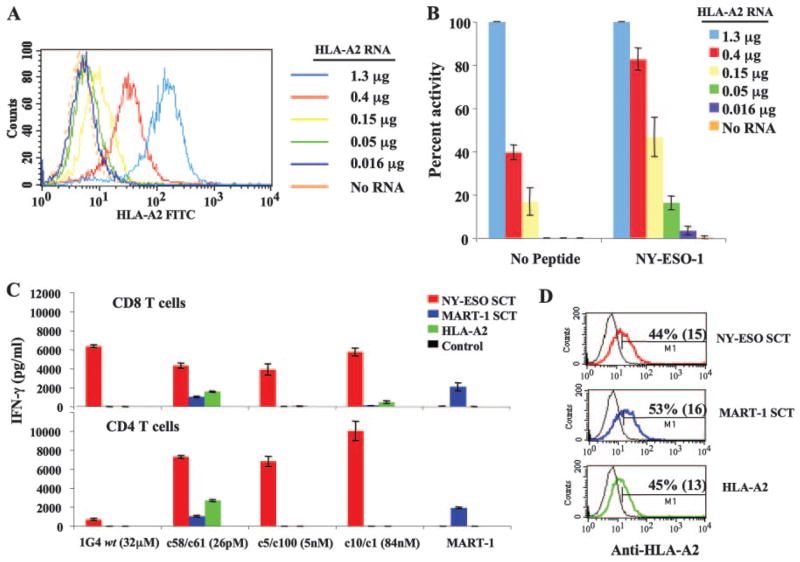
The observation that RCCs that expressed relatively low levels of HLA-A2 failed to stimulate cross-reactive cytokine production by some of the high-affinity TCR-transfected CD8+ T cells (Table II) suggested that HLA-A2 expression levels on stimulating cells affected the specific activation of these T cells. To further test this hypothesis, varying amounts of IVT RNA-encoding HLA-A2 were transfected into the HLA-A2-negative cell line C1R (Fig. 6A). Using these cells as targets, we demonstrated that TCR c10/c1 (84 nM) transfected CD8+ T cells manifest nonspecific T cell activation that was directly correlated with the HLA-A2 expression level (Fig. 6B).
FIGURE 6.
Density of MHC on the surface of APC affects the specificity of recognition by high-affinity TCR-expressing T cells. A, Flow cytometry analysis of HLA-A2 expression on C1R cells transfected with increasing amount of HLA-A2 IVT RNA. B, IFN-γ production by high-affinity TCR c10/c1 (84 nM) IVT RNA-transfected CD8+ T cells incubated with NY-ESO-1 peptide-pulsed or nonpulsed C1R cells expressing HLA-A2 at different levels. Data normalized to 100% for 1.3 μg of HLA-A2 RNA (values; 1234 pg/ml no peptide, 2553 pg/ml NY-ESO-1 peptide, representative of two experiments). C, IFN-γ production by purified CD8+ or CD4+ T cells cocultured with NY-ESO-1 or MART-1 SCT IVT RNA-transfected melanoma cell line 888mel with HLA-A2 IVT RNA or no RNA controls. D, HLA-A2 expression of SCT or HLA-A2 RNA-transfected 888mel was shown (percentage of stained cells are shown with mean fluorescent intensity in parentheses). Data representative of three experiments.
