Abstract
Nuclear and mitochondrial transmission to daughter buds of Saccharomyces cerevisiae depends on Mdm1p, an intermediate filament-like protein localized to numerous punctate structures distributed throughout the yeast cell cytoplasm. These structures disappear and organelle inheritance is disrupted when mdm1 mutant cells are incubated at the restrictive temperature. To characterize further the function of Mdm1p, new mutant mdm1 alleles that confer temperature-sensitive growth and defects in organelle inheritance but produce stable Mdm1p structures were isolated. Microscopic analysis of the new mdm1 mutants revealed three phenotypic classes: Class I mutants showed defects in both mitochondrial and nuclear transmission; Class II alleles displayed defective mitochondrial inheritance but had no effect on nuclear movement; and Class III mutants showed aberrant nuclear inheritance but normal mitochondrial distribution. Class I and II mutants also exhibited altered mitochondrial morphology, possessing primarily small, round mitochondria instead of the extended tubular structures found in wild-type cells. Mutant mdm1 alleles affecting nuclear transmission were of two types: Class Ia and IIIa mutants were deficient for nuclear movement into daughter buds, while Class Ib and IIIb mutants displayed a complete transfer of all nuclear DNA into buds. The mutations defining all three allelic classes mapped to two distinct domains within the Mdm1p protein. Genetic crosses of yeast strains containing different mdm1 alleles revealed complex genetic interactions including intragenic suppression, synthetic phenotypes, and intragenic complementation. These results support a model of Mdm1p function in which a network comprised of multimeric assemblies of the protein mediates two distinct cellular processes.
Cytoplasmic organelles are propagated by growth and division of preexisting organelles (Palade, 1983; Yaffe, 1991; Warren and Wickner, 1996), so an essential feature of cell proliferation is the inheritance of organelles by daughter cells. Organelle inheritance is thought to depend on functions of the cytoskeleton. Such a role for cytoskeletal components has been suggested by microscopic studies that revealed colocalization of organelles with microtubules (Heggeness et al., 1978; Ball and Singer, 1982; Couchman and Rees, 1982), intermediate filaments (David-Ferreira and David-Ferreira, 1980; Mose-Larsen et al., 1982; Chen, 1988), or actin microfilaments (Wang and Goldman, 1978; Kachar and Reese, 1988) in various types of cells. In addition, studies in vitro have indicated possible functions of microtubule-based motor proteins (Vale, 1987) or unconventional myosins (Adams and Pollard, 1986; Allan, 1995) in facilitating organelle movement. However, many details of the activity and roles of particular cytoskeletal components in mediating organelle movement and distribution in living cells remain obscure.
Nuclear and mitochondrial inheritance in the yeast Saccharomyces cerevisiae depends on Mdm1p, an intermediate filament-like protein that defines a series of punctate structures distributed throughout the yeast cytoplasm (McConnell and Yaffe, 1992, 1993). The punctate Mdm1p structures disappear at 37°C in cells harboring the temperature-sensitive mdm1-1 mutation (McConnell and Yaffe, 1992), and this disappearance coincides with a failure to transmit mitochondria from the mother portion of the cell into the growing bud. Additionally, the mdm1-1 lesion causes a disorientation of the mitotic spindle such that nuclear division occurs entirely within the mother portion of the cell (McConnell et al., 1990). These defects indicate that the Mdm1p network has a central function in facilitating organelle inheritance; however, the mechanism of Mdm1p function is unknown (Berger and Yaffe, 1996).
To explore Mdm1p function further, we have generated new mdm1 mutant alleles that cause defects in organelle inheritance but yield stable Mdm1p punctate structures even during incubation of cells at the nonpermissive temperature. These novel alleles have facilitated a genetic dissection of Mdm1p functions in nuclear and mitochondrial inheritance.
Materials and Methods
Yeast Strains and Genetic Methods
S. cerevisiae strains used in this study are listed in Table I. Strain MYY404 is a diploid in which one copy of MDM1 is replaced by the LEU2 gene and was derived from MYY298 as described (McConnell and Yaffe, 1992). Strain MYY404-1b was created by transforming MYY404 with plasmid YCp50-MDM1 (McConnell and Yaffe, 1992), followed by sporulation and recovery of a spore that was MATa, Leu+, Ura+. Strains MYY700 through MYY721 were generated in the MYY290 (Smith and Yaffe, 1991) background by replacing the wild-type, chromosomal copy of MDM1 with different mutant alleles, as described below. Strains MYY725 through MYY746 were derived as temperature-sensitive, MATα spores from crosses of strain MYY291 with strains MYY700 through MYY721. Yeast culture media were prepared, and standard genetic manipulations were carried out as previously described (Rose et al., 1990). Plasmid DNA was prepared in Escherichia coli strain DH5α.
Table I.
Yeast Strains
| Strain | Genotype | Source | ||
|---|---|---|---|---|
| MYY290 | MATa, MDM1, his3, leu2, ura3 | Smith and Yaffe, 1991 | ||
| MYY291 | MATα, MDM1, his3, leu2, ura3 | Smith and Yaffe, 1991 | ||
| MYY298 | MATa/α, MDM1/MDM1, his3/his3, leu2/leu2, ura3/ura3 | Smith and Yaffe, 1991 | ||
| MYY403 | MATa, mdm1-1 | McConnell and Yaffe, 1992 | ||
| MYY404 | MATa/α, MDM1/mdm1::LEU2, his3/his3, leu2/leu2, ura3/ura3 | McConnell and Yaffe, 1992 | ||
| MYY404-1b | MATa, mdm1::LEU2, his3, leu2, ura3, YCp50-MDM1 | This Study | ||
| MYY700 | MATa, mdm1-20, his3,leu2, ura3 | This Study | ||
| MYY701 | MATa, mdm1-199, his3, leu2, ura3 | This Study | ||
| MYY702 | MATa, mdm1-200, his3, leu2, ura3 | This Study | ||
| MYY704 | MATa, mdm1-202, his3, leu2, ura3 | This Study | ||
| MYY709 | MATa, mdm1-204, his3, leu2, ura3 | This Study | ||
| MYY710 | MATa, mdm1-217, his3, leu2, ura3 | This Study | ||
| MYY715 | MATa, mdm1-227, his3, leu2, ura3 | This Study | ||
| MYY717 | MATa, mdm1-228, his3, leu2, ura3 | This Study | ||
| MYY720 | MATa, mdm1-251, his3, leu2, ura3 | This Study | ||
| MYY721 | MATa, mdm1-252, his3, leu2, ura3 | This Study | ||
| MYY725 | MATα, mdm1-20, his3, leu2, ura3 | This Study | ||
| MYY726 | MATα, mdm1-199, his3, leu2, ura3 | This Study | ||
| MYY727 | MATα, mdm1-200, his3, leu2, ura3 | This Study | ||
| MYY729 | MATα, mdm1-202, his3, leu2, ura3 | This Study | ||
| MYY734 | MATα, mdm1-204, his3, leu2, ura3 | This Study | ||
| MYY735 | MATα, mdm1-217, his3, leu2, ura3 | This Study | ||
| MYY740 | MATα, mdm1-227, his3, leu2, ura3 | This Study | ||
| MYY742 | MATα, mdm1-228, his3, leu2, ura3 | This Study | ||
| MYY745 | MATα, mdm1-251, his3, leu2, ura3 | This Study | ||
| MYY746 | MATα, mdm1-252, his3, leu2, ura3 | This Study |
Construction of New mdm1 Alleles
Plasmid pMDM1 was constructed by subcloning the 2.1-kb SalI–EcoRV fragment containing the MDM1 gene from plasmid YCp50-MDM1 into the SalI and EcoRV sites of plasmid pRS423 (Sikorski and Hieter, 1989). Plasmid pMDM1 was mutagenized in vitro with hydroxylamine as described by Sikorski and Boeke (1991).
