Abstract
This article presents a novel methodology for the analysis of ethanolamine glycerophospholipid (PE) and lysoPE molecular species directly from lipid extracts of biological samples. Through brief treatment of lipid extracts with fluorenylmethoxylcarbonyl (Fmoc) chloride, PE and lysoPE species were selectively derivatized to their corresponding carbamates. The reaction solution was infused directly into the ion source of an electrospray ionization mass spectrometer after appropriate dilution. The facile loss of the Fmoc moiety dramatically enhanced the analytic sensitivity and allowed the identification and quantitation of low-abundance molecular species. A detection limitation of attomoles (amoles) per microliter for PE and lysoPE analysis was readily achieved using this technique (at least a 100-fold improvement from our previous method) with a >15,000-fold dynamic range. Through intrasource separation and multidimensional mass spectrometry array analysis of derivatized species, marked improvements in signal-to-noise ratio, molecular species identification, and quantitation can be realized. The procedure is both simple and effective and can be extended to analyze many other lipid classes or other cellular metabolites by adjustments in specific derivatization conditions.jlr Thus, through judicious derivatization, a new dimension exploiting specific functional reactivities in each lipid class can be used in conjunction with shotgun lipidomics to penetrate farther into the low-abundance regime of cellular lipidomes.
Keywords: electrospray ionization-mass spectrometry, fluorenylmethoxylcarbonyl derivatization, lipidome, lipidomics, lysophosphatidylethanolamine, multidimensional mass spectrometry, plasmalogen
Recently, a powerful technique for the direct analysis of global cellular lipidomes [i.e., shotgun lipidomics using intrasource separation and multidimensional electrospray ionization-mass spectrometry (ESI-MS)] has emerged (see 1-3 for recent reviews). In shotgun lipidomics, lipid classes in a crude lipid extract are first separated at the ion source (i.e., intrasource separation) through judicious selection of ion-pairing reagents based on the electrical properties of the lipid classes described previously (2, 4). The basic principles of ESI intrasource separation of lipids extend the charge separation principle of ESI, which is analogous to an electrophoretic cell, as described previously (5, 6). The involvement of ion pairing of intrasource separation is also analogous to ion-exchange chromatography, which facilitates lipid class separations (7). For electrically neutral lipids, selectivity during separations is accomplished through differential ion ratios of lipid classes in the source, depending upon the ion-pairing conditions used.
Cellular lipidomes contain thousands to tens of thousands of individual molecular species of lipids. Most of these biological lipid species are linear combinations of aliphatic chains, backbones, and/or head groups, each of which represents a building block of the molecular species under consideration. Therefore, the identification of individual molecular species of the cellular lipidome can be achieved through the determination of the associated building blocks of cellular lipids. Shotgun lipidomics fulfills this task through multidimensional ESI-MS array analysis under various instrumental conditions, such as changes in ionization conditions (e.g., source temperature and spray voltage) and in fragmentation conditions [e.g., collision gas pressure, collision energy, collision gas, mass loss in neutral loss (NL) mode, and monitored ions in precursor ion (PI) mode] (3). Besides the basic two-dimensional (2D) mass spectral unit, in which the second dimension is constructed with specific lipid building blocks, each series of ramped changes in instrumental condition facilitates the generation of an additional dimension, each of which can potentially be used in multidimensional mass spectrometric analyses (3).
Quantitation in shotgun lipidomics is performed by a two-step process (3, 8, 9). First, the abundant molecular species in a class of polar lipids are quantitated by direct comparison of the ion peak intensities with that of a preselected amount of internal standard for the lipid class in the first-dimensional mass spectrum after correction for the 13C isotopomer differences, as described previously (2, 10, 11). The foundation of this measurement is based on the observation that the ion abundance of a lipid molecular species is linearly correlated with its concentration in the sprayed solution in the low-concentration region. This observation has been validated by many independent studies (3, 9, 12-18). Next, the determined concentrations of these abundant molecular species are used as endogenous standards, in addition to the preselected internal standard, for ratiometric comparisons to quantitate or refine the mass content of low-abundance individual molecular species from at least one of the representative tandem mass scans for the lipid class of interest. By using tandem mass scans, the baseline fluctuations caused by chemical noise can be reduced dramatically. Thus, the measure of the linear dynamic range of ion peak intensity ratios between the selected internal standard and the unknowns of interest is expanded to penetrate into the low-abundance region of lipid molecular species directly from organic extracts. Because of the effects of differential fragmentation kinetics on different molecular species in a class (2), multiple standards, representative of the different physical properties (subclass, acyl chain length, and degree of unsaturation), have to be selected for quantitation based on tandem MS (MS/MS), as demonstrated previously (19-23). Thus, the determined set of individual constituents from step one represent the endogenous standards that are selected for their well-distributed changes in acyl chain length and unsaturation to appropriately cover the lipid class of interest in most cases. Of course, additional standards can be added if necessary, but in most cases, the naturally occurring distribution of acyl chain length and unsaturation is usually sufficient.
In shotgun lipidomics, molecular species containing a head group of phosphoethanolamine, such as ethanolamine glycerophospholipid (PE) and lysoPE, are analyzed after the addition of a small amount of LiOH in negative ion mode (2, 4). These molecular species carry a weakly zwitterionic head group and thus become anionic under alkaline conditions. Unfortunately, under these experimental conditions, many low-abundance PE molecular species and almost the entire lysoPE class are buried in the baseline noise. Moreover, those PE species having high molecular weight can potentially overlap with molecular species from other lipid class(es) (e.g., phosphatidylinositol) and are difficult to identify. Furthermore, at present, there is no highly sensitive and representative MS/MS method that can be used to profile very low-abundance PE and lysoPE molecular species under the experimental conditions developed. Therefore, quantitation of very low-abundance PE molecular species and all lysoPE species is often difficult by shotgun lipidomics using the aforementioned two-step procedure.
In this report, we describe a technique to identify and quantify PE and lysoPE molecular species from lipid extracts of biological tissues after a one-step derivatization in situ with 9-fluorenylmethoxylcarbonyl chloride (Fmoc-Cl) or other reagents. After derivatization, Fmoc-PE and Fmoc-lysoPE molecular species are rendered anionic and can be analyzed directly in the negative ion mode with enhanced sensitivity. Moreover, derivatization with Fmoc shifts PE molecular species out of the region where they potentially overlap with other lipid classes. Finally, product ion analysis of Fmoc-PE and Fmoc-lysoPE molecular species demonstrates a very abundant fragment corresponding to the facile NL of Fmoc. Thus, mass spectrometric analysis by NL of the Fmoc moiety from Fmoc-PE and Fmoc-lysoPE species can be used to identify and quantify lysoPE and very low-abundance PE molecular species. Collectively, this technique provides a new tool for shotgun lipidomics to penetrate farther into the low-abundance regime of PEs directly from organic extracts.
MATERIALS AND METHODS
Materials
Synthetic phospholipids, including phosphatidylcholine (1,2-dimyristoleoyl-sn-glycero-3-phosphocholine; 14:1-14:1 PtdCho), phosphatidylethanolamine (1,2-diarachidonoyl-sn-glycero-3-phosphoethanolamine; 20:4-20:4 PtdEtn), phosphatidylglycerol (1,2-dipentadecanoyl-sn-glycero-3-phosphoglycerol; 15:0-15:0 PtdGro), and 1-dimyristoyl-sn-glycero-3-phosphoethanolamine (14:0 lysoPE) (used as internal standards for the corresponding lipid classes), were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). Fmoc-Cl and dimethylaminopyridine (DMAP) were from Sigma-Aldrich Chemical Co. (St. Louis, MO). 2-(2-Naphthyl)acetyl chloride (NAC) was purchased from Ryan Scientific, Inc. (Isle of Palms, SC). All solvents used for sample preparation and for mass spectrometric analysis were obtained from Burdick and Jackson (Honeywell International, Inc., Burdick and Jackson, Muskegon, MI). Anhydrous chloroform was purchased from Fisher Scientific, Inc. (Philadelphia, PA).
Sample preparation
Male mice (C57BL/6, 4 months of age) were purchased from The Jackson Laboratory (Bar Harbor, ME). Mice were killed by inhalation of carbon dioxide before tissue collection. Mouse eyes were enucleated and hemisected at the ora serrata. The cornea, iris, lens, and vitreous were removed, and the retinas were detached from the eyecups mechanically. The retina lipids were extracted by the Bligh and Dyer procedure (24) with modifications as described previously (2, 4, 8). Briefly, to a small volume of mouse retina homogenate containing ∼100 μg of protein (from one pair of eyes obtained from a single mouse), internal standards including 20:4-20:4 PtdEtn (48.0 nmol/mg protein) and 14:0 lysoPtdEtn (1.0 nmol/mg protein) were added just before extraction. Lipids from each mouse retina sample were extracted against 1.8ml of 50 mM LiCl twice, back extracted against 1.8 ml of 10 mM LiCl twice, filtered with a 0.2 μm polytetrafluoroethylene syringe filter, and finally resuspended and stored in 1:1 (v/v) chloroform-methanol at an approximate concentration of 2 ml/mg protein.
