Abstract
We describe a novel mammalian protein kinase related to two newly identified yeast and fly kinases—Ipl1 and aurora, respectively—mutations in which cause disruption of chromosome segregation. We have designated this kinase as Ipl1- and aurora-related kinase 1 (IAK1). IAK1 expression in mouse fibroblasts is tightly regulated temporally and spatially during the cell cycle. Transcripts first appear at G1/S boundary, are elevated at M-phase, and disappear rapidly after completion of mitosis. The protein levels and kinase activity of IAK1 are also cell cycle regulated with a peak at M-phase. IAK1 protein has a distinct subcellular and temporal pattern of localization. It is first identified on the centrosomes immediately after the duplicated centrosomes have separated. The protein remains on the centrosome and the centrosome-proximal part of the spindle throughout mitosis and is detected weakly on midbody microtubules at telophase and cytokinesis. In cells recovering from nocodazole treatment and in taxol-treated mitotic cells, IAK1 is associated with microtubule organizing centers. A wild-type and a mutant form of IAK1 cause mitotic spindle defects and lethality in ipl1 mutant yeast cells but not in wild-type cells, suggesting that IAK1 interferes with Ipl1p function in yeast. Taken together, these data strongly suggest that IAK1 may have an important role in centrosome and/ or spindle function during chromosome segregation in mammalian cells. We suggest that IAK1 is a new member of an emerging subfamily of the serine/threonine kinase superfamily. The members of this subfamily may be important regulators of chromosome segregation.
Intricate controls have evolved to regulate the process of cell division to ensure the creation of two daughter cells with identical DNA contents. Once the cell has duplicated its DNA, it must physically separate the chromosome pair into opposite poles of the cells and then undergo cytokinesis. Failure to faithfully segregate chromosomes equally into the daughter cells can result in aneuploidy and may often lead to cell death. Such failure can also have a profound impact on the fate of the daughter cells. In multicellular organisms, aneuploidy has been associated with tumorigenesis, as well as having drastic effects on development.
Ordered chromosome segregation requires the regulated interaction of many cellular components, including the centrosome, the chromosomes, the kinetochores, microtubule arrays, and probably other less-understood cellular components. Before chromosome segregation, in early pro-metaphase, the centrosome duplicates and begins nucleation of a radial array of microtubules, termed asters. The duplicated centrosomes migrate around the nucleus to establish the spindle poles. These then act as the microtubule organizing centers (MTOCs)1 for the developing mitotic spindle. Studies carried out in a variety of different organisms have identified some of the estimated 150–200 proteins that make up the centrosome and mitotic spindle, and some of these proteins have been found to be evolutionarily conserved (Kalt and Schliwa, 1993). Gradually, the functions of some of these components are being elucidated.
Increasing evidence suggests that key functions of the centrosomes and spindle may be regulated by reversible phosphorylation (Verde et al., 1990). Early evidence for this idea came from the generation of monoclonal antibodies that recognized phosphorylated epitopes present in the centrosome, kinetochores, and midbody of dividing cells (Vandre et al., 1984, 1986). Subsequently, a number of kinases have been identified that become localized to the centrosome and spindle during mitosis (see below). Entry into and exit from mitosis is regulated by a complex formed by association of a regulatory subunit, cyclin B, and a cyclin-dependent kinase, p34cdc2. This complex, termed M-phase promoting factor (MPF), is an active histone H1 kinase, and its activity regulates entry into M-phase, while MPF inactivation, brought about by cyclin destruction through the ubiquitin pathway, is required for exit from M-phase. Interestingly, a fraction of the cellular p34cdc2 pool is associated with the centrosomes, suggesting that MPF may phosphorylate centrosomal proteins (Bailly et al., 1989). Indeed, it has recently been demonstrated that phosphorylation by p34cdc2 of a human kinesin-related microtubule motor protein, Eg5, regulates its association with the mitotic spindle (Blangy et al., 1995). A number of other kinases, including casein kinase I (Brockman et al., 1992), casein kinase II (Krek et al., 1992), Ca2+/calmodulin-dependent protein kinase II (Ohta et al., 1990), polo-like kinase-1 (Golsteyn et al., 1995), and the cAMP-dependent protein kinase II (Browne et al., 1980; Nigg et al., 1985), have also been localized to the centrosome or spindle, although none of them are exclusively localized to these structures. Although these kinases have been localized biochemically or immunologically to the centrosome or mitotic spindle, the functional roles they play remain unclear. On the other hand, genetic analysis has unveiled a number of components whose activity is required for proper centrosome and mitotic spindle function and for ordered chromosome segregation. For example, two related kinases, Ipl1 and aurora, have recently been isolated from budding yeast and flies, respectively, and mutational inactivation of these kinases causes chromosome missegregation and disruption of centrosome (or spindle pole body) separation (Francisco et al., 1994; Glover et al., 1995). Despite the fact that the subcellular localization of these kinases has not been reported, this information unequivocally demonstrates the importance of these genes in regulating centrosomal and microtubule functions leading to ordered chromosome segregation and cell division.
Many of the components of the cell cycle machinery are evolutionarily conserved. For example, p34cdc2, originally identified in Schizosaccharomyces pombe, has been identified in all eukaryotes so far characterized. Similarly, the dual specificity phosphatase, cdc25, which was also originally identified in S. pombe and is a key regulator of MPF activity, is also widely conserved. Multiple members of this family have been identified in flies, mice, rats, and humans and have been found to regulate both the G1/S and G2/M transitions (Millar and Russell, 1992; Jinno et al., 1994). It seems likely, therefore, that genes identified as regulators of centrosome and spindle function and of chromosome segregation in lower eukaryotes may have mammalian counterparts. In this paper, we describe the identification and characterization of a novel mammalian kinase, Ipl1- and aurora-related kinase 1 (IAK1), related to yeast and fly proteins recognized as chromosome segregation regulators. Expression of IAK1 is tightly regulated during the cell cycle; IAK1 transcripts first appear in S-phase, are elevated at M-phase, and disappear rapidly after completion of mitosis. Western blot analysis reveals that the IAK1 protein is first detected in S-phase, peaks at M-phase, and rapidly disappears at the completion of mitosis.
IAK1 protein is first identified immunocytochemically on the centrosomes just after centrosome duplication. It remains on the centrosome throughout division and is also localized to spindle microtubules from metaphase onwards. Unlike other kinases, which associate with the centrosome and spindle during mitosis, IAK1 associates uniquely with these structures and is undetectable immunocytochemically on other structures in mitotic cells or in interphase cells. We also present data demonstrating that IAK1 can interact with components of the yeast chromosome segregation pathway. Taken together, these data strongly suggest that IAK1 is likely to have an important function during centrosome duplication, spindle organization, and chromosome segregation in mammalian cells. We further suggest that IAK1 may be a member of an emerging subfamily of the serine/threonine kinase superfamily.
Materials and Methods
DNA Amplification and Cloning
1 μg of total RNA extracted from adult mouse testes was reverse transcribed with an oligo dT-adaptor primer 5′ AATTCTGCAGTGAT-ATC(T)17 3′ using muMLV reverse transcriptase (GIBCO BRL, Gaithersburg, MD). The cDNA was used in a subsequent 3′ rapid amplification of cDNA ends (RACE) with the same oligo dT-adaptor primer and a serine/threonine kinase domain-specific degenerate primer 5′ GCGCA-TCC (C/A) GI GA(C/T) (C/T)TI AA(A/G) CCI (C/G)AI AA 3′ (where I denotes inosine) derived from the conserved RDLKPEN domain (Hanks et al., 1988). The PCR parameters were 94°C for 1 min, 50°C for 1 min, and 72°C for 10 min, repeated for 35 cycles. The PCR products were treated with T4 DNA polymerase in the presence of dGTP and dCTP to create an EcoRI overhang on one end and a blunt end on the other (Stoker, 1990), and the PCR fragments were cloned into EcoRI-SmaI double-digested pBluescript KS+ (Stratagene, La Jolla, CA). A degenerate 20 mer 5′ CTI GCI GA(T/C) TTT GGI (T/C)TI 3′ corresponding to the amino acids LADFGLA was labeled with [γ-32P]ATP (Amersham Corp., Arlington Heights, IL) and polynucleotide kinase (Boehringer Mannheim Corp., Indianapolis, IN) and used to screen the PCR library. Hybridization was carried out in 6× SSC, 5% Denhardt's, 0.1% SDS, and 100 μg/ml Salmon sperm DNA overnight at 37°C. The filters were washed in 2× SSC and 0.1% SDS twice at room temperature and finally at 37°C for 10 min each. The clones designated testis-derived serine/threonine kinases (T/STKs), which hybridized to this primer, were analyzed further by sequencing.
cDNA Cloning and Sequencing
Duplicate filters containing ∼500,000 recombinants from an adult mouse testis library (5′ stretch cDNA library in λgt 11; Clontech Labs, Palo Alto, CA) were screened with an 850-bp EcoRI fragment derived from the PCR clone designated T/STK 5. The fragment was labeled with [α32P]dCTP (Amersham Corp.) using a random prime labeling kit (Boehringer Mann-heim Corp.) to a specific activity of 3 × 109 cpm/μg. Hybridization was carried out overnight at 65°C in 5× SSPE, 5% Denhardt's, 0.1% SDS, and 100 μg/ml of Salmon sperm DNA. The hybridized filters were washed twice with 2× SSC and 0.1% SDS at room temperature followed by a stringent wash at 65°C with 0.2% SSC and 0.1% SDS. Plaques giving signals in duplicate filters were further purified by hybridization. cDNA inserts were subcloned into pBluescript KS+ as EcoRI fragments. Sequencing of plasmids containing inserts was carried out in both directions using the T3 and T7 primers (Stratagene) and subsequently by primer walking using other internal primers designed from the sequenced region. The 5′ UTR sequence of IAK1 was isolated by the 5′ RACE using adult mouse testes marathon ready cDNA (Clontech Labs) according to the manufacturer's protocol. The sequence of the first strand primer and the nested primer are 5′ CCA GCT GTG TTT TAA ACA GCA CCT TCA 3′ (nucleotides 582–609) and 5′ ATG TCA CCC CGA CGC CAC ACA GC 3′ (nucleotides 108–130), respectively. All the recombinant DNA techniques described above were essentially from Sambrook et al. (1989). Sequencing was carried out using Sequenase version 2 kit (United States Biochemical Corp., Cleveland, OH) according to the manufacturer's protocol.