Mutagenized, plasmid-borne copies of MDM1 that conferred temperature-sensitive growth on cells that harbored no other copy of MDM1 were identified by a “plasmid shuffling” protocol similar to that described by Sikorski and Boeke (1991). Briefly, MYY404-1b cells were transformed with the pool of mutagenized pMDM1 DNA. Loss of the plasmid YCp50-MDM1 containing the URA3 gene and the wild-type copy of MDM1 was selected by culturing on medium containing 5-fluoro-orotic acid (FOA).1 Cells resistant to FOA were tested for growth at 37°C, and 75 independently derived, temperature-sensitive isolates were identified. Plasmids encoding various MDM1 alleles were recovered and amplified in bacterial cells.
New mutant versions of MDM1 were integrated at the chromosomal MDM1 locus of haploid cells by a two-step gene replacement strategy. DNA sequences encoding mdm1 alleles were excised from plasmid pMDM1 by digestion with SphI and NheI and subcloning into the SphI and NheI sites in the yeast integrating vector YIp5 (Struhl et al., 1979). The YIp5 constructs were linearized at the unique BglII site in the MDM1 gene; linearized plasmids were transformed into MYY290; and Ura+ transformants were isolated. Correct integration of YIp5 at the MDM1 locus was confirmed by PCR analysis and by Southern blotting.
Excision of YIp5 and wild-type MDM1 sequences was selected by plating cells on medium containing FOA. The resulting Ura− isolates were tested for growth on YPD medium at 37°C, and temperature-sensitive strains were identified as candidate mutants. Temperature-sensitive growth of the resulting strains was shown to be caused by mutations in MDM1 by the following analyses. Recessive mutants were complemented by transformation with unmutagenized MDM1 (pMDM1). Dominant mutants were mated to MYY403 (mdm1-1); the resulting diploids were sporulated; and all meiotic progeny were shown to be temperature sensitive for growth. Further, a 2:2 ratio of segregation of temperature sensitivity was observed in meiotic progeny derived from a cross of mdm1 mutants to the wild-type strain MYY291. The presence of a single copy of MDM1 (as well as the absence of YIp5 sequences) was verified by PCR and Southern blot analyses of genomic DNA isolated from each mutant.
New mutant alleles of MDM1 were characterized by DNA sequence analysis as follows. Genomic DNA was isolated from mutant cells harboring each allele. Asymmetric PCR (McCabe, 1990) was performed using different ratios of the primers 5′-CTCAGCTCTTTGGTTCAG-3′ and 5′-GGCTGAAGAAGTGTACC-3′ flanking the MDM1 ORF to create predominantly single-stranded products corresponding to either the coding or noncoding strand of MDM1. Nucleotide sequences of the PCR products were determined by dideoxy sequencing. All mutations were verified by sequencing both coding and noncoding strands derived from independent PCR reactions.
Phenotypic Analysis
Fluorescence and immunofluorescence microscopy to visualize mitochondria, microtubules, nuclei, and Mdm1p structures was performed as previously described (McConnell et al., 1990; McConnell and Yaffe, 1992). To visualize bud scars in conjunction with indirect immunofluorescence, cells were incubated in a solution of 10 μg/ml Calcofluor (Sigma Chemical Co., St. Louis, MO) for 10 min before the final wash step. In some experiments, cell populations were synchronized in the cell cycle by treatment with the yeast mating pheromone α-factor as previously described (McConnell et al., 1990). Electron microscopy was performed as described previously (Sogo and Yaffe, 1994).
Results
Isolation of New mdm1 Alleles
Yeast strains harboring new mdm1 mutant alleles were generated by in vitro mutagenesis of the cloned MDM1 gene followed by replacement of the chromosomal, wild-type MDM1 gene with a mutated copy. This was accomplished in two steps (described in detail in Materials and Methods). First, mutagenized plasmids containing the MDM1 gene were screened for their ability to confer temperature-sensitive growth on cells that contained no other copy of MDM1. Second, candidate mdm1 alleles were integrated at the chromosomal MDM1 locus using a two-step allele replacement strategy. Correct integration and gene replacement were verified by PCR analysis, Southern blotting, and characterization of meiotic progeny derived from crosses of the new mutant strains to the mdm1-1 mutant. 75 independently isolated plasmids conferring temperature-sensitive growth were identified. Of these, mdm1 sequences from 13 were integrated at the MDM1 locus, and 10 mutant alleles that appeared to represent the range of phenotypic variations observed (as described below) were characterized in detail.
The dominant or recessive character of the new mdm1 alleles was determined by examining the growth of heterozygous (MDM1/mdm1) diploids obtained by crossing the mutant strains to the wild-type parent of opposite mating type. Six of the heterozygous diploid strains failed to grow at 37°C and therefore harbored dominant mutations, while the remaining four diploid strains grew at 37°C and therefore contained recessive alleles (Table II). The growth defects conferred by the recessive haploid alleles could be complemented by transformation of the strains with a centromere-based vector containing the wild-type MDM1 gene.
Table II.
New mdm1 Alleles
| Allele | Character* | Mitochondrial Inheritance‡ | Nuclear Phenotype§ | Class‖ | ||||
|---|---|---|---|---|---|---|---|---|
| mdm1-20 | R | − | a | Ia | ||||
| mdm1-199 | R | − | a | Ia | ||||
| mdm1-200 | D | + | a | IIIa | ||||
| mdm1-202 | D | − | + | II | ||||
| mdm1-204 | D | − | b | Ib | ||||
| mdm1-217 | R | + | a | IIIa | ||||
| mdm1-227 | D | + | b | IIIb | ||||
| mdm1-228 | R | + | b | IIIb | ||||
| mdm1-251 | D | − | a | Ia | ||||
| mdm1-252 | D | − | + | II |
R, recessive; D, dominant.
+, normal mitochondrial distribution and morphology. −, defective mitochondrial distribution and morphology. §+, normal nuclear distribution. a, nucleus/nuclear division in mother. b, nucleus/nuclear division in bud.
Roman numeral refers to summary of organelle inheritance: Ia and Ib, mitochondrial distribution and morphology defect and nuclear transmission defects. II, mitochondrial distribution and morphology defect, normal nuclear transmission. IIIa and IIIb, nuclear transmission defect, normal mitochondrial distribution and morphology. lower case a or b refers to nuclear transmission phenotype.