Next, each of the lipid extracts was individually derivatized with Fmoc-Cl as follows. Each lipid extract in 100 μl of 1:1 (v/v) chloroform-methanol containing ∼10 nmol of PE was transferred to a test tube, and the solvents were evaporated under nitrogen gas. Freshly prepared Fmoc-Cl solution in anhydrous chloroform (100 μl) was added to the test tube to make a 1:1 molar ratio of PE to Fmoc-Cl, flushed with nitrogen gas, capped, and mixed with lipids by vortexing. When other molar ratios of PE to Fmoc-Cl were used as indicated, Fmoc-Cl solutions with corresponding concentrations were also prepared before the addition of 100 μl of Fmoc-Cl solution to the reaction vessels. The test tube was kept in the dark (wrapped with foil) at room temperature for the indicated times with occasional vortexing. The reaction solution was diluted 20-fold with 1:1 (v/v) chloroform-methanol before direct infusion into an ESI mass spectrometer for lipid analysis.
Samples that were derivatized with other reagents such as NAC were also prepared similarly. Briefly, freshly prepared NAC solution in anhydrous chloroform (100 μl) was added to the test tube to make a 1:1 molar ratio of PE to NAC and mixed with lipids by vortexing. When other molar ratios of PE to NAC were used, NAC solutions with corresponding concentrations were prepared. The reaction solution was diluted 20-fold with 1:1 (v/v) chloroform-methanol before direct infusion into an ESI mass spectrometer for lipid analysis.
ESI-MS of lipids
ESI-MS analyses were performed using a triple-quadrupole mass spectrometer (TSQ Quantum Ultra; ThermoFinnigan, San Jose, CA) equipped with an electrospray ion source, as described previously (4, 8). Typically, a 1 min period of signal averaging in the profile mode was used for each MS spectrum and up to a 2 min period of signal averaging was used for each MS/MS scan. For MS/MS analysis of Fmoc-PE, the collision gas pressure was set at 1.0 mTorr and a collision energy of 30 eV or indicated values was used for NL scanning of 222.2 u; alternatively, an energy of 25 eV was used for PI scanning of fatty acyl carboxylates. Other MS/MS analysis conditions were used as described previously (4). As usual, plasmenylethanolamine (PlsEtn) molecular species are distinguished from plasmanylethanolamine species by comparing the mass spectra acquired from the samples before and after acid treatment (25, 26). Alternatively, direct identification of PlsEtn molecular species in the positive ion mode is also possible (27, 28). It was found in this study (see Results) that PlsEtn species could be redundantly profiled by PI scanning of lysoPlsEtn ions using a collision energy of 30 eV. Identification of each ion peak corresponding to anionic lipids or Fmoc-PE species was performed in a 2D mass spectrometric array format as described previously (3, 4). Quantitation of each identified PE molecular species was conducted in a two-step procedure as described previously (3, 8, 9).
Miscellaneous
Quantitative data from biological samples were normalized to the protein content of the tissues, and all data are presented as means ± SD of samples from 10 animals. Protein concentration was determined with a BCA protein assay kit (Pierce, Rockford, IL) using BSA as a standard.
RESULTS
Analysis of lipids in conventional shotgun lipidomics
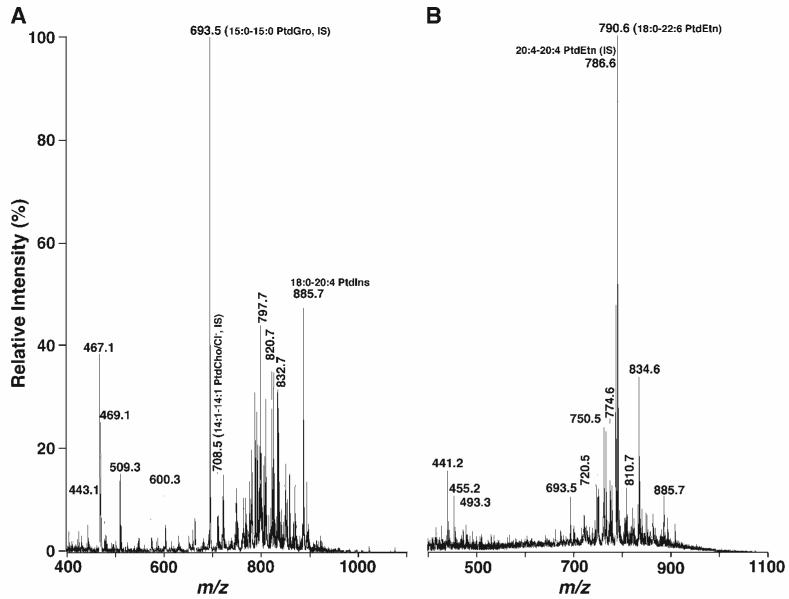
Because it is well known that the lipid profile of retinas is quite complicated (29, 30), we used lipid extracts of mouse retinas as a model system to demonstrate the power of in situ derivatization for the analysis of PE and lysoPE molecular species directly from a lipid extract. ESI-MS analysis of a diluted mouse retina lipid extract directly in the negative ion mode displayed many intense ion peaks between m/z 400 and 1100 (Fig. 1A). 2D MS analysis demonstrated that most of these ions belong to anionic lipids (e.g., 15:0-15:0 PtdGro at m/z 693.5, an internal standard for anionic lipid analysis, and 18:0-20:4 phosphatidylinositol at m/z 885.6), chlorinated choline glycerophospholipid species (chlorinated 14:1-14:1 PtdCho internal standard at m/z 708.6), and chlorinated cerebrosides (e.g., chlorinated per-deuterated N-octadecanoyl cerebroside at m/z 797.7, an internal standard) in the lipid extract (Fig. 2). The 2D mass spectrometric analysis also demonstrated some PE molecular species (e.g., 20:4-20:4 PtdEtn at m/z 786.6 and 18:0-22:6 PtdEtn at m/z 790.6) in modest abundance. Abundant molecular ions corresponding to PE molecular species in the identical diluted mouse retina lipid extract were also demonstrated in the negative ion ESI mass spectrum when the sample was analyzed after the addition of a small amount of LiOH before infusion of the diluted lipid extract (Fig. 1B).
Fig. 1.
Representative negative ion electrospray ionization-mass spectrometry (ESI-MS) spectra of a lipid extract from mouse retinas. Mouse retina lipid extracts were prepared by a modified method of Bligh and Dyer (24) as described in Materials and Methods. ESI mass spectra were acquired in negative ion mode by direct infusion of lipid solution after dilution to a total lipid concentration of ∼50 pmol/μl with 1:1 chloroform-methanol (v/v) (A) or from the identical diluted lipid solution after the addition of 50 nmol LiOH/mg protein (B). The indicated molecular species were identified by two-dimensional (2D) MS (see Fig. 2). IS, internal standard; PtdCho, phosphatidylcholine; PtdEtn, phosphatidylethanolamine; PtdGro, phosphatidylglycerol; PtdIns, phosphatidylinositol.
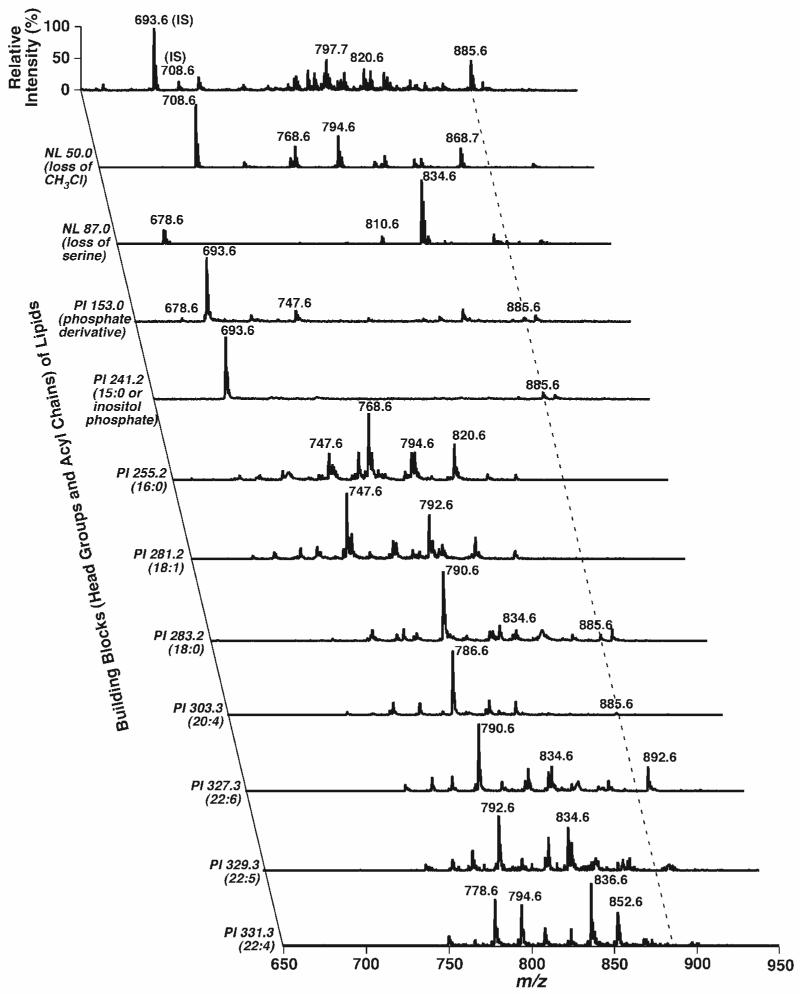
Fig. 2.