RNA Isolation and Northern Analysis
Total RNA from adult tissues was isolated essentially as described by Chomczynski and Sacchi (1987). For Northern analysis, 30 μg of total RNA was separated on a 1.5% agarose-formaldehyde gel, transferred to Nytran filters (Schleicher and Schuell, Inc., Keene, NH), and baked. Blots were hybridized according to Church and Gilbert (1984) with the 850-bp EcoRI fragment derived from IAK1 PCR clone labeled using a random prime labeling kit. This probe does not cross-react with the related kinase expressed in germ cells since the transcript sizes are distinct, and moreover, the probes for each gene map to distinct locations in the mouse genome. Blots were stripped and reprobed with either mouse β-actin or mouse glyceraldehyde 3-phosphate dehydrogenase (GAPDH) to quantitate RNA loading.
Bacterial Expression and Purification of Recombinant IAK1
Recombinant wild-type IAK1 protein was expressed in Escherichia coli under the control of the inducible T7 promoter. The expression vector pRSET C (Invitrogen, San Diego, CA) was used to synthesize a histidine-tagged IAK1 protein fused in frame with gene 10 of phage T7. When cloned at the EcoRI site of pRSET C, the recombinant protein is 5 kD larger due to the NH2-terminal fusion.
Because of the presence of an internal EcoRI site in the IAK1 coding region, to make a wild-type construct, a three-piece ligation containing EcoRI-HindIII–digested pRSET C, EcoRI-SphI fragment containing the 5′ end of IAK1 cDNA, and SphI-HindIII fragment containing the remaining 3′ end of IAK1 cDNA was carried out. This construct, designated pRSET C/W, comprises the entire IAK1 coding region and 250 bp of 3′ UTR.
Construction of IAK1 Mutants
Mutation in the metal binding site of serine/threonine kinases typically abolishes the kinase activity (Gibbs and Zoller, 1991). To construct recombinant plasmids expressing a mutant IAK1 protein containing a D287N mutation, a primer, 5′ GGAGCATGCACCACCCGAAGTTTG 3′, incorporating the necessary nucleotide change was used in combination with 5′ vector-specific primer for PCR on the pRSET C/W template. The 860-bp PCR product was digested with EcoRI and SphI, and a three-piece ligation containing EcoRI-HindIII–digested pRSET C, the mutated EcoRI-SphI PCR–derived fragment, and 3′ SphI-HindIII fragment of the IAK1 cDNA was carried out. This construct was designated pRSET C/D.
The constructs IAK1WT (wild-type) and IAK1D287N (D287N mutant) expressing wild-type and mutant recombinant IAK1 under GAL10 promoter were made as follows: Wild-type and mutant IAK1 cDNAs were derived from PRSET C/W and pRSET C/D, respectively (see above), as XhoI-HindIII fragments and were used to replace the SEP1 gene (XhoI-HindIII fragment) in pRDK249 (Johnson and Kolodner, 1991).
For routine propagation, all the recombinant plasmids were transformed into E. coli DH5α. For bacterial expression of the recombinant proteins, the plasmids were transformed into E. coli BL 21(DE3) pLysS (Novagen, Inc., Madison, WI). In all cases, the synthesis of fusion proteins was induced with 1 mM IPTG.
To purify the bacterially expressed fusion proteins, the induced cells were lysed in a buffer containing 10 mM Tris-HCl, pH 8, 300 mM NaCl, and 0.1% Triton X-100. The lysate was centrifuged, and the supernatant was mixed with Ni-NTA agarose (Qiagen, Chatsworth, CA) and incubated with shaking at 4°C for 2 h. The agarose beads were washed several times with a wash buffer (10 mM Tris-HCl, pH 8, 300 mM NaCl, and 20 mM imidazole) to remove unbound proteins. The bound recombinant protein was eluted from the beads with 10 mM Tris-HCl, pH 8, containing 200 mM imidazole.
In vitro translation reactions were carried out with IAK1 cDNA cloned at the EcoRV site pCDNA3 (Invitrogen) using the TNT T7 Quick Coupled Transcription/Translation System (Promega Corp., Madison, WI) according to manufacturer's specifications. Plasmid containing luciferase gene is used for control translation.
Cell Culture and Synchronization
NIH 3T3 cells were obtained from the American Type Culture Collection (Rockville, MD) and routinely cultured in DME supplemented with 10% BCS in a humidified 5% CO2 atmosphere at 37°C. Mitotic cells were obtained as described in Lake and Jelinek (1993), with minor modifications. Cells were growth arrested in 0.5% serum–supplemented medium for 48 h. After resuming growth in medium containing 10% serum for 12 h, nocodazole (0.4 μg/ml) was added to arrest the cells at metaphase. After 18 h, mitotic cells were collected by mechanical shake off, washed three times in serum-free medium, and replated in DME with 10% BCS without nocodazole. Cells were also blocked at the G1/S phase boundary with aphidicolin. Briefly, cells were growth arrested as described above and released from growth arrest into medium containing aphidicolin (5 μg/ml) for 16 h. These cells were then washed three times with medium without aphidicolin, fed with fresh medium, and allowed to resume cell cycle progression. Samples were collected at hourly intervals after release from aphidicolin block. The synchrony of cells was monitored by the analysis of their DNA content at different time points after release from nocodazole or aphidicolin blocks by staining with propidium iodide and by flow cytometry.
Microtubule Disassembly/Reassembly and Stabilization In Vivo
NIH 3T3 cells were grown on fibronectin-coated coverslips to 80% confluency and were treated with 1–10 μg/ml nocodazole (Sigma Chemical Co., St. Louis, MO) for 4 h. The drug was removed by washing the cells twice in drug-free medium, and the cells were fixed for immunostaining at timed intervals after release from nocodazole block. For taxol treatment, cells were grown as described above and then incubated with 10 μM taxol for 5 h before fixation.
Antibodies
An anti-IAK1 antiserum was made by immunizing rabbits with a synthetic peptide (CGHTSKEPTSKSS) corresponding to the 12 COOH-terminal amino acids of the IAK1 protein that was linked to KLH. This peptide sequence is distinct from that of the other kinases in this family, including STK1 (Niwa et. al., 1996) and the germ cell-specific kinase we have identified (our own unpublished data). Rabbits were immunized with peptide– protein conjugate. Another IAK1 rabbit antiserum was generated against the histidine-tagged full-length recombinant protein produced in E. coli and gave results identical to those obtained with the COOH-terminal peptide antiserum.
In Vitro Kinase Assay
NIH 3T3 cells from different stages in the cell cycle were lysed in lysis buffer consisting of PBS containing 1% NP-40, 1% BSA, 25 mM sodium fluoride, and a cocktail of protease inhibitors (Boehringer Mannheim Corp.), and the lysate was centrifuged at 12,000 rpm for 10 min at 4°C. The supernatants were precleared at 4°C for 1 h with normal rabbit serum, and then the IAK1 specific antiserum was added at 1:500 dilution and incubated at 4°C with shaking for 2 h. The immunoprecipitates were collected by incubation with protein A–Sepharose for 1 h and the beads were washed three times with lysis buffer, three times in lysis buffer without BSA, and three times with kinase buffer (20 mM Tris, pH 7.4, 10 mM MgCl2, and 25 mM β-glycerophosphate). 25 μl of kinase buffer containing 2 μg of myelin basic protein and 20 μCi (3,000 Ci/mmol) of [γ-32P]ATP was added to the beads and the kinase assay was carried out at 30°C for 20 min. An equal volume of SDS containing sample buffer was added, and the samples were boiled for 5 min and separated on a 12% SDS–polyacryl-amide gel. The incorporation of [γ-32P]ATP into myelin basic protein was analyzed by both gel analysis and autoradiography, as well as scanning with a Molecular Dynamics phosphoimager (Sunnyvale, CA).
In experiments to assess the kinase activity of the wild-type and D287N mutant version of IAK1, flag-tagged wild-type and mutant versions of the IAK1 were transiently expressed in NIH 3T3 cells under the control of the cytomegalovirus promoter. Around 24 h after transfection, cells were treated with nocodazole (0.2 μg/ml), and cells were collected 48 h after transfection. The flag-tagged proteins were captured on anti–flag M2 affinity gel (Kodak, Rochester, NY) and used for kinase assays as described before. The activity of the wild-type and D287N mutant was compared to that of the control mock-transfected cells treated in the same manner.