Uncoupling of Nuclear and Mitochondrial Inheritance Defects
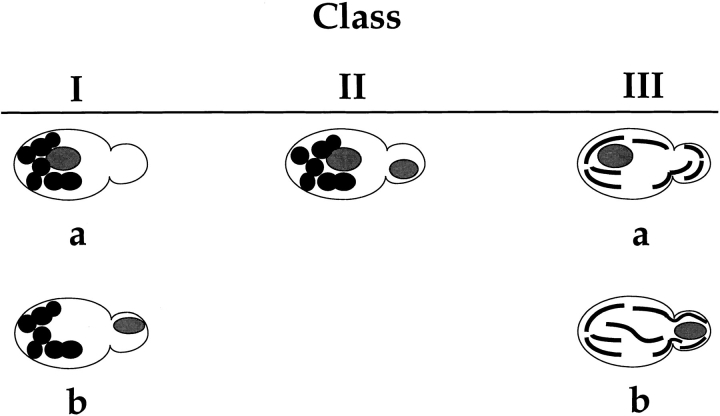
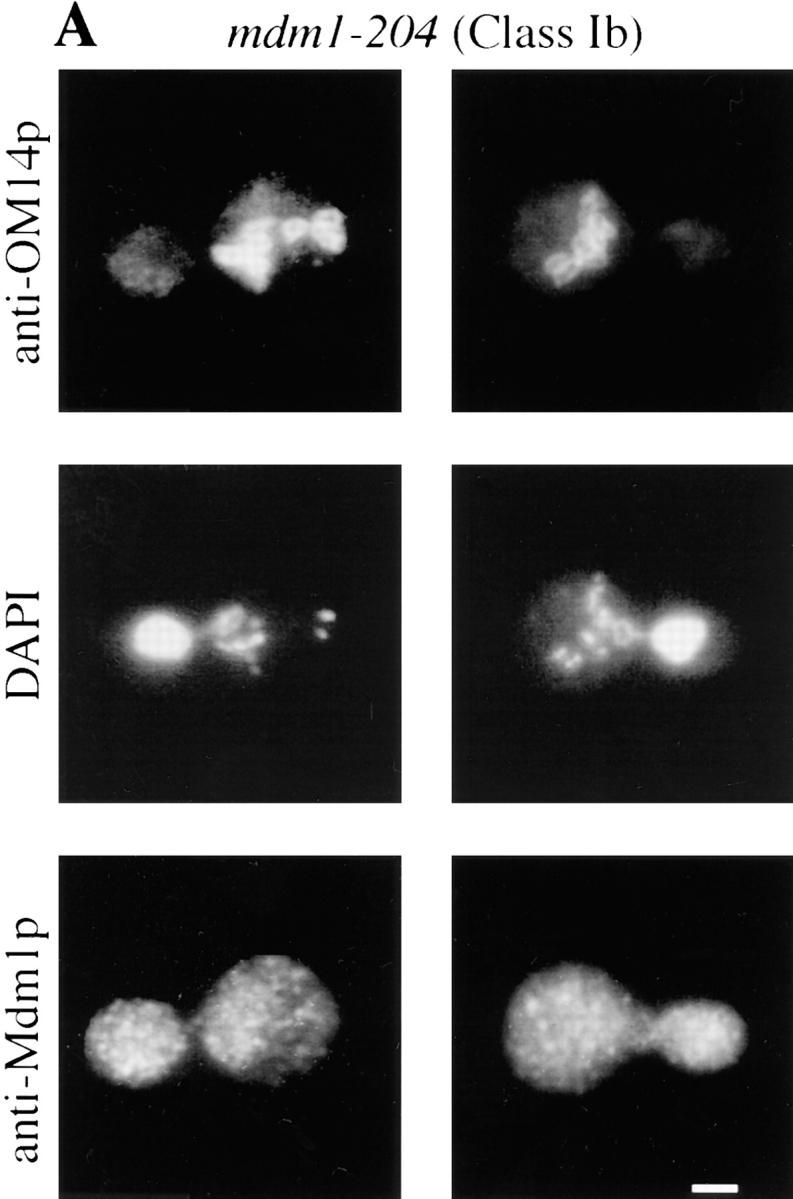
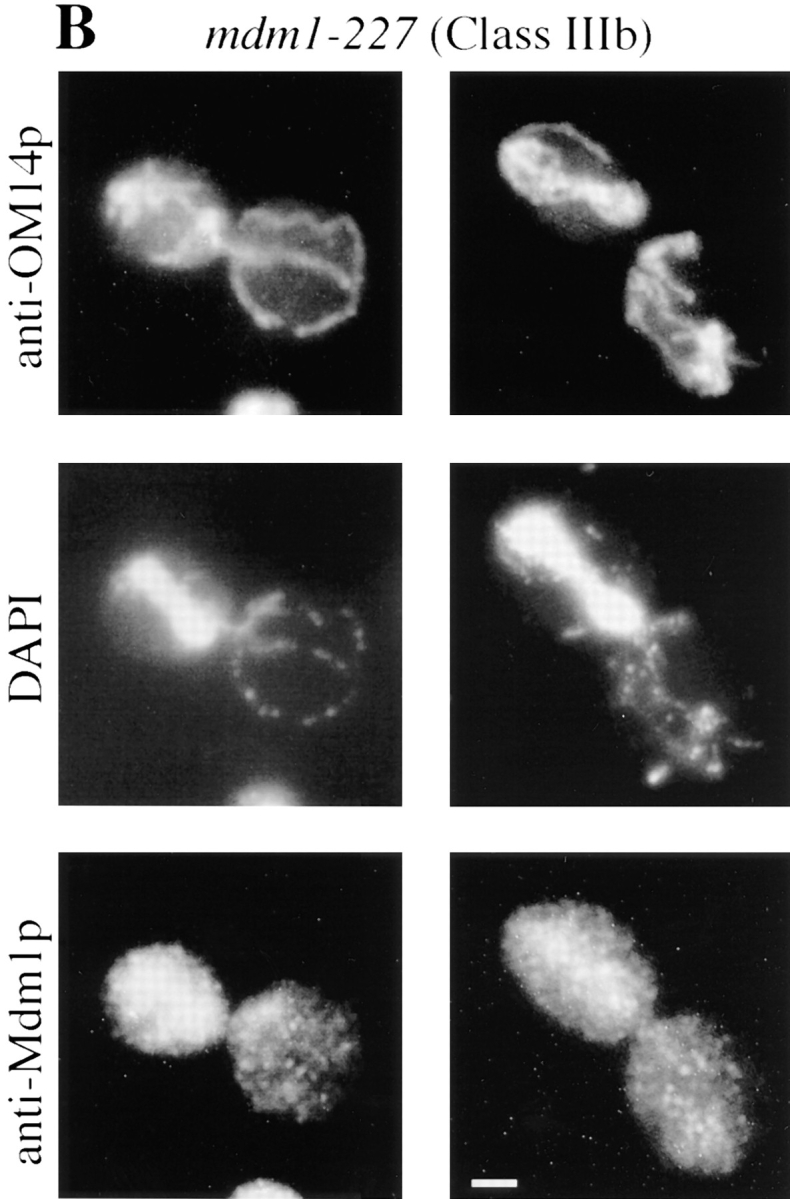
To characterize the effects of the new mutations on organelle inheritance, cells were analyzed by fluorescence and indirect immunofluorescence microscopy (Fig. 1). An initial examination of cells treated with the mitochondria-specific, vital dye, 2-(4-dimethylaminostyryl)-1-methylpyridinium iodide (DASPMI), revealed that only a subset of the new alleles caused defects in mitochondrial distribution. Further analysis of both nuclei and mitochondria by indirect immunofluorescence microscopy permitted the assignment of mutant alleles to three phenotypic classes (Table II). Whereas wild-type cells incubated at 37°C displayed normal organelle distribution and stable Mdm1p structures (Fig. 1 A), Class I mutants displayed defects in both mitochondrial and nuclear transmission to buds (Fig. 1 B), similar to the original mdm1-1 allele. Class II alleles appeared to affect only mitochondrial inheritance, with normal nuclear behavior (Fig. 1 C). Finally, Class III mutants displayed aberrant nuclear transmission but normal mitochondrial distribution (Fig. 1 D). Both dominant and recessive Class I and Class III alleles were identified, but both Class II alleles obtained were dominant. All of the mdm1 mutant cells possessed stable Mdm1p-punctate structures (Fig. 1, B–D), in contrast to the Mdm1p structures observed in the original mdm1-1 strain, which disappeared after incubation at 37°C (McConnell and Yaffe, 1992). Additionally, immunoblot analysis of cellular extracts revealed no difference in Mdm1p levels between wild-type and mutant cells grown at the permissive or nonpermissive temperature (data not shown), indicating that the new mutant phenotypes were not caused by unstable forms of Mdm1p. In some cells there appeared to be a concentration of Mdm1p staining at or around the nucleus; this staining was observed in both wild-type and mutant cells and did not appear to correspond to a particular phase of the cell cycle. Like the original mdm1-1 mutant, none of the new mutant alleles appeared to alter actin distribution (to cortical patches and cables) at either permissive or nonpermissive temperature (data not shown).