Representative 2D ESI mass spectra of a chloroform extract of mouse retinas in negative ion mode. A conventional ESI mass spectrum was acquired in the negative ion mode directly from a diluted mouse retina lipid extract (see Fig. 1A) before analysis of lipid building blocks in the second dimension by precursor ion (PI) scanning and neutral loss (NL) scanning as indicated. Each mass spectral scan was acquired as described previously (4). All mass spectral traces were displayed after normalization to the base peak in each spectrum. IS, internal standard; m:n, acyl chain containing m carbons and n double bonds.
ESI-MS analysis of Fmoc-derivatized lipids
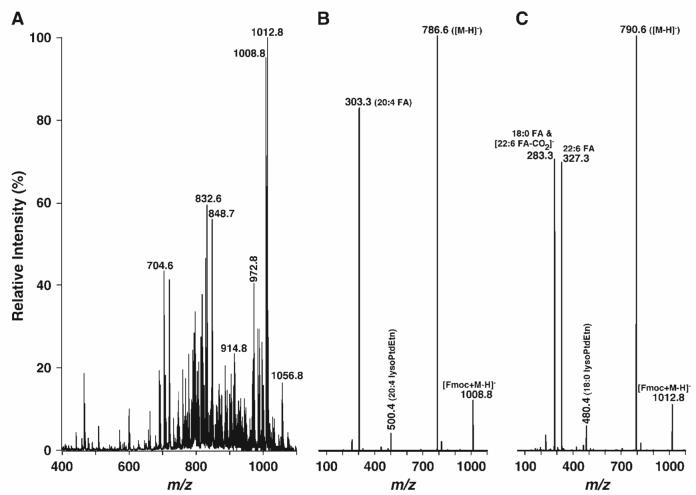
When the identical lipid extract was incubated with Fmoc-Cl at a 1:1 molar ratio of Fmoc-Cl and PE for as short as 5 min and then analyzed by ESI-MS under similar conditions, the negative ion ESI mass spectrum of lipids demonstrated a totally different ion profile (Fig. 3A). Many very abundant clusters of ion peaks corresponding to the Fmoc-PE molecular species appeared in the higher mass range (m/z > 900) (Fig. 3A), whereas the abundant ion peaks corresponding to anionic lipids in Fig. 1 appeared in low or modest abundance (Fig. 3A), such as the peaks at m/z 693.5 (corresponding to 15:0-15:0 PtdGro). Many additional ion peaks were also shown in the region between m/z 700 and 850, corresponding to the chlorinated choline glycerophospholipid or cerebroside molecular species, because of the increase in chloride concentration during Fmoc-Cl treatment.
Fig. 3.
Representative negative ion ESI-MS and product ion ESI mass spectra of a lipid extract of mouse retinas after derivatization with fluorenylmethoxylcarbonyl chloride (Fmoc-Cl). An appropriate amount of Fmoc-Cl in anhydrous chloroform was added to the identical mouse retina lipid extract used in Fig. 1 in a ratio of 1:1 [Fmoc-Cl to ethanolamine glycerophospholipid (PE) content in the extract]. The mixture was incubated at room temperature for 5 min and diluted directly with 1:1 chloroform-methanol to a concentration of ∼50 pmol/μl total lipids. The negative ion ESI mass spectrum (A) was acquired as described in Materials and Methods. Product ion ESI-MS analyses of Fmoc-derivatized pseudomolecular ions at m/z 1008.8 (B) and 1012.8 (C) as shown in A were performed by selection of the pseudomolecular ion in the first quadrupole, collision activation in the second quadrupole with a collision energy of 30 eV and gas pressure of 1 mTorr, and analysis of the resulting product ions in the third quadrupole.
It should be pointed out that the profile of Fmoc-PE species was slightly different from the profile of PE species acquired from the diluted lipid extract without derivatization (compare Fig. 3A with Fig. 1B, Table 1). Generally, ion peaks corresponding to the Fmoc-PE molecular species containing short acyl chains showed a slightly higher instrument response than those containing long acyl chains. This small difference between these two profiles was still present even after the lipid solution was treated with Fmoc-Cl for up to 4 h or with a higher concentration of Fmoc-Cl (up to a ratio of 1:100 PE to Fmoc-Cl). In contrast, residual ion peaks corresponding to PE species were always present under these derivatization conditions, and the peak intensities of these PE ions were reduced when ionization conditions became milder. These results suggest that the small difference between the two profiles is likely attributable to the differential in-source fragmentation of facile loss of the Fmoc moiety from Fmoc-PE. However, the impact of this small difference between two profiles on the quantitation of low-abundance molecular species under mild ionization conditions is very small, as demonstrated in columns 5 and 7 of Table 1 (see below for details).
TABLE 1.
Mass content of PE molecular species in mouse retinas analyzed by ESI-MS after one-step in situ derivatization
| Molecular Species | m/z ([M-H]-) | m/z ([M’-H]-)a | Mass Contentb | Content of Minor Speciesec | Mass Contentd | Content of Minor Speciese | Content from NL Analysisf |
|---|---|---|---|---|---|---|---|
| P14:0-18:1/P14:1-18:0 | 672.6 | 894.8 | 2.4 ± 0.3 | 2.5 ± 0.4 | 0.1 ± 0.0 | ||
| D16:1-16:2 | 684.6 | 906.8 | 0.1 ± 0.0 | 0.2 ± 0.0 | 0.1 ± 0.1 | ||
| D16:0-16:0 | 690.6 | 912.8 | 0.5 ± 0.2 | 0.5 ± 0.2 | 0.2 ± 0.1 | ||
| P14:1-20:4 | 692.6 | 914.8 | 0.4 ± 0.1 | 0.4 ± 0.0 | 0.2 ± 0.1 | ||
| P16:0-18:2 | 698.6 | 920.8 | 0.8 ± 0.1 | 0.8 ± 0.2 | 0.5 ± 0.1 | ||
| P16:0-18:1/P18:1-16:0 | 700.6 | 922.8 | 3.2 ± 0.2 | 3.1 ± 0.2 | 1.7 ± 0.3 | ||
| D16:0-18:2/D16:1-18:1 | 714.6 | 936.8 | 1.1 ± 0.2 | 1.2 ± 0.2 | 0.8 ± 0.1 | ||
| D16:0-18:1/D16:1-18:0 | 716.6 | 938.8 | 3.2 ± 0.3 | 3.2 ± 0.2 | 2.3 ± 0.2 | ||
| P16:1-20:4 | 720.6 | 942.8 | 0.2 ± 0.0 | 0.2 ± 0.1 | 0.1 ± 0.0 | ||
| P16:0-20:4 | 722.6 | 944.8 | 4.3 ± 0.2 | 5.4 ± 0.4 | 3.8 ± 0.2 | ||
| P18:1-18:2 | 724.6 | 946.8 | 0.1 ± 0.1 | 0.1 ± 0.1 | 0.1 ± 0.1 | ||
| P18:1-18:1/P18:0-18:2 | 726.6 | 948.8 | 2.8 ± 0.4 | 3.8 ± 0.2 | 2.0 ± 0.3 | ||
| P18:0-18:1/P16:0-20:1 | 728.6 | 950.8 | 0.6 ± 0.2 | 0.6 ± 0.3 | 0.4 ± 0.1 | ||
| D16:1-20:4 | 736.6 | 958.8 | 0.5 ± 0.1 | 0.5 ± 0.1 | 0.4 ± 0.1 | ||
| D16:0-20:4 | 738.6 | 960.8 | 2.5 ± 0.4 | 3.2 ± 0.1 | 2.2 ± 0.2 | ||
| D18:1-18:2 | 740.6 | 962.8 | 0.1 ± 0.1 | 0.1 ± 0.1 | 0.1 ± 0.1 | ||
| D18:1-18:1/D18:0-18:2 | 742.6 | 964.8 | 1.7 ± 0.3 | 1.7 ± 0.2 | 1.5 ± 0.1 | ||
| D18:0-18:1 | 744.6 | 966.8 | 3.2 ± 0.2 | 3.2 ± 0.3 | 2.9 ± 0.2 | ||
| P16:0-22:6 | 746.6 | 968.8 | 6.0 ± 0.4 | 8.1 ± 0.5 | 7.1 ± 0.1 | ||
| P18:1-20:4/P16:0-22:5 | 748.6 | 970.8 | 6.7 ± 0.5 | 7.6 ± 0.6 | 7.2 ± 0.7 | ||
| P18:0-20:4/P16:0-22:4 | 750.6 | 972.8 | 10.6 ± 0.7 | 16.3 ± 0.8 | 15.8 ± 0.5 | ||
| D16:1-22:6 | 760.6 | 982.8 | 2.8 ± 0.2 | 0.6 ± 0.1 | 0.5 ± 0.1 | ||
| D16:0-22:6/D18:2-20:4 | 762.6 | 984.8 | 10.0 ± 0.5 | 14.2 ± 0.8 | 12.9 ± 0.4 | ||
| D16:0-22:5/D18:1-20:4 | 764.6 | 986.8 | 0.4 ± 0.1 | 0.4 ± 0.1 | 0.4 ± 0.1 | ||