Western Blot Analysis
Affinity-purified recombinant bacterial proteins and lysates from nocodazole- blocked NIH 3T3 cells were separated on 10% SDS-PAGE and blotted onto nitrocellulose. After 1 h of blocking in 5% nonfat dry milk, the blot was incubated overnight at 4°C with preimmune (1:2,000) or the COOH-terminal peptide antibody (1:2,000). Unbound primary antibodies were removed by several washes in PBS containing 0.5% Tween-20. Incubation with secondary peroxidase-coupled goat anti–rabbit IgG (Boehr-inger Mannheim Corp.) was carried out for 1 h at room temperature. After several washes with PBS containing 0.5% Tween-20, the secondary antibody was detected using the enhanced chemiluminescence detection system (Amersham Corp.) according to the manufacturer's protocol.
Immunocytochemistry
For immunocytochemical staining, dividing NIH 3T3 cells were grown on glass coverslips in wells of a 24-well plate. The cells were fixed in methanol (−20°C) for 10 min at room temperature. After fixation, the cells were washed three times in PBS and then incubated in blocking buffer (10% heat-inactivated, normal goat serum, 4% BSA Fraction V in PBS) for 10 min at room temperature. Incubation with the primary antibody diluted in blocking buffer was carried out for 1 h at room temperature, and the coverslips were then washed three times in PBS. The cells were then incubated with a rhodamine-conjugated, goat anti–rabbit antibody (Boehringer Mannheim Corp.) for 1 h at room temperature, washed three times in PBS, and mounted on slides. The slides were observed on a microscope (model Microphot FXA; Nikon, Inc., Melville, NY) equipped with epifluorescence optics. When cells were double stained for microtubules using a mouse monoclonal anti–β-tubulin antibody (Sigma Chemical Co.), the cells were incubated simultaneously with both primary antibodies, washed, and then incubated simultaneously with the goat anti–rabbit antiserum (see above) and FITC-conjugated goat anti–mouse antiserum (Sigma Chemical Co.). Preliminary experiments showed that neither the primary nor secondary antibodies cross-reacted or inhibited the binding of the other antibody. Labeled cells were examined using a confocal laser scanning microscope (Carl Zeiss, Inc., Oberkochen, Germany) essentially as described by Tsfarty et al. (1992). The instrument was calibrated with fluorescent beads to ensure that the filters excluded other wavelengths. Images were captured under fixed parameters using a 40× objective. For immunocytochemical staining of yeast cells, formaldehyde-fixed yeast cells were treated with zymolyase and then processed for fluorescent staining of microtubules and DNA as previously described (Pringle et al., 1989).
Yeast Strains, Media, and Genetic Techniques
The yeast strains used are CCY98-3D-1 (α hom3 his3-Δ200 ura3-52 lys2- Δ101::HIS3::lys2-Δ102) and CCY98-3D-1-1 (α hom3 his3-Δ200 ura3-52 lys2-Δ101::HIS3::lys2-Δ102 ipl-14), which was derived from CCY98-3D-1 by a two-step gene replacement procedure (Francisco et al., 1994). Therefore, these two strains are absolutely isogenic.
Synthetic minimal medium (SD) supplemented with casamino acids and adenine and with glucose, raffinose, or galactose as sole carbon source was prepared as described elsewhere (Rose et al., 1990).
Results
Molecular Cloning of IAK1
In an attempt to clone novel serine/threonine kinases, a 3′ RACE reaction was performed on adult mouse testis total RNA using a degenerate primer whose design was based on the conserved amino acid residues in the catalytic domain of serine/threonine kinases (Hanks et al., 1988). The RACE products were cloned into pBluescript and were further selected by hybridization to another kinase domain–specific oligonucleotide (see Materials and Methods). Positive clones were analyzed by sequencing and further characterized by comparison with other kinases in the database (GenBank) using TFASTA (Pearson and Lipman, 1988) and BLAST (Altschul et al., 1990) programs. One of the clones (designated T/STK 5) was unique and novel. It encodes the protein we have named IAK1. To isolate a full-length cDNA for IAK1, an 850-bp EcoRI fragment of the PCR clone was used to screen a testis cDNA library in λgt11. 15 positive clones were identified, and sequence analysis revealed that they all represent portions of the same cDNA. The longest cDNA isolated from this screening had the complete coding region. To isolate more 5′ sequence (5′ UTR), a 5′ RACE was performed. A composite sequence from the longest cDNA clone and the 5′ RACE clone is presented in Fig. 1. The predicted IAK1 polypeptide has 417 amino acid residues with a calculated molecular mass of 46 kD. The putative catalytic kinase domain lies towards the COOH-terminal portion of the protein and contains all the residues critical for protein kinase function. The protein lacks a signal peptide and a putative transmembrane domain, suggesting that it is likely to be not secreted or membrane associated. At the NH2-terminus of the kinase domain lies the consensus motif Gly-X-Gly-XX-Gly, which constitutes part of the ATP-binding site. The residues in the subdomain VI, Arg-Asp-Leu-Lys-Pro-Glu-Asn, and subdomain VIII, Gly-Thr-Leu-Asp-Tyr-Leu-Pro-Pro-Glu, strongly suggest that IAK1 is a bona fide serine/threonine kinase (Hanks et al., 1988). IAK1 protein contains two sequences that match the protein kinase A (PKA) consensus phosphorylation site R/K-R/K-X-S/T (Pearson and Kemp, 1991). These sites are positioned close to each other and are located in the kinase domain (Fig. 1). One of these PKA sites is in the catalytic domain activation loop and is therefore unlikely to be phosphorylated by PKA.
Figure 1.
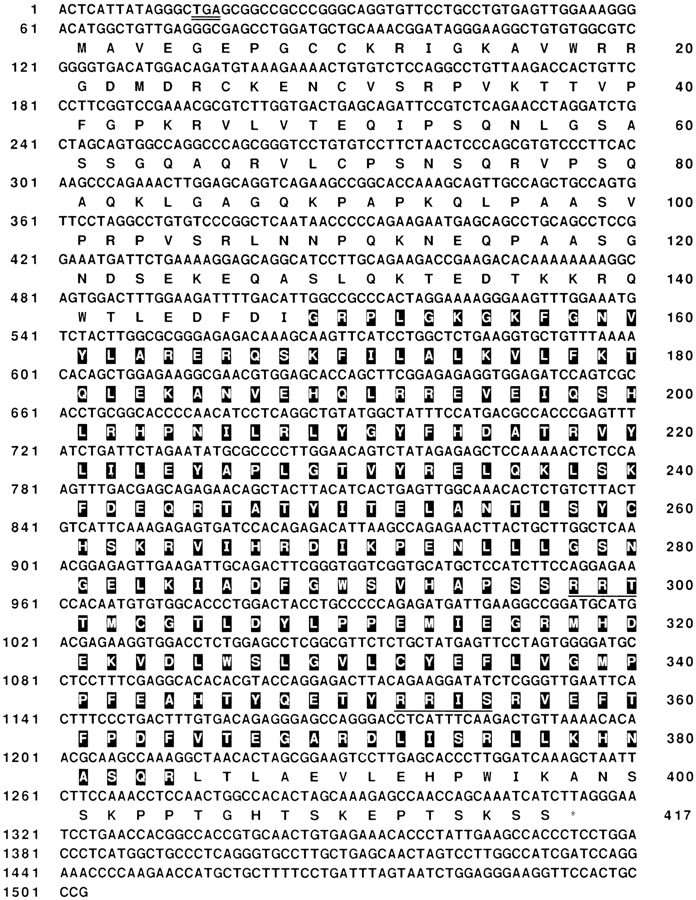
Nucleotide and deduced amino acid sequences of mouse IAK1 cDNA. The composite nucleotide sequence of cDNA clone and the RACE clone is shown. Sequence information is derived from the RACE clone spanning the first 130 bp and the cDNA clone spanning 62–1,503 bp. The in-frame stop codon is doubly underlined. The deduced amino acid sequence of IAK1 kinase is also shown. The putative catalytic domain is displayed in bold face. The PKA consensus phosphorylation sites are underlined.
IAK1 Encodes a Protein Similar to Ipl1 and Aurora Kinases
Southern blot analysis of genomic DNA isolated from a variety of organisms indicates that IAK1 is highly evolutionarily conserved. We were able to detect hybridization of the IAK1 cDNA to genomic DNA isolated from human, chicken, Xenopus, and budding yeast (data not shown). Comparison of IAK1 sequence to other kinases in the database revealed that IAK1 is related to PKA and protein kinase C families. However, IAK1 is more closely related to Ipl1 and aurora, and they form a distinct subfamily of the serine/threonine kinases. The Ipl1 gene was originally isolated in studies designed to identify genes involved in chromosome disjunction in Saccharomyces cerevisiae (Chan and Botstein, 1993). Temperature-sensitive (ts) ipl1 mutant cells missegregate chromosomes severely and die at elevated temperatures. Subsequent isolation and characterization of the Ipl1 gene revealed that Ipl1 encodes a putative serine/threonine kinase and, furthermore, that abolition of Ipl1 gene function results in severe nondisjunction (Francisco et al., 1994). aurora was isolated in a search for mutations that affect the centrosome cycle in Drosophila. The loss of function of the kinase encoded by aurora results in failure of centrosome separation leading to the formation of monopolar spindles (Glover et al., 1995). Fig. 2 shows a comparison of the amino acid sequence between IAK1, Ipl1p, and aurora and two other related kinases. The sequence homology between these kinases extends beyond the highly conserved COOH-terminal catalytic domain. Ipl1p shows an overall identity of 47% over the entire protein and 49% identity in the kinase domain to IAK1. Similarly, aurora is 56% identical to IAK1 over the entire protein and 58% identical in the kinase domain. IAK1 also shows a high degree of homology (74%) to a newly described cell cycle–regulated kinase of unknown function, STK1, identified from mouse testis (Niwa et al., 1996). It also shares 75% homology with Eg2, a frog homologue also of unknown function, whose sequence data are available from GenBank/EMBL/DDBJ under accession number Z17207 (Fig. 2).