Figure 1.


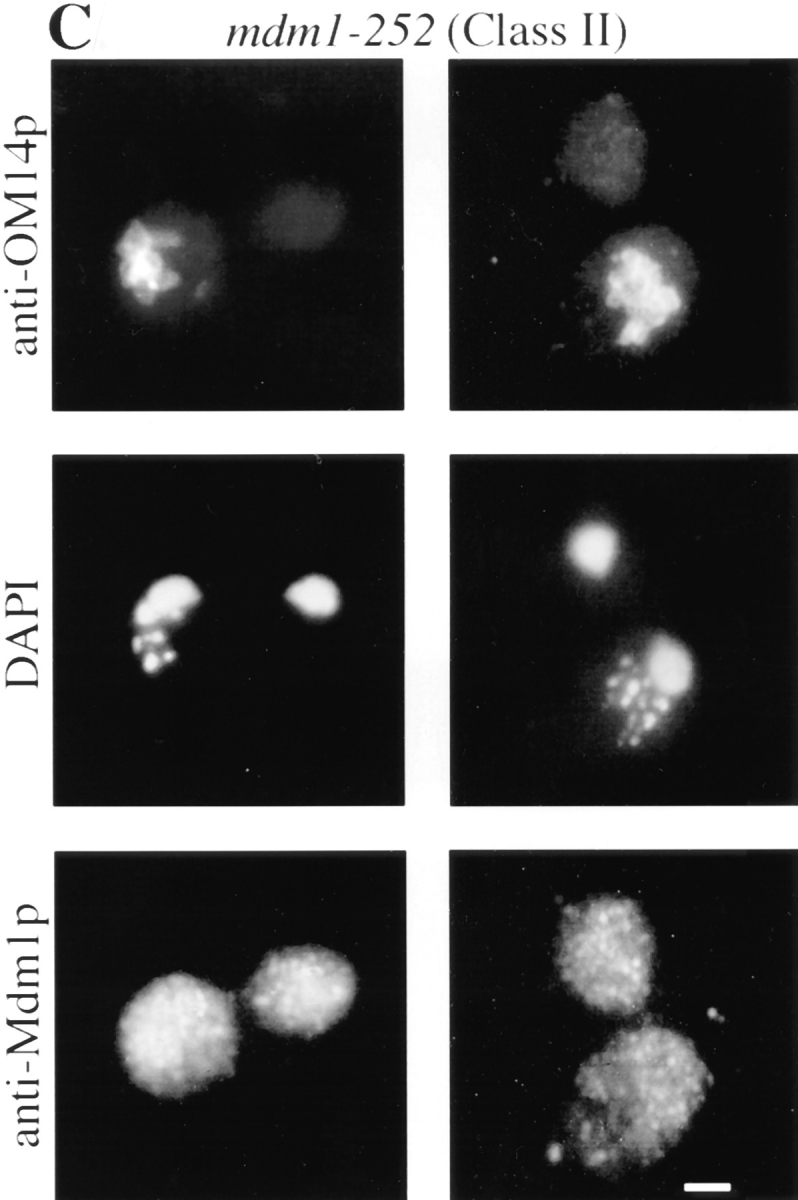
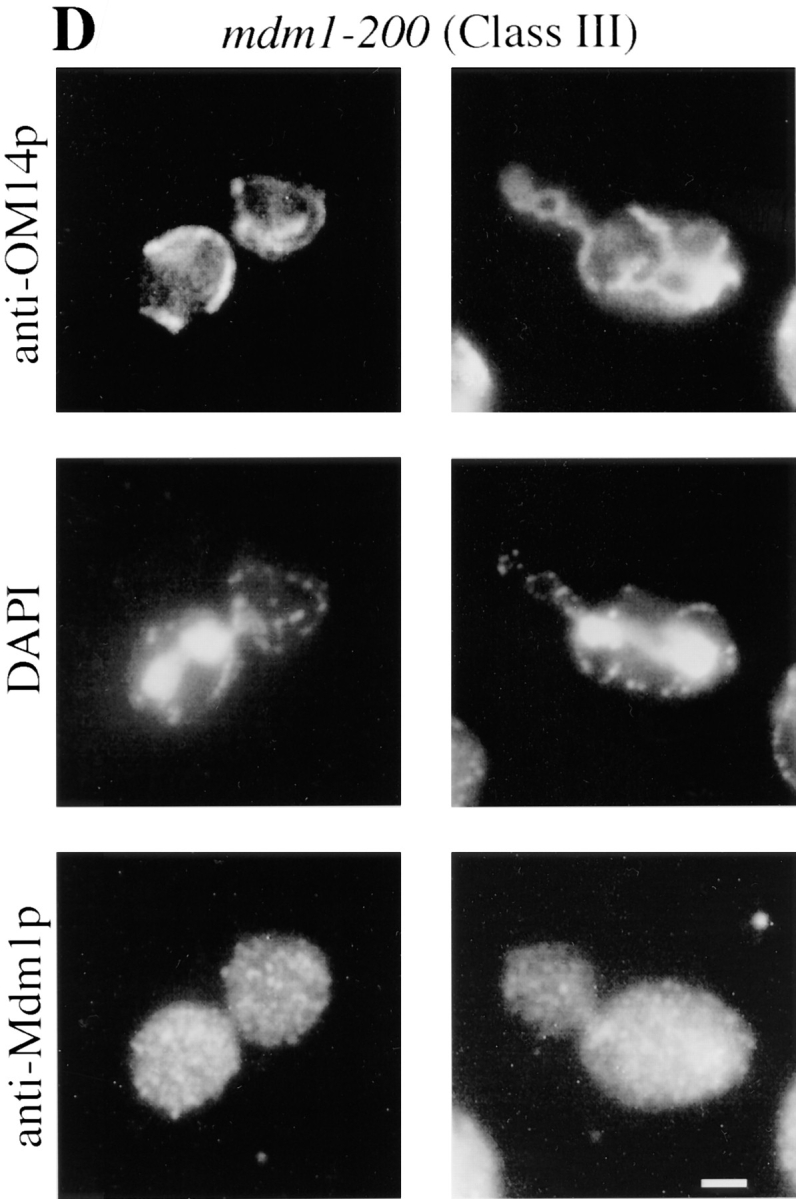
New mdm1 alleles define three phenotypic classes. MYY290 (MDM1) (A), MYY720 (mdm1-251) (B), MYY721 (mdm1-252) (C), and MYY702 (mdm1-200) (D) cells were grown on YPD medium at 23°C, incubated at 37°C for 4 h, fixed with formaldehyde, and processed for indirect immunofluorescence microscopy. Mitochondria were detected with a mouse monoclonal antibody against OM14 (a mitochondrial outer-membrane protein) followed by fluorescein-conjugated goat anti–mouse IgG. Nuclear and mitochondrial DNAs were visualized by DAPI staining. Mdm1p structures were detected using affinity-purified, anti-Mdm1p antibodies followed by rhodamine-conjugated donkey anti–rabbit IgG. For each strain, two representative cells are shown. Bars, 2 μm.