| D18:0-20:4/D16:0-22:4 | 766.6 | 988.8 | 11.7 ± 0.8 | 13.2 ± 0.6 | 13.2 ± 0.1 | ||
| P18:2-22:6 | 770.6 | 992.8 | 0.1 ± 0.1 | 0.1 ± 0.1 | 0.1 ± 0.1 | ||
| P18:1-22:6 | 772.6 | 994.8 | 2.9 ± 0.4 | 2.2 ± 0.1 | 2.2 ± 0.3 | ||
| P18:0-22:6/P18:1-22:5 | 774.6 | 996.8 | 13.2 ± 0.7 | 12.3 ± 0.6 | 14.2 ± 0.7 | ||
| P18:0-22:5/P18:1-22:4 | 776.6 | 998.8 | 2.0 ± 0.3 | 2.0 ± 0.3 | 2.3 ± 0.4 | ||
| P20:0-20:4/P18:0-22:4 | 778.6 | 1,000.8 | 7.8 ± 0.6 | 5.0 ± 0.8 | 3.6 ± 0.3 | ||
| A20:0-20:4 | 780.6 | 1,002.8 | 0.1 ± 0.0 | 0.1 ± 0.1 | 0.1 ± 0.1 | ||
| D18:1-22:6/D20:3-20:4 | 788.6 | 1,010.8 | 0.1 ± 0.0 | 0.1 ± 0.1 | 0.1 ± 0.0 | ||
| D18:0-22:6 | 790.6 | 1,012.8 | 51.3 ± 2.5 | 48.0 ± 2.1 | 53.3 ± 1.1 | ||
| D18:0-22:5/D18:1-22:4 | 792.6 | 1,014.8 | 0.1 ± 0.0 | 0.1 ± 0.1 | 0.1 ± 0.1 | ||
| D20:0-20:4/D18:0-22:4 | 794.6 | 1,016.8 | 3.9 ± 0.4 | 1.5 ± 0.2 | 1.6 ± 0.2 | ||
| D20:4-22:6 | 810.6 | 1,032.8 | 2.3 ± 0.6 | 3.2 ± 0.2 | 2.1 ± 0.5 | ||
| D22:6-22:6 | 834.6 | 1,056.8 | 14.4 ± 1.0 | 8.4 ± 0.2 | 7.4 ± 0.1 | ||
| Total | 157.5 ± 10.1 | 16.5 ± 1.3 | 162.9 ± 8.8 | 11.2 ± 0.8 | 163.0 ± 5.4 | ||
| Grand total | 174.0 ± 11.4 | 174.1 ± 7.9 | 163.0 ± 5.4 |
ESI, electrospray ionization; MS, mass spectrometry; NL, neutral loss; PE, ethanolamine glycerophospholipid. Lipids of mouse retina samples were extracted by a modified method of Bligh and Dyer (24). PE molecular species were identified and quantified by two-dimensional MS or NL analysis of the fluorenylmethoxylcarbonyl (Fmoc) moiety (mass 222.2 u) from Fmoc-derivatized PE (Fmoc-PE) species as described in the text. The results are expressed in nanomoles per milligram of protein and represent means ± SD of lipid extracts from multiple separate animal preparations. Many very low-abundance PE molecular species [<0.02 mol% of each, such as the ion peaks at m/z 924.8, 940.8, and 952.8 (see inset C in Fig. 5), corresponding to 16:0-18:0 plasmenylethanolamine (PlsEtn), 16:0-18:0 phosphatidylethanolamine (PtdEtn), and 18:0-18:0 PlsEtn, respectively] are not listed in the table. The prefix “D” denotes diacyl PE species, “P” represents PlsEtn species, and “A” denotes alkyl-linked plasmanyl PE species.
M’ represents Fmoc-PE.
The mass content of abundant PE molecular species was quantitated from ESI-MS analyses of PE species in the negative ion mode after the addition of a small amount of LiOH (Fig. 1B).
The mass content of minor PE molecular species was quantitated by ESI-MS/MS with the NL of 222.2 u from Fmoc-PE (Fig. 5) using quantified abundant PE species (column 4) as endogenous standards.
The mass content of abundant PE molecular species was quantitated from ESI-MS analyses of Fmoc-PE species in the negative ion mode (Fig. 3A).
The mass content of minor PE molecular species was quantitated by ESI-MS/MS with the NL of 222.2 u from Fmoc-PE (Fig. 5) using quantified abundant Fmoc-PE species (column 6) as endogenous standards.
The mass content of both major and minor PE molecular species was quantitated directly by ESI-MS/MS with the NL of 222.2 u from Fmoc-PE (Fig. 5) using the exogenously added 20:4-20:4 PtdEtn as an internal standard.
Characterization of Fmoc-PE by product ion ESI-MS
Product ion ESI-MS analyses of Fmoc-PE species displayed two types of very abundant fragments (Fig. 3B, C). The first corresponds to the NL of 222.2 u (i.e., Fmoc moiety) from the selected pseudomolecular ion, such as m/z 1008.8 or 1012.8, resulting in an ion corresponding to a deprotonated PE molecular ion such as m/z 786.6 in Fig. 3B or 790.6 in Fig. 3C, respectively. The second type of product ions correspond to fatty acyl carboxylate(s) resulting from the collision-induced dissociation of fatty acyl chains from the selected pseudomolecular ions. For example, the product ion at m/z 303.2 corresponds to arachidonate released from the 20:4-20:4 Fmoc-PtdEtn ion (Fig. 3B), whereas the fragments at m/z 283.3 and 327.3 in Fig. 3C correspond to 18:0 and 22:6 carboxylates fragmented from the 18:0-22:6 Fmoc-PtdEtn ion. It should be recognized that the fragment at m/z 283.3 is also an isobaric ion peak corresponding to a polyunsaturated alkene ion resulting from the loss of carbon dioxide from the 22:6 carboxylate, as described previously (31, 32). In addition to these two types of abundant fragments, some fragments corresponding to lysoPE ions or lysoPE derivatives were also present in low or very low abundance, as a result of the facile secondary loss of the fatty acyl carboxylates.
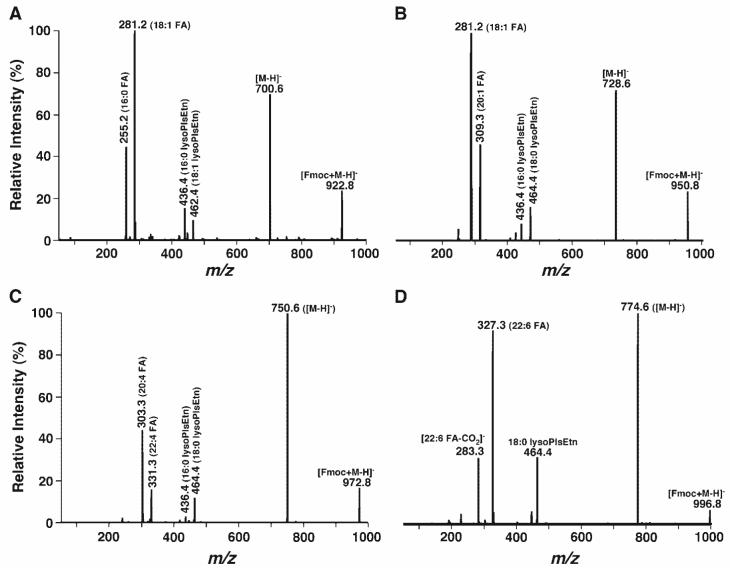
Product ion ESI-MS analyses of Fmoc-PE species also demonstrated that molecular ions corresponding to Fmoc-derivatized PlsEtn species yielded an identical fragmentation pattern to that from PtdEtn species but showed only one fragment ion corresponding to fatty acyl carboxylate (Fig. 4). Furthermore, the fragment ion corresponding to lysoPlsEtn was much more abundant than that of lysoPtdEtn, likely because there was no further or only minimal loss of the alkenyl chain in the lysoPlsEtn fragment (Fig. 4). Intriguingly, most of the ion peaks corresponding to PlsEtn species were isobaric. For example, the peak at m/z 922.8 was composed of 16:0-18:1 Fmoc-PlsEtn and 18:1-16:0 Fmoc-PlsEtn (Fig. 4A), the peak at m/z 950.8 was composed of 18:0-18:1 Fmoc-PlsEtn and 16:0-20:1 Fmoc-PlsEtn (Fig. 4B), and the peak at m/z 972.8 was composed of 18:0-20:4 Fmoc-PlsEtn and 16:0-22:4 Fmoc-PlsEtn (Fig. 4C). Again, a fragment ion at 283.3 resulting from the loss of carbon dioxide from 22:6 carboxylate was also present in the product ion ESI mass spectrum of Fmoc-PE at m/z 996.8, corresponding to the 18:0-22:6 Fmoc-PlsEtn pseudomolecular ion (Fig. 4D). The assignment of all of these PlsEtn molecular species was redundantly confirmed by the presence of abundant fragments of lysoPlsEtn and their disappearance from the spectrum acquired from the lipid solution after treatment with acidic vapor (25, 26). Other identified PlsEtn molecular species in mouse retina lipid extracts are listed in Table 1.