Figure 2.

Amino acid sequence alignment of IAK1 with those of the STK1, Eg2, Ipl1, and aurora kinases. Identical amino acids are shaded. Where amino acids are conserved in three or more of these related kinases, shading is in black. Where amino acids are conserved in only two members, shading is in gray. To maximize alignment, gaps represented by dots were introduced. Amino acid alignments were carried out using the University of Wisconsin Genetics Computer Group Program PILEUP.
IAK1 Is Highly Expressed in Testis and Ovary and Is Cell Cycle Regulated
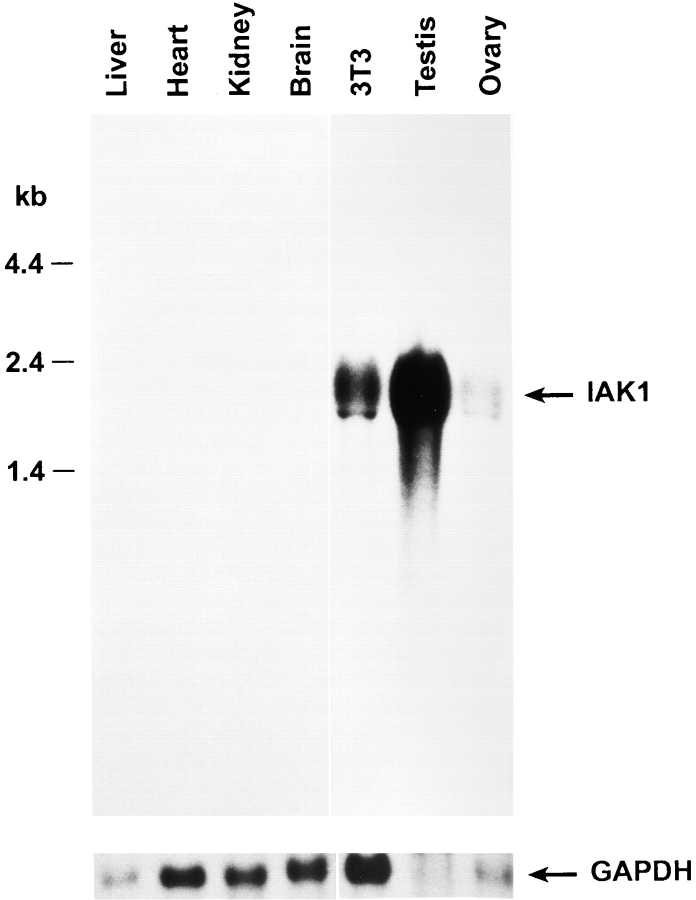
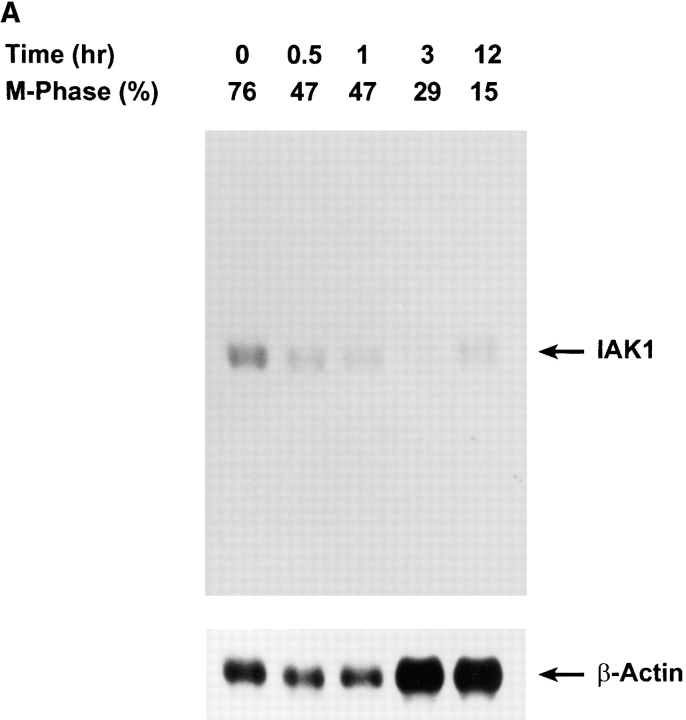
Northern blot analysis of IAK1 expression in adult mouse tissues was carried out using an 850-bp fragment comprising the entire 3′ UTR and part of the COOH-terminal coding region as a probe. We identified transcripts of ∼2 kb in testis and ovary but not in the other adult tissues examined (Fig. 3). The multiple transcripts observed probably result from differential splicing or differential polyadenylation rather than identification of other members of this family since the probe used shows less than 20% homology to other members of this gene family. IAK1 transcripts were also detected in RNA from NIH 3T3 cells (Fig. 3) as well as in CCE embryonic stem cells, F9 embryonal carcinoma cells, adult spleen, and thymus (data not shown). These data suggest that IAK1 is expressed in most dividing cells. The failure to detect IAK1 transcripts in many adult tissues probably reflects the low level of cell division in many adult tissues. To further study whether the expression of IAK1 is cell cycle regulated, we blocked NIH 3T3 cells in M-phase with nocodazole and analyzed IAK1 RNA expression after release from block and progression through the cell cycle. In nocodazole-treated cells, high levels of IAK1 transcripts were observed. As cells progressed into G1 phase of the cell cycle, transcript levels declined and by 3 h after release from nocodazole block, IAK1 transcripts were virtually undetectable. About 12 h after release from nocodazole block, cells entered S-phase, and IAK1 transcripts again became detectable (Fig. 4 A). Similar cell cycle–dependent accumulation of transcripts was obtained by block and release experiments using other blocking agents, such as aphidicolin. In aphidicolin block and release experiments, IAK1 transcripts were present at the G1/S boundary (0 h), reached highest levels around M-phase (7 h), and started to decline as cells entered the next G1 phase (10 h) (Fig. 4 B). These data strongly suggest that the accumulation of IAK1 transcripts is regulated in a cell cycle–dependent manner.
Figure 3.
Northern blot analysis of IAK1 mRNA expression in adult mouse tissues. Total RNA (30 μg) from adult mouse tissues was separated on a formaldehyde-agarose gel and blotted onto nytran membranes. The blot was hybridized with an 850-bp fragment derived from the PCR clone. The blot was stripped and re-probed with GAPDH to quantitate RNA loading.
Figure 4.
Cell cycle analysis of IAK1 expression by Northern blot analysis. (A) Nocodazole block and release: Cells were serum-starved for 48 h in 0.5% β serum–containing medium. After resuming growth in a medium containing 10% serum for 12 h, cells were incubated in a medium containing 0.4 μg/ml of nocodazole for 18 h. Mitotic cells were collected by mechanical shake-off, replated into medium without nocodazole, and allowed to progress though the cell cycle. (B) Aphidicolin block and release: NIH 3T3 cells were serum-starved for 48 h in medium supplemented with 0.5% serum. The starved cells were released into the medium containing 10% serum and 5 μg/ml of aphidicolin for 18 h. After the aphidicolin block, the cells were washed three times in serum-free medium and released into medium containing 10% serum without aphidicolin. Cells were collected at different times after the release, and total RNA was isolated. Northern blot analysis of total RNA isolated from aphidicolin- and nocodazole-blocked and released cells were performed as described above except blots were reprobed with a β-actin probe to quantitate loading. The degree of synchrony of NIH 3T3 cells was determined by propidium iodide staining and flow cytometry and is shown as the percentage of cells of G2/M phase.