Class I and Class II Mutants Display Aberrant Mitochondrial Morphology
In addition to a defect in mitochondrial distribution, microscopic analysis of Class I and Class II mutants revealed altered mitochondrial morphology. Instead of the wild-type, tubular mitochondrial profiles (Fig. 1 A), mitochondria in these mdm1 mutants appeared to be round structures of fairly uniform size (Fig. 1, B and C). This morphology was confirmed for a Class II mutant, mdm1-252 (MYY721), by electron microscopy (Fig. 2). The round mitochondria frequently appeared clumped together in the mother portion of a dividing cell and were often localized to the end opposite the emerging bud. Wild-type mitochondrial morphology was observed in Class II mutant strains cultured at permissive temperature (data not shown). After the shift of a Class II mutant culture to 37°C, cells with empty buds began appearing in the population after 1 h; cells with small, round mitochondria were apparent after 2 h; and the majority of cells displayed both empty buds and small, round mitochondria by 4 h (data not shown).
Figure 2.

Class II alleles display altered mitochondrial morphology and defects in mitochondrial inheritance. MYY290 (MDM1) (A) and MYY721 (mdm1-252) (B) cells were grown in YPG medium overnight at 23°C, incubated at 37°C in YPG for 6 h, prefixed with glutaraldehyde, fixed in KMnO4, and stained in uranyl acetate. Sections were stained with lead citrate. Two representative cells of each strain are shown. Bar, 1 μm.
Class I and Class III Mutant Alleles Produce Two Distinct Nuclear Transmission Defects
Characterization of the different Class I and Class III alleles revealed two types of defect in nuclear transmission. Some alleles (Class Ia and Class IIIa) were defective for the movement of nuclei into daughter buds (Fig. 1, B and D). Additionally, nuclear division occurred in a fraction of these cells, yielding two nuclei in the mother portion of the cell. These phenotypes are similar to those described for the mdm1-1 mutant (McConnell et al., 1990).
A distinct, novel nuclear phenotype was displayed by several other alleles (Class Ib and Class IIIb). In these strains, a significant fraction of the cells incubated at the nonpermissive temperature exhibited all of the nuclear DNA localized to the bud (Fig. 3, A and B). Double-label experiments in which cells were stained with both DAPI (to reveal DNA) and Calcofluor (to label bud scars on the mother portion of the cells) confirmed that daughter buds were, in fact, receiving all of the nuclear DNA in these cells (Fig. 3 C). Visualization of microtubules in Class Ib and Class IIIb cells by indirect immunofluorescence microscopy revealed that many of the nuclei in the buds possessed unelongated mitotic spindles (Fig. 3 C), indicating aberrant positioning of the premitotic spindle apparatus.
Figure 3.
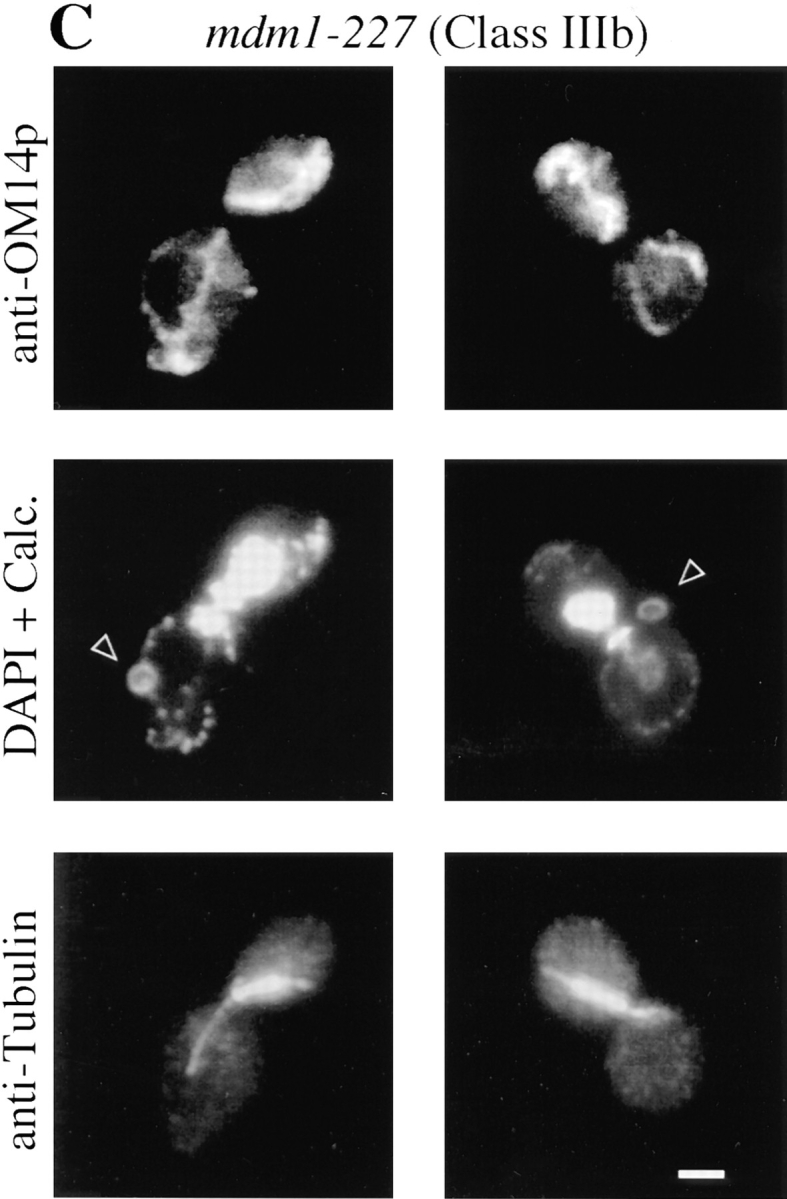
Class Ib-mdm1 and Class IIIb-mdm1 cells display complete transfer of nuclear DNA to daughter buds. MYY709 (mdm1-204) (A) and MYY715 (mdm1-227) (B and C) cells were grown at 23°C, incubated for 4 h at 37°C, and fixed and processed as described for Fig. 1. (A and B) Cells were stained with anti-OM14 followed by fluorescein-conjugated goat anti–mouse IgG, DAPI, and anti-MDM1p followed by rhodamine-conjugated donkey anti–rabbit IgG. (C) Cells were stained with anti-OM14 followed by fluorescein-conjugated goat anti–mouse IgG, DAPI and calcofluor (Calc.; to visualize bud scars), and anti-tubulin followed by rhodamine-conjugated donkey anti–rat IgG. Two representative cells are shown for each strain. Bars, 2 μm.
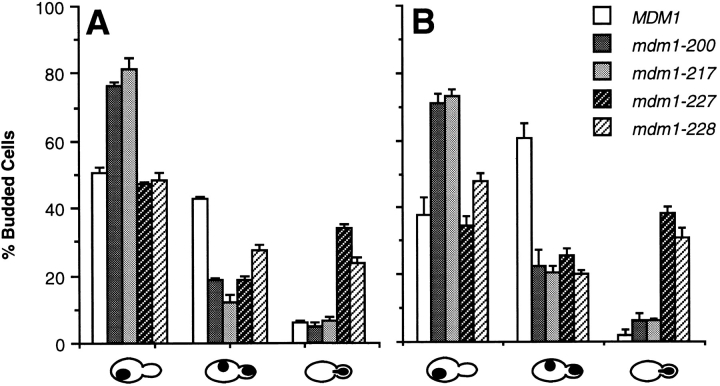
To characterize the Class IIIb phenotype in greater detail, mutant cells were synchronized in the G1 (unbudded) stage of the cell cycle, and nuclear distribution was analyzed 2 and 4 h after release from the cell cycle block at the non permissive temperature. After both 2 and 4 h, 70–82% of budded cells harboring either of two different Class IIIa alleles (mdm1-200 or mdm1-217) possessed nuclei confined to the mother portion of the cell (Fig. 4). In contrast, a substantial fraction (24–34%) of budded cells harboring either of two Class IIIb alleles (mdm1-227 or mdm1-228) displayed nuclear DNA entirely transferred to the bud after 2 h (Fig. 4), and this fraction increased further (to 31– 38%) by 4 h. Correspondingly, the fraction of cells possessing properly divided nuclei distributed between the mother and bud was significantly reduced in both Class IIIa and Class IIIb cultures compared to wild-type cells.