Fig. 4.
Product ion ESI-MS analyses of Fmoc-derivatized plasmenylethanolamine (PlsEtn) molecular species in the lipid extracts of mouse retinas. PlsEtn molecular species were identified by treatment of the mouse retina lipid extracts with acidic vapor, under which these species disappeared. Product ion ESI-MS analyses of Fmoc-derivatized pseudo-PlsEtn molecular ions at m/z 922.8 (A), 950.8 (B), 972.8 (C), and 996.8 (D) in the ESI mass spectrum of Fmoc-PE (Fig. 3A) were performed as described in the legend to Fig. 3. The presence of both fatty acyl carboxylates and lysoPlsEtn ions indicated the structures of Fmoc-PlsEtn.
Profiling Fmoc-PE and Fmoc-lysoPE molecular species by NL of the Fmoc moiety
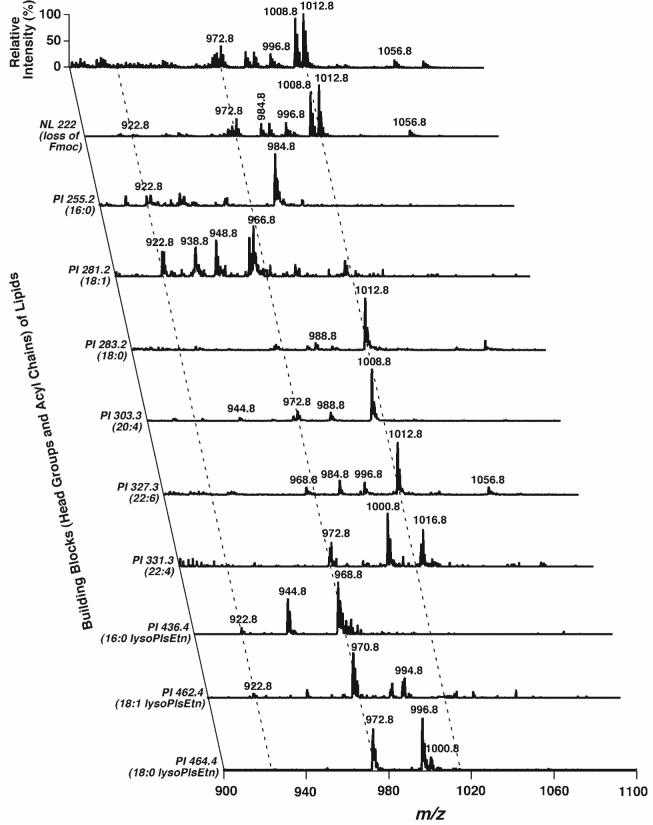
Next, the most abundant fragment representing the facile loss of the Fmoc moiety from the Fmoc-PE ion (i.e., NL of 222.2 u) was exploited to profile the molecular species of PE and other related species (Fig. 5). Nearly 40 ion peaks could be recognized in the Fmoc-PE region over m/z 900 (Fig. 5, inset A), whereas only ∼10 abundant ion peakscould be easily identified in the mass spectrum of non-derivatized PE (Fig. 1B). 2D MS analyses were performed to identify the acyl chain constituents through the analysis of all potential building blocks of the Fmoc-PE, including the Fmoc moiety itself, all naturally occurring fatty acyl chains, and lysoPlsEtn (Fig. 6). Therefore, the isobaric species of the ions and the regiospecificity of each molecular species were determined from the presence of multiple cross-peaks and the analysis of intensity ratios of the related cross-peaks, as described previously (33). 2D ESI-MS analysis demonstrated the identification of more than 50 PE molecular species in mouse retina lipid extracts; these are listed in Table 1. For example, a very low-intensity ion peak at m/z 922.8 was crossed with fragment peaks in the scans of NL222.2, PI255.2, PI281.2, PI436.4, and PI462.4 (along the left broken line in Fig. 6), indicating the presence of 16:0-18:1 Fmoc-PlsEtn and 18:1-16:0 Fmoc-PlsEtn isobaric ions at m/z 922.8. The ratios of fragment intensities of both carboxylates and lysoPlsEtn were redundantly used to estimate the composition of these isobaric species. Similarly, isobaric species of 16:0-22:4 Fmoc-PlsEtn and 18:0-20:4 Fmoc-PlsEtn at m/z 972.8 (along the middle broken line in Fig. 6) were also identified (Table 1).
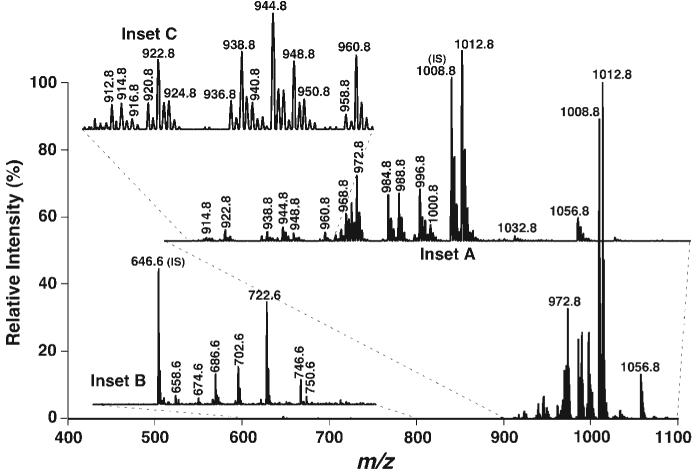
Fig. 5.
Tandem mass spectrum of Fmoc-derivatized phosphoethanolamine-containing lipids by NL of the Fmoc moiety. Phosphoethanolamine-containing species were derivatized by the addition of an equimolar amount of Fmoc-Cl as described in the legend to Fig. 3. NL tandem mass spectra of Fmoc-PE (inset A) and Fmoc-lysoPE (inset B) were acquired by coordinately scanning both the first and third quadrupoles with a mass difference (i.e., NL) of 222.2 u, corresponding to the NL of a Fmoc moiety, while collision activation was performed in the second quadrupole at collision energy of 30 eV and collision gas pressure of 1 mTorr. Inset C indicates the presence of many very low-abundance PE molecular species in the region. IS, internal standard.
Fig. 6.
Representative 2D ESI mass spectra of Fmoc-PE of a chloroform extract of mouse retinas in the negative ion mode. A conventional ESI mass spectrum of Fmoc-PE was acquired in the negative ion mode directly from a diluted mouse retina lipid extract after derivatization with Fmoc-Cl (Fig. 3A) as described in the legend to Fig. 3. Analyses of Fmoc-PE building blocks in the second dimension, including the Fmoc moiety, fatty acyl carboxylates, and lysoPlsEtn ions by PI scanning and NL scanning, were performed as described in Materials and Methods. All mass spectral traces were displayed after normalization to the base peak in each spectrum.
Surprisingly, a cluster of ion peaks in the Fmoc-lysoPE region (m/z 600-800) was also present in the NL mass spectrum (NL222.2) (Fig. 5, inset B). Each of these ion peaks was identified as a Fmoc-lysoPE molecular species (Table 2). This mass spectrum represents the first profile of lysoPE molecular species directly from a lipid extract of a biological sample. The mass content of each lysoPE species was assessed by comparison of the individual peak intensity with that of the selected internal standard (i.e., 14:0 lysoPtdEtn) after correction for 13C isotopomer intensity differences, as described previously (2, 10, 11) (Table 2). These results demonstrated a >15,000-fold dynamic range [from the most intense peak at m/z 1012.8 (51 nmol/mg protein; Table 1) to the lowest abundant ion peaks at m/z 656.6 (3 pmol/mg protein; Table 2)]. These results also indicated that the detection limitation of the descriptive technique was in the attomoles (amoles) per microliter range, because the total concentration of the infused lipid solution was <50 pmol/μl. In addition, no other abundant molecular species in the examined region were found in the mass spectrum of NL222.2 (Fig. 5), suggesting that the derivatization was relatively specific to phosphoethanolamine-containing molecular species under the experimental conditions used. It should be noted that the ion peak at m/z 722.6 (corresponding to 20:4 lysoPtdEtn) was prominent in the spectrum. We found that this high mass content of 20:4 lysoPtdEtn was the result of contamination from the 20:4-20:4 PtdEtn internal standard. The mass content of endogenous 20:4 lysoPtdEtn was determined from samples without the addition of 20:4-20:4 PtdEtn (Table 2).