IAK1 Protein Levels and Kinase Activity Are Cell Cycle Regulated
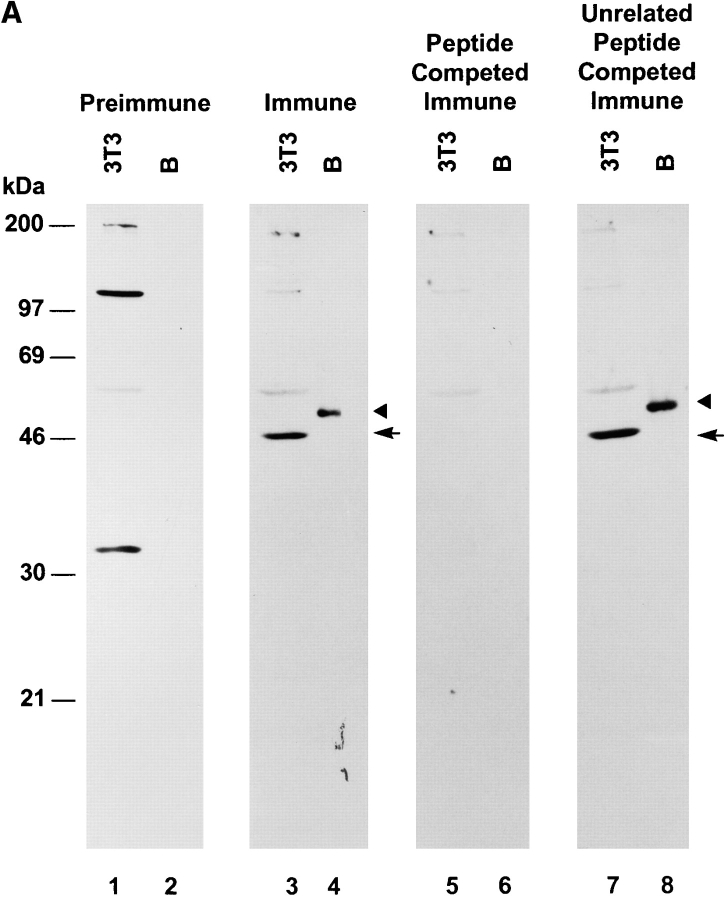
To study the expression pattern of the IAK1 protein and its subcellular localization, we raised a polyclonal antiserum to the 12 COOH-terminal amino acids of the IAK1 protein. This peptide sequence is distinct from that of other members of this family including STK1 and the germ cell–specific kinase related to IAK1. In Western blot analysis of nocodazole-treated NIH 3T3 cells, this antiserum recognized a major protein of ∼46 kD, as well as some additional minor bands (Fig. 5 A, lane 3). Antiserum binding to the 46-kD band (but not the other minor bands) was completely competed with the immunogenic peptide (Fig. 5 A, lane 5). A recombinant, histidine-tagged IAK1 protein produced in bacteria was also recognized by this antiserum (Fig. 5 A, lane 4), and again the binding could be completely competed out with the immunogenic peptide (Fig. 5 A, lane 6). Antibody binding was not inhibited by another, unrelated peptide, suggesting that peptide competition was specific (Fig. 5 A, lanes 7 and 8). In vitro translation of the full-length IAK1 cDNA in a reticulocyte lysate resulted in two protein bands (Fig. 5 B, lane 2), in which the faster migrating one is predicted to result from the translational initiation from the downstream methio-nine at position 23 of IAK1 protein. Interestingly, the endogenous IAK1 protein (Fig. 5 B, lane 3) corresponds to the faster migrating band, and we do not detect any endogenous protein corresponding to our full-length IAK1 cDNA. This suggests that the cDNA we isolated could be a splice variant of IAK1 and that the protein product corresponding to this variant is less abundant or less stable in NIH 3T3 cells. Taken together, these data suggest that this antiserum specifically recognizes the IAK1 protein in NIH 3T3 cells.
Figure 5.
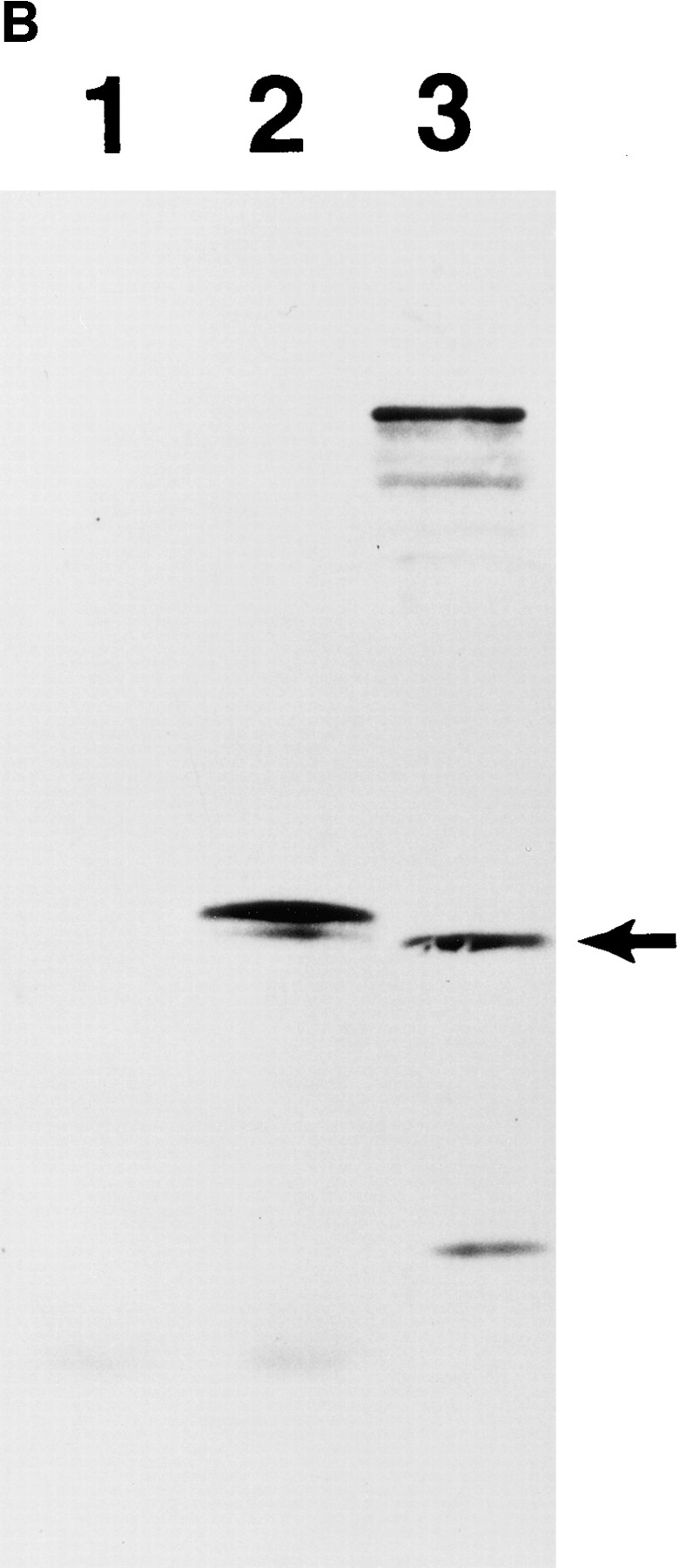
(A) Characterization of an IAK1-specific COOH-terminal peptide antiserum. A COOH-terminal peptide antiserum was characterized by Western blot analysis against a bacterially produced recombinant IAK1 protein (lanes B) and extracts of mitotic NIH 3T3 cells (lanes 3T3). For peptide competition, the antiserum was preincubated with 5 μl of a 1 mg/ml solution of the peptide immunogen or an unrelated peptide for 1 h at room temperature before incubation with the blot. The arrowhead indicates bacterially expressed recombinant IAK1, and the arrow indicates the endogenous IAK1 in NIH 3T3 cells. (B) In vitro translation of IAK1. Full-length IAK1 cDNA cloned in pcDNA3 was used for in vitro translation using the TNT T7 Quick Coupled Transcription/Translation system. An aliquot of the translated products was analyzed along with cell extract from nocodazole-blocked cells on 10% polyacrylamide gel. A similar plasmid containing luciferase gene in the place of IAK1 cDNA was used for the control. The gel was blotted and probed with IAK1-specific peptide antiserum as described in the Materials and Methods section. Lane 1, Control luciferase cDNA translated in vitro; lane 2, IAK1 cDNA translated in vitro; lane 3, nocodazole-treated NIH 3T3 cell lysate.
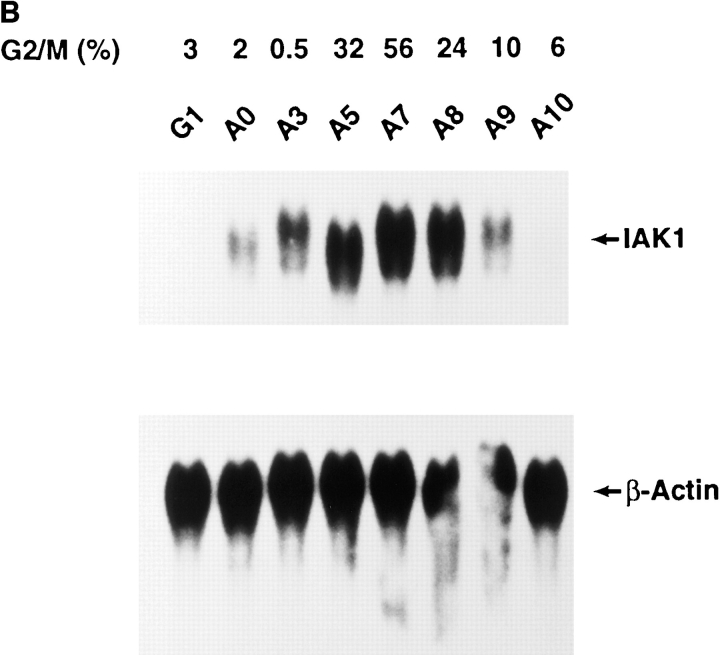
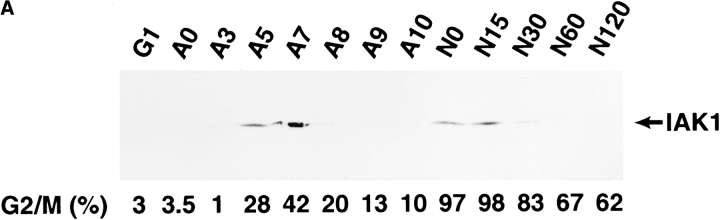
Using this antiserum, we analyzed IAK1 protein levels by Western blot analysis in cells at different stages of the cell cycle (Fig. 6 A). In cells isolated 7 h after release from serum-starvation (G1 phase), we were unable to detect IAK1 protein, suggesting that the protein is not present during this stage of the cell cycle. Cells were also blocked with aphidicolin at the G1/S phase boundary, and after release of cells from this block, we gradually saw accumulation of IAK1 protein. The peak of IAK1 expression occurred ∼7 h after release from aphidicolin block, at which time a large percentage of cells were in G2/M-phase as judged by FACS® analysis. We also blocked cells at M-phase with nocodazole and followed IAK1 protein levels after release from the nocodazole block. Consistent with data from the aphidicolin block and release experiments, we detected high levels of IAK1 protein in M-phase cells. As cells released from nocodazole block reentered and progressed through the cell cycle into the G1 phase, IAK1 protein levels rapidly declined. These data demonstrate that IAK1 protein levels, like IAK1 transcripts, are regulated in a cell cycle–dependent manner.