The cross-reactivity displayed by high-affinity TCR-transfected CD8 T cells suggested that high-affinity TCRs could exhibit increased binding to some undefined endogenous Ag(s). To investigate this possibility, we transfected target cells with single-chain peptide-β2 microglobulin-MHC trimers (SCT), which results in the preferential expression of a single peptide in association with the linked HLA-A2 molecule (25). HLA-A2− melanoma cell line 888mel was transfected, and similar levels of SCT expression were observed on the cell surface (Fig. 6D). The 888mel expressing a SCT for NY-ESO-1 p157–165 peptide was efficiently recognized by both 1G4 wt and by three high-affinity TCR-transfected CD8+ T cells (Fig. 6C). In CD4+ cells, the three mutated TCRs produced similar levels of cytokine when cultured with the NY-ESO-1 SCT-expressing cells, while the wt 1G4 TCR demonstrated minimal reactivity. Cells transfected with the c5/c100 (5 nM) and c10/c1 (84 nM) TCRs did not recognize control-transfected cells expressing MART-1 SCT or HLA-A2 alone in either CD8+ or CD4+ cells. Nonspecific activation was observed for the highest affinity TCR c58/c61 (26 pM) in both CD8+ and CD4 T+ cells stimulated with both MART-1 SCT and HLA-A2 alone (Fig. 6C), but IFN-γ secretion was always less with the MART-1 SCT than with the HLA-A2-transfected cells. This suggests the existence of endogenous agonist peptide-HLA-A2 Ag(s) that were recognized by the very highest affinity TCR c58/c61 (26 pM) in the high-avidity cell-to-cell context.
Discussion
The results from our present study support the hypothesis that raising the affinity of a TCR by directing the free energy of binding toward the MHC helices leads to a loss in peptide Ag specificity as may be seen in negative selection in the thymus (14, 29). Previous study using a soluble form of the highest affinity TCR, c58/c61 (26 pM), showed that this TCR bound to the surface of T2 APC cells in a totally MHC-specific and peptide-dependent manner (18). Our pMHC-tetramer-staining experiments also demonstrated specific binding to CD8+ T cells expressing this TCR (Fig. 1) and yet there was a near complete lose of specificity when this same TCRs was put in the context of a native T cell (Table I). This dramatic difference in sensitivity and specificity between soluble TCR molecules binding to target cells and T cell-mediated recognition may be due to the cumulative effects of the avidity of multiple TCR-pMHC interactions, as well as to other additional interactions between T cells and target cells.
Our data indicate that the density of MHC molecules on target cell and the presence of the CD8 coreceptor on the effector T cell were important variables in the activation of T cells transfected with these TCRs. Multiple studies have indicated that CD8 molecules play an important role in T cell recognition by stabilizing binding of the TCR to pMHC complexes, as well as by enhancing intracellular signaling, and lowering the threshold for T cell activation (27, 30–32). In the current study, incubation with an anti-CD8 Ab increased the specificity of CD8+ T cell transfected with the high-affinity TCRs (Fig. 4A). In addition, CD4+ T cells that expressed the high-affinity 1G4 TCRs specifically recognized NY-ESO-1+ /HLA-A2+ target cells, where CD8+ T cells expressing the same TCR demonstrated a lack of specificity (Fig. 5). These observations provide evidence that the CD8 molecule plays an important role in mediating the cross-reactivity observed with the high-affinity 1G4 TCR.
The finding that enhanced Ag-specific recognition and cross-reactivity to self Ags, or even peptide-independent T cell triggering, could be achieved by increasing MHC class I density on target cells (Table II) indicates that TCR avidity for the MHC molecule is a significant arbiter of T cell specificity, and that TCR affinities for MHC and peptide maybe interchangeable (33, 34). This is of biological importance, because different sites and cells in the thymus express MHC at different levels to effect positive and negative selections (35, 36). The study by Sandberg et al. (34) on the immunization of β2m-deficient transgenic mice, which expressed subnormal levels of MHC class I, with an H-2Db-restricted immunodominant lymphocytic choriomeningitis virus gp33 peptide, showed that resulting CTL were specific for the gp33 epitope when loaded onto cells with low MHC class I expression. However, unlike gp33-specific CTLs generated from normal MHC class I-expressing mice, these CTLs also killed cells expressing high levels of self-MHC class I in the absence of specific peptide. Their study identified peptide-independent triggering of a CTL clone with high avidity for self-MHC (34).