TABLE 2.
Mass content of lysoPE molecular species in mouse retinas analyzed by ESI-MS/MS after one-step in situ derivatization
| Molecular Species | m/z([M-H]-) | m/z ([M’-H]-)a | Mass Content |
|---|---|---|---|
| P16:1 | 434.4 | 656.6 | 3.0 ± 0.5 |
| P16:0 | 436.4 | 658.6 | 53.2 ± 5.3 |
| 16:1 | 450.4 | 672.6 | 6.1 ± 2.3 |
| 16:0 | 452.4 | 674.6 | 43.0 ± 4.7 |
| P18:1 | 462.4 | 684.6 | 38.2 ± 2.6 |
| P18:0 | 464.4 | 686.6 | 198.8 ± 23.6 |
| 18:1 | 478.4 | 700.6 | 30.1 ± 1.3 |
| 18:0 | 480.4 | 702.6 | 268.9 ± 9.0 |
| 20:4 | 500.4 | 722.6 | 40.7 ± 1.8 |
| 22:6 | 524.4 | 746.6 | 189.8 ± 5.4 |
| 22:5 | 526.4 | 748.6 | 21.0 ± 2.7 |
| 22:4 | 528.4 | 750.6 | 55.0 ± 6.7 |
| Total | 947.5 ± 71.3 |
LysoPE, ethanolamine glycerolysophospholipid. Lipids of mouse retina samples were extracted by a modified method of Bligh and Dyer (24). LysoPE molecular species were identified and quantified by the NL of the Fmoc moiety (mass 222.2 u) from Fmoc-lysoPE as described in the text. The results are expressed in picomoles per milligram of protein and represent means ± SD of lipid extracts from multiple separate animal preparations. The prefix “P” denotes lysoPlsEtn species.
M’ represents Fmoc-lysoPE.
Quantitative analysis of PE and lysoPE molecular species in mouse retina lipid extracts
Finally, the two-step procedure (3, 8, 9) was used to quantitate PE molecular species in mouse retina lipids. First, the abundant PE molecular species were accurately quantitated by comparisons of ion peak intensities in ESI-MS analysis without fragmentation or derivatization (Fig. 1B, Table 1, column 4). Then, using these quantified abundant PE species as endogenous standards plus the exogenously added internal standard, all of the low-abundance PE molecular species shown in the scan of NL222.2 (Fig. 5) were quantitated by ratiometric comparisons of the peak intensity of each low-abundance PE molecular species with those of the nearest neighboring abundant PE molecular species after correction for 13C isotopomer intensity differences. The quantitative results of these low-abundance PE molecular species are listed in column 5 of Table 1. It was found that the low-abundance PE molecular species in mouse retinas accounted for only 16.5 nmol/mg protein and thus represented ∼10 mol% of the total PE mass content.
To assess the effect of Fmoc derivatization on the quantitation of PE species, two additional analyses were performed. In the first analysis, the mass content of abundant PE molecular species was assessed using the ESI mass spectra after Fmoc derivatization (Fig. 3A, Table 1, column 6), then the low-abundance PE molecular species were analyzed using these newly determined sets of PE species as endogenous standards for comparisons, as described above (Table 1, column 7). The results showed that the adverse effects of Fmoc derivatization on the quantitation of PE species were quite small under our experimental conditions, although mass differences between those obtained before and after derivatization for some individual molecular species might be present. In the second analysis, the mass content of both abundant and low-abundance PE species was assessed by NL scan of 222.2 u (Fig. 5) and by comparison with the exogenous internal standard (Table 1, column 8). This revealed an ∼10% difference using this approach from the results obtained using the two-step procedure (Table 1).
It should be pointed out that there were some differences in the mass content of some PE molecular species containing very long acyl chains using the three analytical methods described (Table 1). In the mass region of PE molecular species corresponding to very long acyl chains, there is an increased likelihood that other lipid classes and/or isotopomers derived from there could contribute to some of the observed ion intensity measured directly, as stated in the introduction. These potential overlaps can be estimated through ratiometric comparisons from 2D mass spectrometric array analysis after appropriate consideration of fragmentation kinetic factors. However, through modification with Fmoc as described herein, PE molecular species were shifted to a higher mass region in which no overlap was present, and NL analysis of the Fmoc moiety further specifically eliminated any minor overlaps present in the new mass region. Accordingly, shotgun lipidomics after derivatization with Fmoc can accurately quantitate PE molecular species directly from a lipid extract of a biological sample.
Factors affecting the analysis of phosphoethanolamine-containing compounds during derivatization
Four additional experiments were performed to examine the effects of the derivatization conditions on the profile of PE species. First, the effects of molar ratios of Fmoc-Cl to PE in the lipid extracts on PE derivatization were examined by varying the ratios of reagents and targets, including 0.5:1, 1:1, 10:1, and 100:1 ratios. It was found that a ratio of ∼1:1 yielded the best result. When a lower ratio was used, incomplete derivatization was present and analysis was less sensitive. When a higher ratio was used, three side effects on lipid analysis were present. One was an increase in chemical noise during ESI-MS analysis. The second was the appearance of other derivatized lipid species besides PE species, such as those from phosphatidylserine. The third was the degradation of PlsEtn molecular species as a result of the increased acidity when a high concentration of Fmoc-Cl was used.
The second experiment examined the effects of incubation time on the analyses of phosphoethanolamine-containing compounds. It was found that the derivatization reaction was very fast and that the derivatized lipid could be used to analyze phosphoethanolamine-containing species in as short as 5 min after the addition of Fmoc-Cl solution to the reaction vessel. No differences in the mass spectra were found when the lipid solution was analyzed within 30 min after the addition of the derivatization reagent to the reaction vessel. However, the peak intensities of ions corresponding to the PlsEtn species were gradually reduced, indicating that the degradation of these species likely was attributable to the acidic environment. This effect can be minimized by the inclusion of DMAP in equimolar ratio to Fmoc-Cl in the reaction vessel.
The third experiment examined the effects of pH conditions on derivatization. It was found that the addition of a small amount of base (e.g., equimolar amounts of DMAP or other weak bases to the PE content) yielded the best derivatization results, including rapid completion of derivatization and minimization of the degradation of PlsEtn species. The former was likely attributable to the increase in the primary amine group of the phosphoethanolamine-containing species, and the latter was likely the result of changes in the acidic environment.
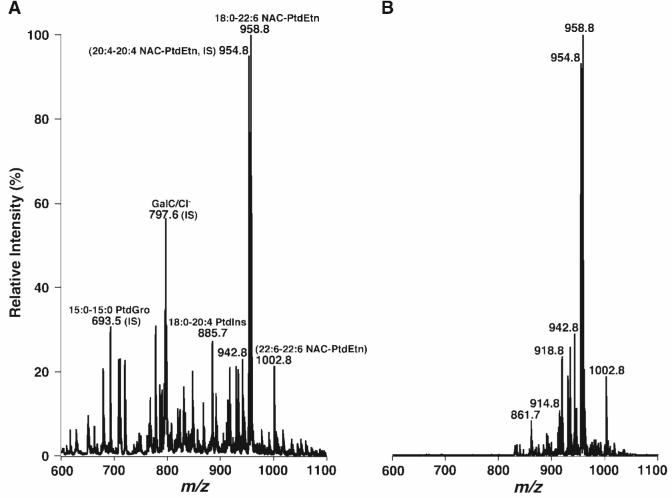
Finally, additional derivatization reagents such as NAC (molecular weight of 204.655) were also exploited for the analysis of PE molecular species. Essentially identical profiles of NAC-derivatized PE species in mouse retina lipid extracts to those obtained from Fmoc derivatization were demonstrated by mass spectrometric analysis (Fig. 7). However, it should be pointed out that because of the differences in chemical structures of the examined derivatization reagents, optimal derivatization and analysis conditions for different reagents were different. For example, longer time was needed for NAC derivatization compared with Fmoc-Cl derivatization. The tighter amide bond in NAC derivatives showed the lower in-source fragmentation than in Fmoc derivatives, but this tighter linkage resulted in a reduction of analysis sensitivity by MS/MS with the NL of the NAC-derivatized moiety. Both the prolonged derivatization time and the resulting tighter linkage in NAC derivatization compared with Fmoc-Cl derivatization also led to the reduction of derivatization specificity. For example, the ion peak at m/z 861.7 resulted from the derivatization of 15:0-15:0 PtdGro (ion peak at m/z 693.5) (Fig. 7).
Fig. 7.
Representative negative ion ESI-MS and NL mass spectra of 2-(2-naphthyl)acetyl chloride (NAC)-derivatized PE molecular species of mouse retina lipid extracts. An appropriate amount of NAC in anhydrous chloroform was added to the identical mouse retina lipid extract used in Fig. 1 in a molar ratio of 1:1 (NAC to PE content in the extract). The mixture was incubated at room temperature for 30 min and diluted directly with 1:1 chloroform-methanol to a concentration of ∼50 pmol/μl total lipids. The negative ion ESI mass spectrum (A) was acquired as described in Materials and Methods. The NL tandem mass spectrum of NAC-derivatized PE molecular species (B) was acquired by coordinately scanning both the first and third quadrupoles with a mass difference (i.e., NL) of 168.2 u, corresponding to the NL of a 2-(2-naphthyl)acetyl moiety, while collision activation was performed in the second quadrupole at collision energy of 40 eV and collision gas pressure of 1 mTorr. IS, internal standard.