Figure 6.
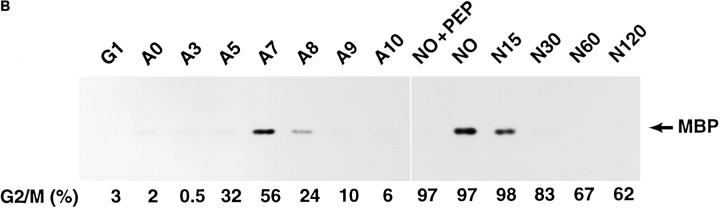
(A) Western blot analysis of IAK1 levels during the cell cycle. NIH 3T3 cells were blocked at various stages of the cell cycle by serum starvation or treatment with aphidicolin or nocodazole. After release from block, cells were harvested at intervals to collect cells at different stages of the cell cycle. Cell lysates prepared from 400,000 cells were then separated by SDS-PAGE and analyzed by Western blot analysis with the IAK1 antiserum. Cells were harvested 7 h after release from serum starvation (G1), 0, 3, 5, 7, 8, 9, and 10 h after release from aphidicolin block (A0–A10), and 0, 15, 30, 60, and 120 min after release from nocodazole block (N0–N120). The percentage of cells at G2/M phase of the cell cycle, as judged by propidium iodide staining and FACS® analysis, is shown below each lane. (B) Activity profile of IAK1 kinase through cell cycle. NIH 3T3 cells at different stages of the cell cycle were collected as described in Fig. 5 C. Cells were lysed, and in vitro kinase activity of IAK1 was determined in cell lysates using myelin basic protein as the exogenous substrate as detailed in Materials and Methods. The percentage of cells in G2/M phase was determined for each sample by flow cytometry and presented for comparison.
To find out whether IAK1 activity is also cell cycle regulated, we measured the activity of IAK1 through the cell cycle. Since our preliminary results did not show any indication of IAK1 autophosphorylation, we used exogenous substrates such as histone H1, β-casein, and myelin basic protein (MBP) for measuring IAK1 kinase activity. Fig. 6 B shows the MBP kinase activity of IAK1 measured in cells released from aphidicolin and nocodazole block as described above. The kinase activity pattern of IAK1 is also cell cycle regulated with its maximum activity at M-phase. The peak of kinase activity occurs 2–3 h after the protein is first detected by Western blot analysis, suggesting that posttranslational modifications may regulate IAK1 kinase activity.
IAK1 Protein Is Localized to the Centrosome and Mitotic Spindle
To localize IAK1 protein in dividing NIH 3T3 cells, we carried out indirect immunocytochemistry (Fig. 7). The localization pattern of IAK1 protein was distinct. We were unable to detect IAK1 protein in interphase cells (Fig. 7 A), and the protein was first detected in the separated, duplicated centrosomes of prophase cells (Fig. 7 B). The protein remained in the duplicated centrosomes during migration around the nucleus (Fig. 7 C) and at metaphase and anaphase was found in the centrosome and on spindle microtubules (Fig. 7, D and E). The protein remains on the centrosome and spindle throughout division. At telophase, IAK1 is still associated with the centrosomes, and weak staining was observed in the newly forming midbody microtubules. During cytokinesis, IAK1 remains associated with the midbody microtubules (Fig. 7 F). Antibody binding could again be competed with immunogenic peptide but not with an unrelated peptide, demonstrating that the pattern described was caused by antibody binding to the IAK1 protein (data not shown). Moreover, an identical staining pattern was observed using a different polyclonal antiserum raised against the whole IAK1 protein (data not shown). Counterstaining of cells with a monoclonal antibody that recognizes microtubules demonstrated that the antibody was indeed binding to the centrosomes and spindle (Fig. 7). The IAK1 staining pattern was observed independently of β-tubulin staining and was not the result of the strong fluorescein signal bleeding through into the rhodamine channel.
Figure 7.
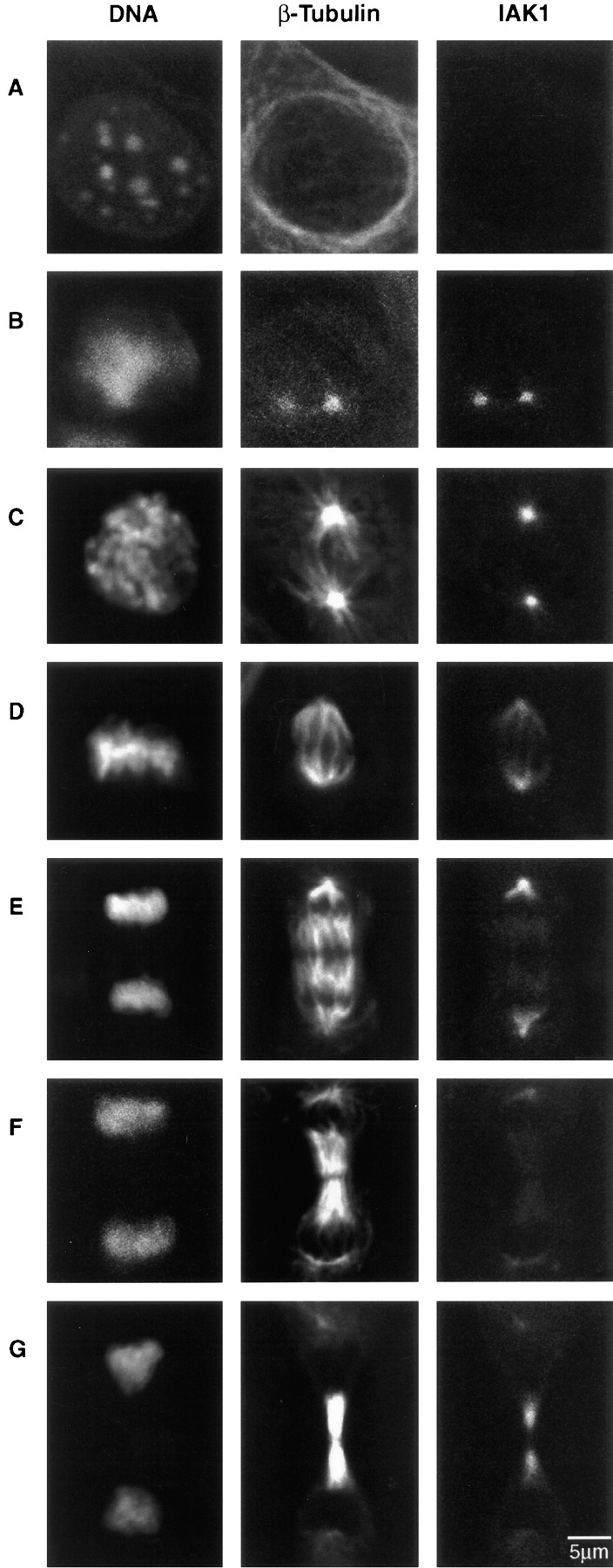
Immunocytochemical localization of IAK1 during the cell cycle. Dividing cultures of NIH 3T3 cells were fixed in ice-cold methanol and then triple-stained with DAPI for DNA, with an anti–β-tubulin monoclonal antibody for microtubules and with an anti-IAK1 antiserum. The cells were visualized with a laser scanning confocal microscope as described in Materials and Methods. Cells are at the following stages of the cell cycle: interphase (A), prophase (B), prometaphase (C), metaphase (D), late anaphase (E), telophase (F), and cytokinesis (G). Bar, 5 μm.
To further investigate the association of IAK1 protein with centrosomes and spindle microtubules, we analyzed IAK1 localization in cells treated with the microtubule-destabilizing drug nocodazole. When log phase cultures of NIH 3T3 cells are treated with nocodazole, cells arrest and accumulate at metaphase. Nocodazole-arrested cells were stained with a monoclonal anti–β-tubulin antibody and the anti-IAK1 antiserum. In nocodazole-treated cells, β-tubulin staining was observed throughout the cytoplasm, and two weak foci were also observed. These foci are likely to be the centrioles, which are known to be resistant to the effects of nocodazole (De Brabander et al., 1980). Fig. 8 A shows such a cell in which only one of these foci is in the focal plane. IAK1 staining in these cells was also observed weakly throughout the cytoplasm and in bright dots coincident with the foci of β-tubulin staining (Fig. 8 A). These data strongly suggest that most of the IAK1 protein in dividing cells is associated with pericentriolar material and tubulin of the mitotic spindle, which is consistent with the observed IAK1 staining pattern in dividing cells. They also suggest that a pool of IAK1 is associated with the centrioles independently of microtubule polymerization. In cells recovering from nocodazole block, microtubules reform from multiple MTOCs, which coalesce to form a bipolar spindle. Under these conditions, IAK1 is found to be associated with each MTOC as well as the mitotic spindle (Fig. 8 A).