Figure 4.
Nuclear distribution defects in Class III-mdm1 cells. MYY290 (MDM1), Class IIIa mutants MYY702 (mdm1-200) and MYY710 (mdm1-217), and Class IIIb mutants MYY715 (mdm1-227) and MYY717 (mdm1-228) were synchronized with α-factor at 23°C, released from the cell cycle block at 37°C, fixed with formaldehyde after 0, 2, or 4 h, and stained with DAPI. For each sample, the distribution of phenotypes among 300 cells was determined. Values represent the mean ± standard deviation for triplicate samples. Between 85 and 95% of cells in 0 h samples were unbudded. Inset shows symbols corresponding to each culture, and each phenotype is represented below the x axis. (A) 2 h after release at 37°C. (B) 4 h after release at 37°C.
Certain Combinations of mdm1 Alleles Exhibit Intragenic Complementation and Suppression
During initial genetic analysis, diploids generated by crosses of all the new mdm1 alleles to the original mdm1-1 mutant strain were found to grow at 37°C, suggesting intragenic complementation (data not shown). However, crosses of new alleles to the wild-type strain from which the mdm1-1 mutant was derived (A364a) (McConnell et al., 1990) revealed that alleles found to be dominant in the MYY290 background behaved as recessive mutations in the hybrid strain. Subsequent attempts to reconstruct the mdm1-1 mutation in the genetic background of the new alleles (strain MYY290) were unsuccessful, and this failure was consistent with the mdm1-1 mutation being lethal in strain MYY290 (data not shown). Because of these strain background variations, subsequent analyses were limited to the new alleles in an otherwise isogenic MYY290 background.
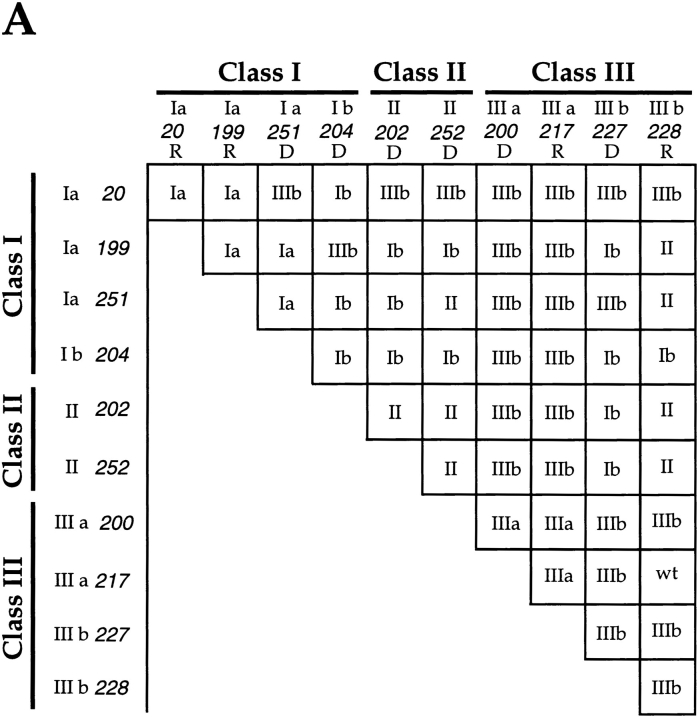
To examine in greater detail the interactions between different mdm1 mutant alleles, all possible diploid combinations of the 10 new alleles were generated, and phenotypes of the resulting diploids were analyzed. Cells were examined for growth at 37°C and by fluorescence microscopy for mitochondrial and nuclear distribution. The consequent mutant phenotypes revealed by this analysis indicated a complex pattern of interactions (Fig. 5 A). First, certain combinations of dominant alleles yielded an apparently additive (and expected) phenotype. For example, the combination of either dominant Class II mutation with the dominant Class IIIb mutation, mdm1-227, produced diploids displaying a Class Ib phenotype. Second, some combinations of alleles yielded unexpected synthetic phenotypes. For example, the diploid obtained by crossing the recessive Class Ia mutant, mdm1-199, with the dominant Class II mutant, mdm1-202, displayed a Class Ib phenotype. Third, certain crosses led to intragenic suppression in which a phenotype caused by a dominant mutation was not observed in the diploid. An example of such intragenic suppression was the cross of the dominant Class II mutant, mdm1-202, with the dominant Class IIIa mutant, mdm1-200, resulting in a Class IIIb diploid that displayed no defect in mitochondrial distribution (suppression of the mitochondrial defect). A final type of interaction was intragenic complementation, in which the combination of two different alleles eliminated mutant phenotypes observed in one or both of the parental strains. An example of such complementation emerged from the mating of recessive Class IIIa mutant, mdm1-217, with recessive Class IIIb mutant, mdm1-228, to yield a diploid strain with wild-type growth and normal organelle distribution (Fig. 5 B).
Figure 5.
Intragenic interaction among mdm1 alleles. Genetic crosses were performed to obtain all possible pairwise combinations of different mdm1 alleles. Organelle inheritance phenotypes were analyzed as described in Fig. 1. (A) Phenotypic classes of mdm1 diploids. Numbers on the x and y axis indicate mdm1 allele of parental strains. Roman numeral refers to phenotypic class, and lower case “a” or “b” refers to nuclear transmission phenotype, as described in Table II and in the text. “R” or “D” indicates the recessive or dominant character of individual alleles. (B) Intragenic complementation is observed between mdm1-217 (Class IIIa) and mdm1-228 (Class IIIb). Haploids and diploids harboring the mdm1-217 and mdm1-228 mutations were tested for growth on YPD at 37°C. (a–c) mdm1-217; (a) MATa, (b) MATα, and (c) MATa/α; (d–f) mdm1-228; (d) MATa, (e) MATα, and (f) MATa/α; (g) mdm1-217/mdm1-228 (a/α); (h) mdm1-228/mdm1-217.
Sequence Analysis
Mutations corresponding to the new mdm1 alleles were determined by nucleotide sequence analysis and are listed in Table III. Each mdm1 allele consists of a single mutation, with the exception of mdm1-251, a Class Ia allele containing R187Q and E378K. The R187Q mutation was identified also in the Class II allele, mdm1-252, and is therefore likely to be responsible for the mitochondrial inheritance defect in mdm1-251. Consistent with this finding, diploids expressing both mdm1-251 and mdm1-252 display a Class II phenotype (Fig. 5 A). These observations suggest that the E378K mutation causes a recessive, type “a” nuclear segregation defect (which is suppressed in the diploid).
Table III.