DISCUSSION
Derivatization has been used previously in the ESI-MS analysis of intact lipids (34-36). The main purpose of the derivatization is to convert nonpolar or less polar lipids into polar lipids that carry an inherent charge, such as by adding a sulfate group to cholesterol (34) or converting oxosteroids into their oximes (35). Through this approach, one can dramatically improve the sensitivity of mass spectrometric analysis of these compounds. A detection limit in the low picogram per microliter range for the analysis of less polar lipids can be readily achieved after derivatization, as described previously (34, 35).
PE molecular species contain a polar head group and can be analyzed by ESI-MS in either the positive or the negative ion mode, particularly after isolation of the lipid class by chromatography (37, 38). However, the analysis of low-abundance species was quite difficult because of the presence of chemical noise during direct infusion (37) (see also introduction) or because of peak overlaps and baseline fluctuations in the case of reverse-phase HPLC coupled with ESI-MS (39). ESI-MS/MS has also been used extensively to profile PE molecular species by either PI scanning of m/z 196 from deprotonated PE molecular ions in the negative ion mode or NL of 141 u from protonated PE ions in the positive ion mode (19, 20, 22). However, the sensitivity of both approaches was not sufficient to penetrate the low-abundance region of PE molecular species directly after extraction.
The current study presents a novel approach for the analyses of PE and lysoPE molecular species, including those in the very low-abundance regime, through derivatization of the primary amine of the phosphoethanolamine-containing species in the lipid extracts and direct infusion of the derivatized and diluted lipid extracts. By this approach, the PE species were shifted to a higher m/z region that could be selected for using the desired derivatization reagent in which overlaps with other endogenous lipid constituents in the lipid extracts were rare. In this approach, derivatization turns zwitterionic phosphoethanolamine-containing species into “mass-shifted” derivatized anionic species. Through this approach, the sensitivity for the analysis of derivatized phosphoethanolamine-containing species is much higher than that realized by a direct approach (as described in the introduction) in which LiOH is added and the introduction of some chemical noise as a result of the addition of LiOH is inevitable.
Furthermore, in the current method, because of the presence of an abundant fragment resulting from the facile loss of the derivatized moiety under very mild conditions of collision-induced dissociation, NL scanning of this moiety is a very sensitive and semiquantitative measure of pseudomolecular ions of PE species. The detection limit of this technique is in the 50-100 amol/μl range, and dynamic ranges of >15,000-fold can be appreciated (see the estimation in the last section). Through this approach, identification and quantitation/refinement of most phosphoethanolamine-containing molecular species, including those with very low abundance, such as lysoPE species, by 2D ESI-MS analysis can be readily achieved.
Although mass content of lysoPE along with other lysophospholipid classes in biological samples has been determined previously by LC-MS (40, 41), to our best knowledge, direct profiling of lysoPE molecular species from lipid extracts has not been reported because of their low abundance and the inherent difficulties in profiling the species. The tandem mass spectra of lysoPE reported here (Fig. 5, inset B) represent the first lysoPE profiles in biological samples. The mass profile of lysoPE species in mouse retina lipid extracts (Table 2) was comparable to the sn-1 aliphatic chain moieties of PE molecular species in the samples (Table 1), suggesting that the lysoPE species in the retina lipid extracts are mainly sn-1 lyso species.
It should be pointed out that the assessment of mass content of each lysoPE molecular species was based solely on MS/MS analysis with one internal standard. Thus, the accuracy of this assessment might vary from one species to another because of the differential fragmentation kinetics of different molecular species, as discussed previously (2). If more accurate quantitation is desired, multiple internal standards of lysoPE species covering the different physical properties should be used (2, 19), or a calibration curve for each salient lysoPE molecular species should be established (40).
Collectively, derivatization of phosphoethanolamine-containing species provides a new dimension at which to analyze PE and lysoPE molecular species directly from organic extracts. This is particularly important because PlsEtn and lysoPlsEtn molecular species are very sensitive to oxidation, silica-catalyzed vinyl ether cleavage, or extensive loss during chromatography. The approach described here unambiguously identifies and quantifies very low-abundance PE molecular species. The procedure is both simple and effective. Therefore, this approach, when used in combination with shotgun lipidomics based on intrasource separation and multidimensional MS, can be used to profile and quantitate the large majority of PE and lysoPE molecular species directly from a lipid extract of a biological sample. In general, derivatization not only increases the sensitivity of analysis of these lipids but also selectively allows the analysis of this class of lipids by specifically shifting the class of lipids to a region with no interference from other lipid classes or by specifically characterizing the class of derivatized lipids by facilitating MS/MS analyses. Therefore, derivatization adds a new dimension to shotgun lipidomics for the global analysis of cellular lipidomes directly from lipid extracts of biological samples.jlr
Acknowledgments
This work was supported by Grant P01HL-57278 and the Neurosciences Education and Research Foundation.
Abbreviations
- 2D
two-dimensional
- DMAP
dimethylaminopyridine
- ESI
electrospray ionization
- Fmoc-Cl
fluorenylmethoxylcarbonyl chloride
- m:n
acyl chain containing m carbons and n double bonds
- MS
mass spectrometry
- NAC
2-(2-naphthyl)acetyl chloride
- NL
neutral loss
- PE
ethanolamine glycerophospholipid
- PI
precursor ion
- PlsEtn
plasmenylethanolamine
- PtdCho
phosphatidylcholine
- PtdEtn
phosphatidylethanolamine
- PtdGro
phosphatidylglycerol
REFERENCES
- 1.Han X, Gross RW. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J. Lipid Res. 2003;44:1071–1079. doi: 10.1194/jlr.R300004-JLR200. [DOI] [PubMed] [Google Scholar]
- 2.Han X, Gross RW. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of the cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom. Rev. 2005;24:367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- 3.Han X, Gross RW. Shotgun lipidomics: multi-dimensional mass spectrometric analysis of cellular lipidomes. Expert Rev. Proteomics. 2005;2:253–264. doi: 10.1586/14789450.2.2.253. [DOI] [PubMed] [Google Scholar]
- 4.Han X, Yang J, Cheng H, Ye H, Gross RW. Towards fingerprinting cellular lipidomes directly from biological samples by two-dimensional electrospray ionization mass spectrometry. Anal. Biochem. 2004;330:317–331. doi: 10.1016/j.ab.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Tang L, Kebarle P. Effect of the conductivity of the electrosprayed solution on the electrospray current. Factors determining analyte sensitivity in electrospray mass spectrometry. Anal. Chem. 1991;63:2709–2715. [Google Scholar]
- 6.Ikonomou MG, Blades AT, Kebarle P. Electrosprayion spray: a comparison of mechanisms and performance. Anal. Chem. 1991;63:1989–1998. [Google Scholar]
- 7.Gross RW, Sobel BE. Isocratic high-performance liquid chromatography separation of phosphoglycerides and lysophosphoglycerides. J. Chromatogr. 1980;197:79–85. doi: 10.1016/s0021-9673(00)80538-5. [DOI] [PubMed] [Google Scholar]
- 8.Han X, Cheng H, Mancuso DJ, Gross RW. Caloric restriction results in phospholipid depletion, membrane remodeling and triacylglycerol accumulation in murine myocardium. Biochemistry. 2004;43:15584–15594. doi: 10.1021/bi048307o. [DOI] [PubMed] [Google Scholar]
- 9.Han X, Cheng H. Characterization and direct quantitation of cerebroside molecular species from lipid extracts by shotgun lipidomics. J. Lipid Res. 2005;46:163–175. doi: 10.1194/jlr.D400022-JLR200. [DOI] [PubMed] [Google Scholar]
- 10.Han X, Gross RW. Quantitative analysis and molecular species fingerprinting of triacylglyceride molecular species directly from lipid extracts of biological samples by electrospray ionization tandem mass spectrometry. Anal. Biochem. 2001;295:88–100. doi: 10.1006/abio.2001.5178. [DOI] [PubMed] [Google Scholar]