Figure 8.
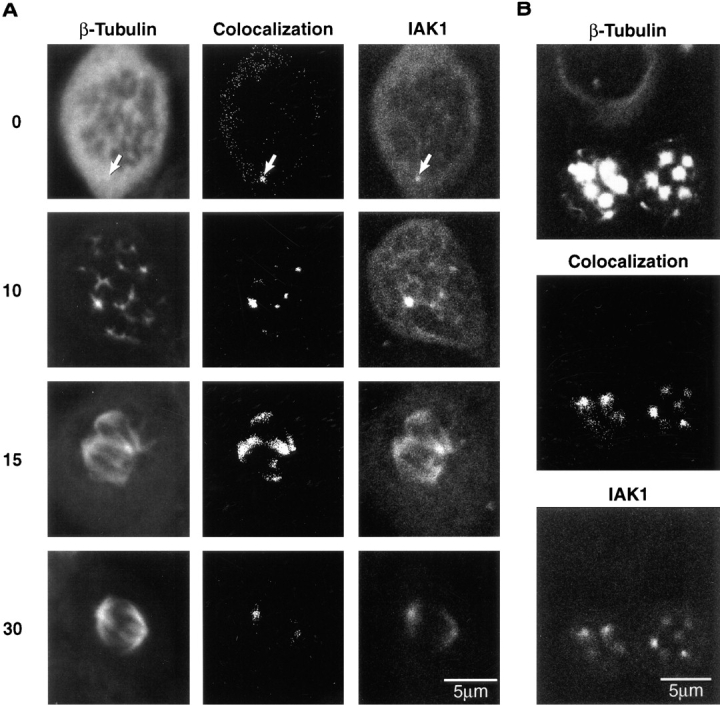
(A) Localization of IAK1 in nocodazole-treated cells. NIH 3T3 cells were treated with nocodazole (5 μg/ml) for 4 h, washed in fresh medium, and then fixed in ice-cold methanol at various times after release from nocodazole block. Cells were stained with a monoclonal anti–β-tubulin antibody or an anti-IAK1 antiserum followed by the appropriate secondary antibodies and visualized by laser scanning confocal microscopy as described in Materials and Methods. Shown are cells at time zero (0), 10 min after release from nocodazole (10), 15 min after release (15), and 30 min after release (30). Bar, 5 μm. (B) Localization of IAK1 in taxol-treated cells. NIH 3T3 cells were treated with taxol (10 μM) for 5 h and then fixed in ice-cold methanol. Cells were stained with antibodies as above for β-tubulin and IAK1. The two lower cells represent M-phase cells, while the upper cell is in interphase. Bar, 5 μm.
To further investigate the association of IAK1 with MTOCs, we treated cells with taxol, which results in the dispersal of pericentriolar material. The redistribution of the pericentriolar material results in the formation of multiple miniasters in each mitotic cell. In NIH 3T3 cells treated with taxol, the miniasters could be stained with the anti–β-tubulin antiserum (Fig. 8 B). Each of these miniasters was also stained with varying intensity for IAK1. In each mitotic cell, two prominent foci were strongly stained with the IAK1 antiserum (Fig. 8 B). These asters may be the only ones that contain centrioles and were formed before nuclear envelope breakdown. The remaining asters likely represent ones that were nucleated by the dispersed pericentriolar material. We did not observe IAK1 staining in interphase cells treated with taxol, which is consistent with our observations on IAK1 protein levels in these cells (Fig. 8 B).
Expression of IAK1 Interferes with Ipl1p Function in Yeast
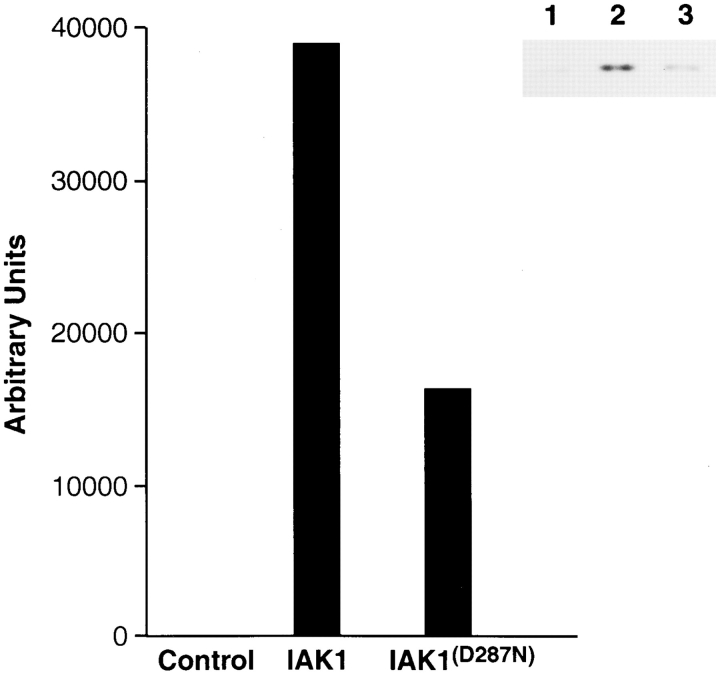
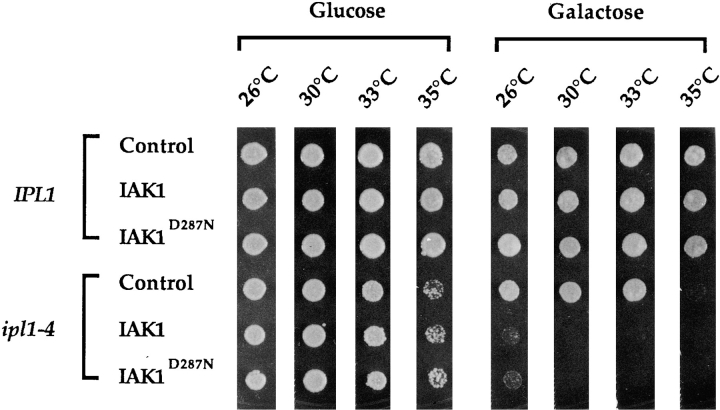
Given the sequence similarity between the mouse IAK1 and yeast Ipl1p protein kinases, we were interested in knowing whether these two kinases might be functionally related. In particular, we wanted to find out whether Ipl1p function could be substituted by the expression of IAK1 in yeast. For this purpose, we constructed plasmids expressing a wild-type or a mutant form (D287N) of IAK1 cDNA under the control of the glucose-repressible and galactose-inducible GAL10 promoter (Johnson and Kolodner, 1991). These same cDNAs were tested for their kinase activity by expressing flag-tagged versions in NIH 3T3 cells. The flag-tagged proteins were immunoprecipitated using an antiflag monoclonal antibody, and their kinase activity was measured, using MBP as the exogenous substrate. These data revealed that the D287N mutation in the metal binding site reduced the kinase activity by 2.5-fold compared to the wild-type kinase activity but did not totally abolish kinase activity (Fig. 9). The yeast expression plasmids (IAK1WT and IAK1D287N) were then introduced into wild-type and isogenic ipl1-4 temperature-sensitive mutant cells (Francisco et al., 1994). Under inducing conditions (on galactose medium), expression of IAK1 and the mutant IAK1D287N form had no effect on the growth of the wild-type yeast cells (Fig. 10). The wild-type construct also failed to suppress the temperature-sensitive phenotype caused by the recessive ipl-4 mutation. Thus, Ipl1p function cannot be substituted by the expression of IAK1. Surprisingly, expression of IAK1 actually resulted in the lethality of ipl1-4 but not wild-type cells at the otherwise permissive growth temperature of 30°C (Fig. 10), even though similar amounts of IAK1 were produced in both cell types (data not shown). Expression of a mutant form of IAK1 (IAK1D287N) having reduced kinase activity also resulted in lethality of ipl1-4 cells but not wild-type cells (Fig. 11). Thus, expression of IAK1 appeared to interfere with, rather than substitute for, Ipl1p function in yeast.
Figure 9.
The mutant IAK1D287N shows reduced kinase activity. Cell extracts from NIH 3T3 cells transiently expressing the flag-tagged IAK1 and IAK1D287N constructs were prepared, and the wild-type and the mutant form of the kinase was selectively isolated using the anti–flag M2 affinity gel as described in the Materials and Methods section. Mock-transfected cells were used as the control. The affinity-purified wild-type and mutant form of the IAK1 kinase was used for the in vitro kinase activity using myelin basic protein as the substrate. The experiment was repeated three times, and the typical result from a single experiment is presented here. The incorporation of [γ-32P]ATP into myelin basic protein was measured using a Molecular Dynamics phosphoimager, and the result is presented in the form of a bar diagram after background correction. The inset shows the autoradiographic representation of the same result. Lane 1, control mock-transfected cells; lane 2, IAK1; lane 3, IAK1D287N.
Figure 10.
Expression of mouse IAK1 causes growth inhibition of ipl1 mutant yeast cells. Suspension of yeast carrying high copy number URA3-plasmid pAJ47 (control), IAKWT (containing wild-type IAK1 cDNA under control of the GAL10 promoter), or IAKD287N (containing mutant IAK1D287N cDNA under control of the GAL10 promoter) were spotted on supplemented minimal SD solid medium (lacking uracil) that contained either glucose or galactose as the sole carbon source. These cells were allowed to grow at the indicated temperatures for 3 d, except that cells spotted on galactose medium were allowed to grow at 26°C for 5 d. The isogenic yeast strains used were CCY98-3D-1 (wild-type IPL1) and CCY98-3D-1-1 (mutant ipl1-4).