A recent crystallographic study by Colf et al. (37) describes the molecular basis for alloreactivity of the 2C TCR and suggest that the main arbitrator of foreign MHC recognition by this TCR was associated with changes in the nature of the interaction between the TCR and the pMHC. A high-affinity version of the 2C TCR was shown to make distinct contacts with the foreign MHC, and such divergent interactions may also be associated with the cross-reactivity observed with the high-affinity receptors studied herein. Increased avidity for MHC can compensate for the lack of avidity for peptide, suggesting that these binding force are functionally interchangeable and can be considered separately (34, 37). Taken together, our results and the reports by Colf et al. and Sandberg et al. (34, 37) support the model that T cell recognition is based on the total avidity contributed by TCR affinity for MHC, affinity for peptide, the number of pMHC complexes, and that high-affinity TCR may use different combinations of these interactions to mediate superior recognition while at the same time, displaying the properties of cross-reactivity. The presence of the CD4 or CD8 coreceptors could amplify cross-reactivity by stabilizing the TCR: MHC interactions.
NY-ESO-1, a member of the cancer/testis class of Ags (38, 39), is an attractive target for tumor immunotherapy because it is expressed in a high percentage (20–80% at the RNA level) of common tumors, including cancers of the breast, lung, bladder, liver, prostate, and ovary. Our previous study showed that human primary T lymphocytes transduced with TCR α- and β-chains isolated from CTL clone ET-8F specific for HLA-A2-restricted NY-ESO-1 p157–165 could recognize and kill diverse human tumor cell lines but the tumor recognition was weak (23). Theoretically, TCRs with high-ligand affinity are predicted to increase the avidity of the T cell thereby requiring lower levels of Ags for efficient target cell recognition. Therefore, an approach to immunotherapy of cancers using TCR-transfected T cells may be enhanced by using high-affinity TCRs that convey the T cells with enhanced efficacy for eradicating tumors. TCRs c12/c2 (450 nM) and wt/c59 (4 μM), which have lower affinities relative to other TCRs generated in this study, are specific for NY-ESO-1, p157–165 peptide and showed superior T cell activities over their wt counterparts 1G4 and ET-8F in transfected CD4+ T cells (Figs. 3 and 5). Unlike their MHC class II-restricted CD4 counterpart, high-affinity MHC class I-restricted TCR-expressing CD4+ T cells may provide additional power to eradicate tumor cells by their direct strong tumor recognition and killing abilities. We have recently shown that the adoptive transfer of autologous PBLs transduced with retroviruses encoding a wt anti-MART-1/HLA-A2-specific TCR can mediate durable cancer regression in some patients (40), and these anti-NY-ESO-1 high-affinity TCRs, when transferred into CD4+ T cells may have similar potential for use in clinical cancer immunotherapy.
Footnotes
This work was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health.
Abbreviations used in this paper: pMHC, peptide-MHC; wt, wild type; RCC, renal cell carcinoma cell line; IVT, in vitro transcribed; βsm, β2-microglobulin; IRES, internal ribosomal entry site; TIL, tumor-infiltrating lymphocyte; MSCV, murine stem cell virus; SCT, single chain trimer.
Disclosures
Alan D. Bennett, Yi Li, Peter E. Molloy, Steven M. Dunn, and Bent K. Jakobsen are employed by MediGene Ltd., Oxon, UK.