- 11.Han X. Characterization and direct quantitation of ceramide molecular species from lipid extracts of biological samples by electrospray ionization tandem mass spectrometry. Anal. Biochem. 2002;302:199–212. doi: 10.1006/abio.2001.5536. [DOI] [PubMed] [Google Scholar]
- 12.Han X, Gross RW. Electrospray ionization mass spectroscopic analysis of human erythrocyte plasma membrane phospholipids. Proc. Natl. Acad. Sci. USA. 1994;91:10635–10639. doi: 10.1073/pnas.91.22.10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han X, Gubitosi-Klug RA, Collins BJ, Gross RW. Alterations in individual molecular species of human platelet phospholipids during thrombin stimulation: electrospray ionization mass spectrometry-facilitated identification of the boundary conditions for the magnitude and selectivity of thrombin-induced platelet phospholipid hydrolysis. Biochemistry. 1996;35:5822–5832. doi: 10.1021/bi952927v. [DOI] [PubMed] [Google Scholar]
- 14.Lehmann WD, Koester M, Erben G, Keppler D. Characterization and quantification of rat bile phosphatidylcholine by electrospray-tandem mass spectrometry. Anal. Biochem. 1997;246:102–110. doi: 10.1006/abio.1996.9941. [DOI] [PubMed] [Google Scholar]
- 15.Kim HY, Wang TC, Ma YC. Liquid chromatography/mass spectrometry of phospholipids using electrospray ionization. Anal. Chem. 1994;66:3977–3982. doi: 10.1021/ac00094a020. [DOI] [PubMed] [Google Scholar]
- 16.DeLong CJ, Baker PRS, Samuel M, Cui Z, Thomas MJ. Molecular species composition of rat liver phospholipids by ESI-MS/MS: the effect of chromatography. J. Lipid Res. 2001;42:1959–1968. [PubMed] [Google Scholar]
- 17.Koivusalo M, Haimi P, Heikinheimo L, Kostiainen R, Somerharju P. Quantitative determination of phospholipid compositions by ESI-MS: effects of acyl chain length, unsaturation, and lipid concentration on instrument response. J. Lipid Res. 2001;42:663–672. [PubMed] [Google Scholar]
- 18.Han X, Gross RW. Specific lipid alterations in Alzheimer’s disease and diabetes: shotgun global cellular lipidome analyses by electrospray ionization mass spectrometry using intrasource separation. In: Feng L, Prestwich GD, editors. Functional Lipidomics. Marcel Dekker, Inc.; New York: 2005. pp. 285–306. [Google Scholar]
- 19.Brugger B, Erben G, Sandhoff R, Wieland FT, Lehmann WD. Quantitative analysis of biological membrane lipids at the low picomole level by nano-electrospray ionization tandem mass spectrometry. Proc. Natl. Acad. Sci. USA. 1997;94:2339–2344. doi: 10.1073/pnas.94.6.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Welti R, Li W, Li M, Sang Y, Biesiada H, Zhou H-E, Rajashekar CB, Williams TD, Wang X. Profiling membrane lipids in plant stress responses. Role of phospholipase Dα in freezing-induced lipid changes in Arabidopsis. J. Biol. Chem. 2002;277:31994–32002. doi: 10.1074/jbc.M205375200. [DOI] [PubMed] [Google Scholar]
- 21.Ekroos K, Chernushevich IV, Simons K, Shevchenko A. Quantitative profiling of phospholipids by multiple precursor ion scanning on a hybrid quadrupole time-of-flight mass spectrometer. Anal. Chem. 2002;74:941–949. doi: 10.1021/ac015655c. [DOI] [PubMed] [Google Scholar]
- 22.Blom TS, Koivusalo M, Kuismanen E, Kostiainen R, Somerharju P, Ikonen E. Mass spectrometric analysis reveals an increase in plasma membrane polyunsaturated phospholipid species upon cellular cholesterol loading. Biochemistry. 2001;40:14635–14644. doi: 10.1021/bi0156714. [DOI] [PubMed] [Google Scholar]
- 23.Liebisch G, Lieser B, Rathenberg J, Drobnik W, Schmitz G. High-throughput quantification of phosphatidylcholine and sphingomyelin by electrospray ionization tandem mass spectrometry coupled with isotope correction algorithm. Biochim. Biophys. Acta. 2004;1686:108–117. doi: 10.1016/j.bbalip.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 24.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 25.Ford DA, Rosenbloom KB, Gross RW. The primary determinant of rabbit myocardial ethanolamine phosphotransferase substrate selectivity is the covalent nature of the sn-1 aliphatic group of diradyl glycerol acceptors. J. Biol. Chem. 1992;267:11222–11228. [PubMed] [Google Scholar]
- 26.Kayganich KA, Murphy RC. Fast atom bombardment tandem mass spectrometric identification of diacyl, alkylacyl, and alk-1-enylacyl molecular species of glycerophosphoethanolamine in human polymorphonuclear leukocytes. Anal. Chem. 1992;64:2965–2971. doi: 10.1021/ac00047a015. [DOI] [PubMed] [Google Scholar]
- 27.Hsu F-F, Turk J. Electrospray ionization/tandem quadrupole mass spectrometric studies on phosphatidylcholines: the fragmentation processes. J. Am. Soc. Mass Spectrom. 2003;14:352–363. doi: 10.1016/S1044-0305(03)00064-3. [DOI] [PubMed] [Google Scholar]
- 28.Berry KAZ, Murphy RC. Electrospray ionization tandem mass spectrometry of glycerophosphoethanolamine plasmalogen phospholipids. J. Am. Soc. Mass Spectrom. 2004;15:1499–1508. doi: 10.1016/j.jasms.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 29.Bell MV, Tocher DR. Molecular species composition of the major phospholipids in brain and retina from rainbow trout (Salmo gairdneri). Occurrence of high levels of di-(n-3)polyunsaturated fatty acid species. Biochem. J. 1989;264:909–915. doi: 10.1042/bj2640909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sellner PA, Clough JA. Fatty acid composition of phospholipids from chick neural retina during development. Exp. Eye Res. 1992;54:725–730. doi: 10.1016/0014-4835(92)90027-p. [DOI] [PubMed] [Google Scholar]
- 31.Hsu FF, Turk J. Charge-remote and charge-driven fragmentation processes in diacyl glycerophosphoethanolamine upon low-energy collisional activation: a mechanistic proposal. J. Am. Soc. Mass Spectrom. 2000;11:892–899. doi: 10.1016/S1044-0305(00)00159-8. [DOI] [PubMed] [Google Scholar]
- 32.Hsu F-F, Turk J. Characterization of phosphatidylinositol, phosphatidylinositol-4-phosphate, and phosphatidylinositol-4,5-bisphosphate by electrospray ionization tandem mass spectrometry: a mechanistic study. J. Am. Soc. Mass Spectrom. 2000;11:986–999. doi: 10.1016/S1044-0305(00)00172-0. [DOI] [PubMed] [Google Scholar]
- 33.Han X, Gross RW. Structural determination of picomole amounts of phospholipids via electrospray ionization tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 1995;6:1202–1210. doi: 10.1016/1044-0305(95)00568-4. [DOI] [PubMed] [Google Scholar]
- 34.Sandhoff R, Brugger B, Jeckel D, Lehmann WD, Wieland FT. Determination of cholesterol at the low picomole level by nano-electrospray ionization tandem mass spectrometry. J. Lipid Res. 1999;40:126–132. [PubMed] [Google Scholar]
- 35.Liu S, Sjovall J, Griffiths WJ. Analysis of oxosteroids by nano-electrospray mass spectrometry of their oximes. Rapid Commun. Mass Spectrom. 2000;14:390–400. doi: 10.1002/(SICI)1097-0231(20000331)14:6<390::AID-RCM882>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 36.Griffiths WJ, Liu S, Alvelius G, Sjovall J. Derivatisation for the characterisation of neutral oxosteroids by electrospray and matrix-assisted laser desorption/ionisation tandem mass spectrometry: the Girard P derivative. Rapid Commun. Mass Spectrom. 2003;17:924–935. doi: 10.1002/rcm.1002. [DOI] [PubMed] [Google Scholar]
- 37.Ramanadham S, Hsu FF, Bohrer A, Nowatzke W, Ma Z, Turk J. Electrospray ionization mass spectrometric analyses of phospholipids from rat and human pancreatic islets and subcellular membranes: comparison to other tissues and implications for membrane fusion in insulin exocytosis. Biochemistry. 1998;37:4553–4567. doi: 10.1021/bi9722507. [DOI] [PubMed] [Google Scholar]
- 38.Ivanova PT, Cerda BA, Horn DM, Cohen JS, McLafferty FW, Brown HA. Electrospray ionization mass spectrometry analysis of changes in phospholipids in RBL-2H3 mastocytoma cells during degranulation. Proc. Natl. Acad. Sci. USA. 2001;98:7152–7157. doi: 10.1073/pnas.131195098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Byrdwell WC. Dual parallel liquid chromatography/dual mass spectrometry (LC2/MS2) of bovine brain total lipid extract. J. Liq. Chromatogr. R. T. 2003;26:3147–3181. [Google Scholar]
- 40.Xiao YJ, Schwartz B, Washington M, Kennedy A, Webster K, Belinson J, Xu Y. Electrospray ionization mass spectrometry analysis of lysophospholipids in human ascitic fluids: comparison of the lysophospholipid contents in malignant vs nonmalignant ascitic fluids. Anal. Biochem. 2001;290:302–313. doi: 10.1006/abio.2001.5000. [DOI] [PubMed] [Google Scholar]
- 41.Fang N, Yu S, Badger TM. LC-MS/MS analysis of lysophospholipids associated with soy protein isolate. J. Agric. Food Chem. 2003;51:6676–6682. doi: 10.1021/jf034793v. [DOI] [PubMed] [Google Scholar]