Sequence Analysis of New mdm1 Alleles
| Allele | Class | Mutation* | Substitution | |||
|---|---|---|---|---|---|---|
| mdm1-20 | Ia | A 541 G | T181A | |||
| mdm1-199 | Ia | G 1062 C | M354I | |||
| mdm1-204 | Ib | G 1067 T | W356L | |||
| mdm1-251 | Ia | G 560 A | R187Q | |||
| G 1132 A | E378K | |||||
| mdm1-202 | II | C 1234 G | R412G | |||
| mdm1-252 | II | G 560 A | R187Q | |||
| mdm1-200 | IIIa | A 1099 G | N367D | |||
| mdm1-217 | IIIa | A 590 G | E197G | |||
| mdm1-227 | IIIb | C 446 G | T149T | |||
| mdm1-228 | IIIb | A 1057 G | N353D |
Nucleotide positions are numbered relative to ‘A' of ATG, which is defined as position 1.
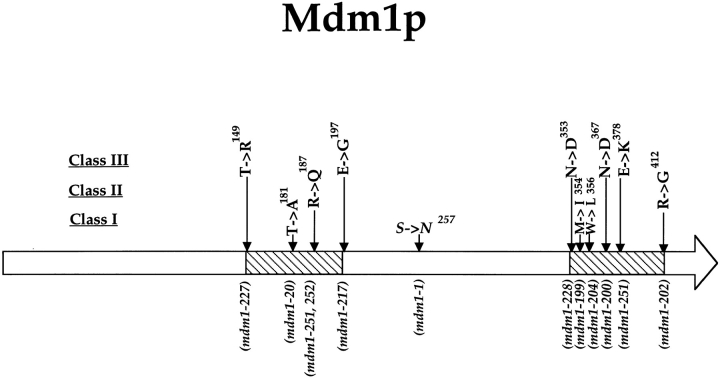
The mutations found in the various mdm1 alleles mapped to two discrete clusters within the MDM1 open reading frame (Fig. 6). These lesions did not appear to cluster with respect to phenotype, as mutations conferring all three phenotypic classes were found in both regions.
Figure 6.
Distribution of mutations in Mdm1p.
Discussion
Previously, Mdm1p was identified as a component of a novel system of cytoplasmic structures essential for nuclear and mitochondrial inheritance in yeast (McConnell et al., 1990; McConnell and Yaffe, 1992, 1993). These earlier investigations revealed that incubation of the mdm1-1 mutant at the nonpermissive temperature led to defects in nuclear and mitochondrial transmission to daughter buds and to the disappearance of cytoplasmic, punctate structures defined by Mdm1p. We have now separated the assembly and stability of these Mdm1p structures from their function in organelle inheritance by the isolation and analysis of new mdm1 alleles. Furthermore, these new alleles define three phenotypic classes (summarized in Fig. 7). At restrictive temperature, cells harboring these new mutant alleles possess stable Mdm1p punctate structures yet display aberrant organelle inheritance. Although currently available techniques do not allow the identification of punctate structures that might be incorrectly assembled, the successful transmission of nuclei in Class II mutants and of mitochondria in Class III mutants implies that some property other than network assembly (e.g., specific binding of an interacting protein) is affected by the new mutations.
Figure 7.
Summary of mdm1 mutant phenotypes.
The specific mutations that give rise to the new mdm1 alleles alter residues in two distinct regions within the Mdm1p protein (Fig. 6). Each of these regions contains mutations yielding each phenotypic class, so our analysis does not indicate that distinct structural domains of Mdm1p are specialized for function in nuclear or mitochondrial inheritance. Rather, various mdm1 mutations may differentially affect the binding affinity of proteins that link mitochondria or nuclei to Mdm1p structures. Thus, the new mdm1 lesions appear to define two distinct binding sites for the interaction of Mdm1p with other cellular structures.
Class I and Class III mdm1 mutations cause aberrant positioning of the mitotic spindle leading to defective nuclear transmission. Different alleles conferring this phenotype display two strikingly different consequences: type “a” alleles (similar to the original mdm1-1 allele) show nuclear DNA entirely confined to the mother portion of the cell; while type “b” alleles show the complete transfer of all nuclear DNA into daughter buds. In a fraction of cells of both types, nuclear division occurs to yield two nuclei confined to either the mother or bud, respectively. Similar nuclear phenotypes have been observed for several other mutations that affect the positioning of the mitotic spindle. Type “a” phenotypes have been reported for cells with specific mutations in β-tubulin (tub2-401; Sullivan and Huffaker, 1992) and actin (act1-4; Palmer et al., 1992), as well as in dynein (dhc1/dyn1 [Eshel et al., 1993; Li et al., 1993; Yeh et al., 1995] and jnm1 [McMillan and Tatchell, 1994]), Act3p/Act5p (Clark and Meyer, 1994; Muhua et al., 1994), and Num1p (Kormanec et al., 1991; Farkasovsky and Kuntzel, 1995). A type “b” phenotype has been described for cells with the esp1 mutation (McGrew et al., 1992). All of these mutations affect proteins that are either components of the mitotic spindle or appear to interact with either spindle poles or astral microtubules. Mdm1p may interact with one or more of these components, and evidence for such interaction may emerge from further genetic analysis of particular Class III alleles.
The intragenic interactions observed among various mdm1 alleles support a model of Mdm1p assembly into multimeric structures and suggest that the nature or composition of these multimeric assemblies influences the activity of the Mdm1p network. Intragenic complementation of some recessive organelle inheritance defects suggests that functions defective in one allele can sometimes be supplied by a second allele in trans. In addition, complex genetic phenomena involving various allelic pairs, such as intragenic suppression and synthetic phenotypes, imply complex physical interactions between Mdm1p subunits, such that a domain of one molecule may influence the activity of a nearby region of a partner subunit in a multimeric complex. This model of Mdm1p as a multimeric complex is consistent with biochemical data indicating the aggregation of Mdm1p in cellular extracts (McConnell and Yaffe, 1992) and the assembly of purified Mdm1p into intermediate filament-like structures in vitro (McConnell and Yaffe, 1993).
One model for Mdm1p function is that this protein forms a matrix or scaffold-like network in the cytoplasm that is used to position and orient cellular structures. In particular, the spindle pole bodies (SPBs) and mitochondria could be tethered to this scaffolding at particular times during the cell cycle. Changes in mitochondrial distribution or spindle position might involve selective binding and release along the Mdm1p network. A mutation that affects organelle distribution might act either by weakening binding or by preventing appropriate release. For example, if positioning of the mitotic spindle normally involves binding of the maternal SPB to Mdm1p structures and release of the daughter SPB to be carried into the bud, different effects of mdm1 mutations on nuclear position could be explained either by failure to release the daughter SPB (type “a” alleles) or failure to bind the maternal SPB (type “b” alleles). Similarly, mitochondrial distribution and morphology might reflect the binding of the mitochondrial outer membrane or outer membrane-associated proteins to Mdm1p structures. Either loss of such binding or an unregulated or strengthened interaction with Mdm1p could produce the changes in mitochondrial morphology and distribution observed in Class I and Class II mdm1 alleles. The isolation and analysis of allele-specific suppressors, particularly of Class II and Class III alleles, should lead to the identification of proteins that interact with Mdm1p to mediate its functions in nuclear and mitochondrial inheritance.
Acknowledgments
We are grateful to Lance Washington for assistance with electron microscopy and to Karen Berger, Randy Hampton, Kelly Shepard, and Mark Nickas for critical reading of the manuscript and their helpful suggestions.
This work was supported by grant GM44614 from the National Institutes of Health and grant MCB-9305338 from the National Science Foundation.