References
- 1.Ashton-Rickardt PG, Bandeira A, Delaney JR, Van Kaer L, Pircher HP, Zinkernagel RM, Tonegawa S. Evidence for a differential avidity model of T cell selection in the thymus. Cell. 1994;76:651–663. doi: 10.1016/0092-8674(94)90505-3. [DOI] [PubMed] [Google Scholar]
- 2.Goldrath AW, Bevan MJ. Selecting and maintaining a diverse T-cell repertoire. Nature. 1999;402:255–262. doi: 10.1038/46218. [DOI] [PubMed] [Google Scholar]
- 3.Davis MM, Boniface JJ, Reich Z, Lyons D, Hampl J, Arden B, Chien Y. Ligand recognition by αβ T cell receptors. Annu Rev Immunol. 1998;16:523–544. doi: 10.1146/annurev.immunol.16.1.523. [DOI] [PubMed] [Google Scholar]
- 4.Williams CB, Engle DL, Kersh GJ, Michael White J, Allen PM. A kinetic threshold between negative and positive selection based on the longevity of the T cell receptor-ligand complex. J Exp Med. 1999;189:1531–1544. doi: 10.1084/jem.189.10.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackman M, Yague J, Kubo R, Gay D, Coleclough C, Palmer E, Kappler J, Marrack P. The T cell repertoire may be biased in favor of MHC recognition. Cell. 1986;47:349–357. doi: 10.1016/0092-8674(86)90591-x. [DOI] [PubMed] [Google Scholar]
- 6.Zerrahn J, Held W, Raulet DH. The MHC reactivity of the T cell repertoire prior to positive and negative selection. Cell. 1997;88:627–636. doi: 10.1016/s0092-8674(00)81905-4. [DOI] [PubMed] [Google Scholar]
- 7.Brocker T. Survival of mature CD4 T lymphocytes is dependent on major histocompatibility complex class II-expressing dendritic cells. J Exp Med. 1997;186:1223–1232. doi: 10.1084/jem.186.8.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirberg J, Berns A, von Boehmer H. Peripheral T cell survival requires continual ligation of the T cell receptor to major histocompatibility complex-encoded molecules. J Exp Med. 1997;186:1269–1275. doi: 10.1084/jem.186.8.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Markiewicz MA, Girao C, Opferman JT, Sun J, Hu Q, Agulnik AA, Bishop CE, Thompson CB, Ashton-Rickardt PG. Long-term T cell memory requires the surface expression of self-peptide/major histocompatibility complex molecules. Proc Natl Acad Sci USA. 1998;95:3065–3070. doi: 10.1073/pnas.95.6.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alam SM, Travers PJ, Wung JL, Nasholds W, Redpath S, Jameson SC, Gascoigne NR. T-cell-receptor affinity and thymocyte positive selection. Nature. 1996;381:616–620. doi: 10.1038/381616a0. [DOI] [PubMed] [Google Scholar]
- 11.Zeh HJ, 3rd, Perry-Lalley D, Dudley ME, Rosenberg SA, Yang JC. High avidity CTLs for two self-antigens demonstrate superior in vitro and in vivo antitumor efficacy. J Immunol. 1999;162:989–994. [PubMed] [Google Scholar]
- 12.Alexander-Miller MA, Leggatt GR, Berzofsky JA. Selective expansion of high- or low-avidity cytotoxic T lymphocytes and efficacy for adoptive immunotherapy. Proc Natl Acad Sci USA. 1996;93:4102–4107. doi: 10.1073/pnas.93.9.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rudolph MG, I, Wilson A. The specificity of TCR/pMHC interaction. Curr Opin Immunol. 2002;14:52–65. doi: 10.1016/s0952-7915(01)00298-9. [DOI] [PubMed] [Google Scholar]