Abbreviations used in this paper
- FOA
5-fluoro-orotic acid
- SPB
spindle pole body
Footnotes
Please address all correspondence to Dr. Michael P. Yaffe, Department of Biology, 0347, University of California, San Diego, La Jolla, CA 92093-0347. Tel.: (619) 534-4769; Fax: (619) 534-4403; E-mail: myaffe@ucsd.edu
References
- Adams RJ, Pollard TD. Propulsion of organelles isolated from Acanthamoeba along actin filaments by myosin-I. Nature (Lond) 1986;322:754–756. doi: 10.1038/322754a0. [DOI] [PubMed] [Google Scholar]
- Allan V. Membrane traffic motors. FEBS (Fed Eur Biochem Soc) Lett. 1995;369:101–106. doi: 10.1016/0014-5793(95)00615-g. [DOI] [PubMed] [Google Scholar]
- Ball EH, Singer SJ. Mitochondria are associated with microtubules and not with intermediate filaments in cultured fibroblasts. Proc Natl Acad Sci USA. 1982;79:123–126. doi: 10.1073/pnas.79.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger KH, Yaffe MP. Mitochondrial distribution and inheritance. Experientia (Basel) 1996;52:1111–1116. doi: 10.1007/BF01952109. [DOI] [PubMed] [Google Scholar]
- Chen LB. Mitochondrial membrane potential in living cells. Annu Rev Cell Biol. 1988;4:155–181. doi: 10.1146/annurev.cb.04.110188.001103. [DOI] [PubMed] [Google Scholar]
- Clark SW, Meyer DI. ACT3: a putative centractin homologue in S. cerevisiaeis required for proper orientation of the mitotic spindle. J Cell Biol. 1994;127:129–138. doi: 10.1083/jcb.127.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couchman JR, Rees DA. Organelle-cytoskeleton relationships in fibroblasts: mitochondria, Golgi apparatus, and endoplasmic reticulum in phases of movement and growth. Eur J Cell Biol. 1982;27:47–54. [PubMed] [Google Scholar]
- David-Ferreira KL, David-Ferreira JF. Association between intermediate-sized filaments and mitochondria in rat Leidyg cells. Cell Biol Int Rep. 1980;4:655–662. doi: 10.1016/0309-1651(80)90204-0. [DOI] [PubMed] [Google Scholar]
- Eshel D, Urrestarazu S, Vissers S, Jauniax J-C, Van Vliet-Reedijk JC, Planta RJ, Gibbons IR. Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc Natl Acad Sci USA. 1993;90:11172–11176. doi: 10.1073/pnas.90.23.11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkasovsky M, Kuntzel H. Yeast Num1p associates with the mother cell cortex during S/G2phase and affects microtubular function. J Cell Biol. 1995;131:1003–1014. doi: 10.1083/jcb.131.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heggeness MH, Simon M, Singer SJ. Association of mitochondria with microtubules in cultured cells. Proc Natl Acad Sci USA. 1978;75:3863–3866. doi: 10.1073/pnas.75.8.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachar B, Reese TS. The mechanism of cytoplasmic streaming in Characean algal cells: sliding of endoplasmic reticulum along actin filaments. J Cell Biol. 1988;106:1545–1552. doi: 10.1083/jcb.106.5.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kormanec J, Schaaff-Gerstenschlager I, Zimmermann FK, Perecko D, Kuntzel H. Nuclear migration in Saccharomyces cerevisiaeis controlled by the highly repetitive 313 kDa NUM1 protein. Mol Gen Genet. 1991;230:277–287. doi: 10.1007/BF00290678. [DOI] [PubMed] [Google Scholar]
- Li Y-Y, Yeh E, Hays T, Bloom K. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc Natl Acad Sci USA. 1993;90:10096–10100. doi: 10.1073/pnas.90.21.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe, P.C. 1990. Production of single-stranded DNA by asymmetric PCR. In PCR Protocols. M.A. Innis, D.H. Gelfand, J.J. Sninsky, and T.J. White, editors. Academic Press, San Diego. 76–83.
- McConnell SJ, Yaffe MP. Nuclear and mitochondrial inheritance in yeast depends on novel cytoplasmic structures defined by the MDM1 protein. J Cell Biol. 1992;118:385–395. doi: 10.1083/jcb.118.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell SJ, Yaffe MP. Intermediate filament formation by a yeast protein essential for organelle inheritance. Science (Wash DC) 1993;260:687–689. doi: 10.1126/science.8480179. [DOI] [PubMed] [Google Scholar]
- McConnell SJ, Stewart LC, Talin A, Yaffe MP. Temperature-sensitive yeast mutants defective in mitochondrial inheritance. J Cell Biol. 1990;111:967–976. doi: 10.1083/jcb.111.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrew JT, Goetsch L, Byers B, Baum P. Requirement for ESP1 in the nuclear division of Saccharomyces cerevisiae. . Mol Biol Cell. 1992;3:1443–1454. doi: 10.1091/mbc.3.12.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan JN, Tatchell K. The JNM1 gene in the yeast Saccharomyces cerevisiaeis required for nuclear migration and spindle orientation during the mitotic cell cycle. J Cell Biol. 1994;125:143–158. doi: 10.1083/jcb.125.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mose-Larsen P, Bravo R, Fey SJ, Small JV, Celis JE. Putative association of mitochondria with a subpopulation of intermediate-sized filaments in cultured human skin fibroblasts. Cell. 1982;31:681–692. doi: 10.1016/0092-8674(82)90323-3. [DOI] [PubMed] [Google Scholar]
- Muhua L, Karpova TS, Cooper JA. A yeast actin-related protein homologous to that in vertebrate dynactin complex is important for spindle orientation and nuclear migration. Cell. 1994;78:669–679. doi: 10.1016/0092-8674(94)90531-2. [DOI] [PubMed] [Google Scholar]
- Palade G. Membrane biogenesis: an overview. Methods Enzymol. 1983;96:29–40. doi: 10.1016/s0076-6879(83)96004-4. [DOI] [PubMed] [Google Scholar]
- Palmer RE, Sullivan DS, Huffaker T, Koshland D. Role of astral microtubules and actin in spindle orientation and migration in the budding yeast, Saccharomyces cerevisiae. . J Cell Biol. 1992;119:583–593. doi: 10.1083/jcb.119.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M.D., F. Winston, and P. Hieter. 1990. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York.
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Boeke JD. In Vitromutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- Smith BJ, Yaffe MP. A mutation in the yeast heat-shock factor gene causes temperature-sensitive defects in both mitochondrial protein import and the cell cycle. Mol Cell Biol. 1991;11:2647–2655. doi: 10.1128/mcb.11.5.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo LF, Yaffe MP. Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of the mitochondrial outer membrane. J Cell Biol. 1994;126:1361–1373. doi: 10.1083/jcb.126.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K, Stinchcomb DT, Scherer S, Davis RW. High frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci USA. 1979;76:1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan DS, Huffaker TC. Astral microtubules are not required for anaphase B in Saccharomyces cerevisiae. . J Cell Biol. 1992;119:379–388. doi: 10.1083/jcb.119.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD. Intracellular transport using microtubule-based motors. Annu Rev Cell Biol. 1987;3:347–378. doi: 10.1146/annurev.cb.03.110187.002023. [DOI] [PubMed] [Google Scholar]
- Wang E, Goldman RD. Functions of cytoplasmic fibers in intracellular movements in BHK-21 cells. J Cell Biol. 1978;79:708–726. doi: 10.1083/jcb.79.3.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G, Wickner W. Organelle inheritance. Cell. 1996;84:395–400. doi: 10.1016/s0092-8674(00)81284-2. [DOI] [PubMed] [Google Scholar]
- Yaffe MP. Organelle inheritance in the yeast cell cycle. Trends Cell Biol. 1991;1:160–164. doi: 10.1016/0962-8924(91)90017-4. [DOI] [PubMed] [Google Scholar]
- Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. . J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]