- 14.Holler PD, Holman PO, Shusta EV, O’Herrin S, Wittrup KD, Kranz DM. In vitro evolution of a T cell receptor with high affinity for peptide/MHC. Proc Natl Acad Sci USA. 2000;97:5387–5392. doi: 10.1073/pnas.080078297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holler PD, Lim AR, Cho BK, Rund LA, Kranz DM. CD8+ T cell transfectants that express a high affinity T cell receptor exhibit enhanced peptide-dependent activation. J Exp Med. 2001;194:1043–1052. doi: 10.1084/jem.194.8.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holler PD, Chlewicki LK, Kranz DM. TCRs with high affinity for foreign pMHC show self-reactivity. Nat Immunol. 2003;4:55–62. doi: 10.1038/ni863. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Moysey R, Molloy PE, Vuidepot AL, Mahon T, Baston E, Dunn S, Liddy N, Jacob J, Jakobsen BK, Boulter JM. Directed evolution of human T-cell receptors with picomolar affinities by phage display. Nat Biotechnol. 2005;23:349–354. doi: 10.1038/nbt1070. [DOI] [PubMed] [Google Scholar]
- 18.Purbhoo MA, Sutton DH, Brewer JE, Mullings RE, Hill ME, Mahon TM, Karbach J, Jager E, Cameron BJ, Lissin N, et al. Quantifying and imaging NY-ESO-1/LAGE-1-derived epitopes on tumor cells using high affinity T cell receptors. J Immunol. 2006;176:7308–7316. doi: 10.4049/jimmunol.176.12.7308. [DOI] [PubMed] [Google Scholar]
- 19.Topalian SL, Solomon D, Rosenberg SA. Tumor-specific cytolysis by lymphocytes infiltrating human melanomas. J Immunol. 1989;142:3714–3725. [PubMed] [Google Scholar]
- 20.Wang QJ, Hanada K, Perry-Lalley D, Bettinotti MP, Karpova T, Khong HT, Yang JC. Generating renal cancer-reactive T cells using dendritic cells (DCs) to present autologous tumor. J Immunother. 2005;28:551–559. doi: 10.1097/01.cji.0000175495.13476.1f. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Y, Zheng Z, Cohen CJ, Gattinoni L, Palmer DC, Restifo NP, Rosenberg SA, Morgan RA. High-efficiency transfection of primary human and mouse T lymphocytes using RNA electroporation. Mol Ther. 2006;13:151–159. doi: 10.1016/j.ymthe.2005.07.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scholten KB, Kramer D, Kueter EW, Graf M, Schoedl T, Meijer CJ, Schreurs MW, Hooijberg E. Codon modification of T cell receptors allows enhanced functional expression in transgenic human T cells. Clin Immunol. 2006;119:135–145. doi: 10.1016/j.clim.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Zhao Y, Zheng Z, Robbins PF, Khong HT, Rosenberg SA, Morgan RA. Primary human lymphocytes transduced with NY-ESO-1 antigen-specific TCR genes recognize and kill diverse human tumor cell lines. J Immunol. 2005;174:4415–4423. doi: 10.4049/jimmunol.174.7.4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morgan RA, Dudley ME, Yu YY, Zheng Z, Robbins PF, Theoret MR, Wunderlich JR, Hughes MS, Restifo NP, Rosenberg SA. High efficiency TCR gene transfer into primary human lymphocytes affords avid recognition of melanoma tumor antigen glycoprotein 100 and does not alter the recognition of autologous melanoma antigens. J Immunol. 2003;171:3287–3295. doi: 10.4049/jimmunol.171.6.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu YY, Netuschil N, Lybarger L, Connolly JM, Hansen TH. Cutting edge: single-chain trimers of MHC class I molecules form stable structures that potently stimulate antigen-specific T cells and B cells. J Immunol. 2002;168:3145–3149. doi: 10.4049/jimmunol.168.7.3145. [DOI] [PubMed] [Google Scholar]
- 26.Hughes MS, Yu YY, Dudley ME, Zheng Z, Robbins PF, Li Y, Wunderlich J, Hawley RG, Moayeri M, Rosenberg SA, Morgan RA. Transfer of a TCR gene derived from a patient with a marked antitumor response conveys highly active T-cell effector functions. Hum Gene Ther. 2005;16:457–472. doi: 10.1089/hum.2005.16.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia KC, Scott CA, Brunmark A, Carbone FR, Peterson PA, Wilson IA, Teyton L. CD8 enhances formation of stable T-cell receptor/MHC class I molecule complexes. Nature. 1996;384:577–581. doi: 10.1038/384577a0. [DOI] [PubMed] [Google Scholar]
- 28.Purbhoo MA, Boulter JM, Price DA, Vuidepot AL, Hourigan CS, Dunbar PR, Olson K, Dawson SJ, Phillips RE, Jakobsen BK, et al. The human CD8 coreceptor effects cytotoxic T cell activation and antigen sensitivity primarily by mediating complete phosphorylation of the T cell receptor ζ chain. J Biol Chem. 2001;276:32786–32792. doi: 10.1074/jbc.M102498200. [DOI] [PubMed] [Google Scholar]
- 29.Bevan MJ. In thymic selection, peptide diversity gives and takes away. Immunity. 1997;7:175–178. doi: 10.1016/s1074-7613(00)80520-8. [DOI] [PubMed] [Google Scholar]
- 30.Davis SJ, van der Merwe PA. TCR triggering: co-receptor-dependent or -independent? Trends Immunol. 2003;24:624–626. doi: 10.1016/j.it.2003.10.009. author reply 626–627. [DOI] [PubMed] [Google Scholar]
- 31.Wooldridge L, van den Berg HA, Glick M, Gostick E, Laugel B, Hutchinson SL, Milicic A, Brenchley JM, Douek DC, Price DA, Sewell AK. Interaction between the CD8 coreceptor and major histo-compatibility complex class I stabilizes T cell receptor-antigen complexes at the cell surface. J Biol Chem. 2005;280:27491–27501. doi: 10.1074/jbc.M500555200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arcaro A, Gregoire C, Bakker TR, Baldi L, Jordan M, Goffin L, Boucheron N, Wurm F, van der Merwe PA, Malissen B, Luescher IF. CD8β endows CD8 with efficient coreceptor function by coupling T cell receptor/CD3 to raft-associated CD8/p56lck complexes. J Exp Med. 2001;194:1485–1495. doi: 10.1084/jem.194.10.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fooksman DR, Gronvall GK, Tang Q, Edidin M. Clustering class I MHC modulates sensitivity of T cell recognition. J Immunol. 2006;176:6673–6680. doi: 10.4049/jimmunol.176.11.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandberg JK, Karre K, Glas R. Recognition of the major histo-compatibility complex restriction element modulates CD8+ T cell specificity and compensates for loss of T cell receptor contacts with the specific peptide. J Exp Med. 1999;189:883–894. doi: 10.1084/jem.189.6.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delaney JR, Sykulev Y, Eisen HN, Tonegawa S. Differences in the level of expression of class I major histocompatibility complex proteins on thymic epithelial and dendritic cells influence the decision of immature thymocytes between positive and negative selection. Proc Natl Acad Sci USA. 1998;95:5235–5240. doi: 10.1073/pnas.95.9.5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang SJ, Ahn S, Park CS, Holmes KL, Westrup J, Chang CH, Kim MG. The quantitative assessment of MHC II on thymic epithelium: implications in cortical thymocyte development. Int Immunol. 2006;18:729–739. doi: 10.1093/intimm/dxl010. [DOI] [PubMed] [Google Scholar]
- 37.Colf LA, Bankovich AJ, Hanick NA, Bowerman NA, Jones LL, Kranz DM, Garcia KC. How a single T cell receptor recognizes both self and foreign MHC. Cell. 2007;129:135–146. doi: 10.1016/j.cell.2007.01.048. [DOI] [PubMed] [Google Scholar]
- 38.Scanlan MJ, Gure AO, Jungbluth AA, Old LJ, Chen YT. Cancer/testis antigens: an expanding family of targets for cancer immunotherapy. Immunol Rev. 2002;188:22–32. doi: 10.1034/j.1600-065x.2002.18803.x. [DOI] [PubMed] [Google Scholar]
- 39.Scanlan MJ, Simpson AJ, Old LJ. The cancer/testis genes: review, standardization, and commentary. Cancer Immun. 2004;4:1. [PubMed] [Google Scholar]
- 40.Morgan RA, Dudley ME, Wunderlich JR, Hughes MS, Yang JC, Sherry RM, Royal RE, Topalian SL, Kammula US, Restifo NP, et al. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. 2006;314:126–129. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]