Figure 11.
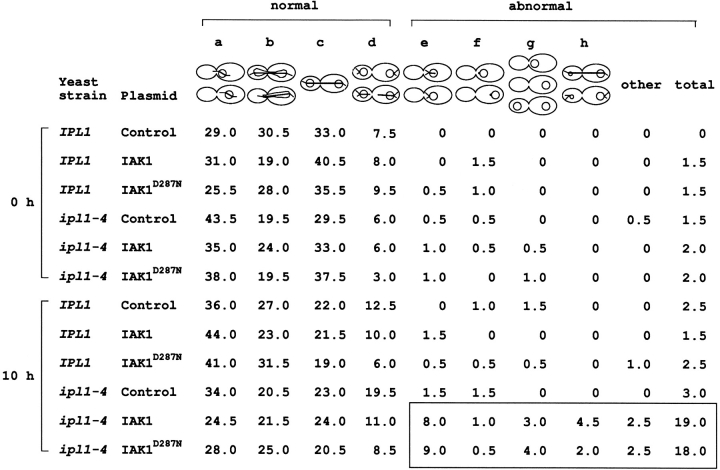
Expression of IAK1 causes microtubule defects in ipl1 mutant yeast cells. Isogenic wild-type IPL1 (CCY98-3D-1) and mutant ipl1-4 (CCY98-3D-1-1) cells carrying the indicated plasmids (see Fig. 10 for description) were grown to early log phase at 30°C in supplemented minimal SD liquid medium (lacking uracil) that contained 2% raffinose as the sole carbon source (noninducing). At 0 h, galactose was added to give a final concentration of 4% (inducing), and the cultures were incubated at 30°C for another 10 h. At the time indicated, cells were fixed with formaldehyde, and the distribution of chromosomal DNA and microtubules in these cells were examined by indirect immunofluorescence microscopy. For each sample, 200 large-budded cells were scored, and the percentage of cells belonging to each cytological class is shown here. The classes represent: (a) cells with unseparated chromatin mass and a short to medium bipolar mitotic spindle; (b) cells with chromatin mass that is not fully separated and with an elongated, bipolar mitotic spindle; (c) cells with evenly separated chromatin masses and an elongated, bipolar mitotic spindle; (d) cells with evenly separated chromatin masses and a partially disassembled mitotic spindle or no mitotic spindle; (e) cells with unseparated chromatin mass, a single unduplicated or unseparated MTOC (spindle pole body), and an apparently monopolar spindle or no mitotic spindle; (f) cells with unseparated chromatin mass, a single unduplicated or unseparated MTOC, and no other microtubule structure; (g) cells with no MTOC or microtubule structure; and (h) cells clearly with unevenly separated chromatin masses. The boxed region highlights the abnormal phenotypes in ipl-4 yeast expressing the wild-type and IAK1D287N mammalian proteins.
To determine how expression of IAK1 might result in lethality in ipl1-4 mutant cells, we examined by immuno-fluorescence microscopy the effect of wild-type or mutant IAK1 expression on the organization of chromosome mass and microtubules in wild-type or ipl1-4 mutant cells. Our results showed that the induction (by galactose) of wild-type or mutant IAK1 expression for 10 h at 30°C did not result in specific cell cycle arrest of wild-type or ipl1-4 mutant cells (data not shown). However, the organization of chromosome mass and microtubules appeared abnormal in a fraction of ipl1-4 mutant cells that expressed either form of IAK1. This abnormal organization was best illustrated in large-budded yeast cells, which normally contain more elaborate microtubule structures (Fig. 8). The most common abnormality observed was the presence of ipl1-4 cells that had unseparated chromosome mass, a single MTOC that is unduplicated or unseparated, and an apparently monopolar spindle or no mitotic spindle (Fig. 11, class e). This abnormal cytological phenotype has recently been observed in some temperature-sensitive ipl1 mutant cells incubated at their restrictive temperatures (Chan, C., unpublished results). Another common abnormality observed was the presence of ipl1-4 cells that had unevenly separated chromosome masses (Fig. 11, class h). A more severe form of this phenotype has previously been reported for ipl1-2 mutant cells incubated at elevated temperatures (Francisco et al., 1994). Thus, expression of wild-type or a mutant form of IAK1 in ipl1-4 mutant cells at the otherwise permissive temperature of 30°C resulted in cytological phenotypes similar, though not identical, to those exhibited by ipl1-2 mutant cells incubated at their restrictive temperatures. This further suggests that expression of IAK1 interferes with Ipl1p function in yeast.
Discussion
Genetic and biochemical studies in yeast, flies, and toads have identified some of the estimated 150–200 components of the centrosome and mitotic spindle, but few such proteins have been identified in mammalian cells (for review see Kalt and Schliwa, 1993). Here we describe a novel mammalian kinase, IAK1, whose expression is tightly regulated, temporally and spatially, in the cell cycle. IAK1 has a distinct expression pattern, being localized to the separated, duplicated centrosomes and mitotic spindle of dividing cells. In cells recovering from nocodazole treatment and taxol-treated cells, IAK1 is associated with each MTOC, suggesting that it may play a role in microtubule formation and/or stabilization. Because of its inherent protein kinase activity and the timing of its expression and association with the centrosome and spindle, IAK1 is clearly a candidate regulator of phosphorylation changes on these structures during mitosis. The localization of IAK1 that we describe here is strikingly similar to another serine/threonine kinase, the Polo-like kinase (Plk1) (Golsteyn et al., 1995). Interestingly, Plk1 was detected in the centrosomes of interphase cells and remained on those structures during prophase and mitosis (Golsteyn et al., 1995). Plk1 and IAK1 may interact to regulate each other's function, but we have been unable to demonstrate physical association of these kinases in immunoprecipitation and Western blot analyses (Chase, D., G. Gopalan, P.J. Donovan, and D. Ferris, unpublished observations). Nevertheless, Plk1 and IAK1 may be members of a common signal transduction cascade. One striking difference between Plk1 and IAK1 is that Plk1, unlike IAK1, is detected in the cytoplasm of interphase cells. Our Western blot data demonstrates that IAK1 protein may be present in the cell in S-phase before its immunocytochemical detection in the duplicated centrosomes at the G2 phase of the cell cycle. This suggests either that IAK1 may be present on the centrosome in S-phase, but that the antigenic epitope recognized by the COOH-terminal antiserum is masked, or that IAK1 may be present in the centrosome or cytoplasm at levels undetectable with our current reagents. Analysis of IAK1 protein levels in subcellular fractions of S-phase cells should address this question.
In cells treated with nocodazole, IAK1 remains associated with the centriole, suggesting that IAK1 association with this structure is independent of the presence of polymerized microtubules. IAK1 localization in MTOCs and spindles of cells recovering from nocodazole is consistent with it playing some role in microtubule polymerization or stabilization. A similar conclusion can be reached from the localization of IAK1 to MTOCs in taxol-treated cells. One potential role of IAK1 might be to stabilize microtubules, which could lead to centrosome separation and microtubule-mediated chromosome movements at mitosis. Alternatively, IAK1 might act on microtubule motor proteins that have been implicated in the process of centrosome separation and chromosome segregation (for review see Walczak and Mitchison 1996). Indeed, Glover and colleagues have suggested that the Drosophila aurora kinase might act on a kinesin-related protein (Glover et al., 1995). In this regard, it is apparent that the localization pattern of IAK1 is very similar to that described for the kinesin-related protein Eg5 in vertebrate cell lines (Houliston et al., 1994). Whether IAK1 interacts with and phosphorylates Eg5 (or other kinesin-related proteins) is being examined. The ability of IAK1 to disrupt mitotic spindle architecture and to affect the segregation of chromosome mass in ts ipl1 mutants strongly suggests that it can interact with a component of the yeast Ipl1 pathway or a closely related pathway. Thus, our current efforts to identify such a protein may facilitate our further understanding of the mode of action of IAK1.
What is the function of IAK1? The recent localization of the Ipl1p protein kinase to the yeast mitotic spindle (Chan, C., unpublished results) demonstrates that IAK1 is similar to Ipl1p not only in sequence but also in subcellular localization. Our data show that IAK1 cannot rescue the ipl1-4 mutant phenotype and therefore may not perform identical functions as Ipl1p. Nevertheless, the data presented here also demonstrate that IAK1 exacerbates ipl1 mutant phenotypes since it causes inviability of ipl1-4 mutant but not wild-type cells (Fig. 7). These data suggest that IAK1 proteins act as dominant-negative proteins whose effect is only seen when Ipl1p function is impaired. Similarly, Nurse and colleagues showed that a kinase-dead Xenopus CDK2 gene disrupted cell cycle progression in a ts cdc2 fission yeast mutant but not in wild-type cells (Paris et al., 1994). This effect is most likely brought about by the ability of the expressed proteins to interfere with the activity of the endogenous kinase by competing for interacting proteins. Expression of other kinases (as well as several other genes or mutations) in ts ipl1 mutants does not cause lethality of the ts ipl1 mutant at the permissive temperature (Chan, C., unpublished observations). However, expression of the Drosophila aurora kinase, like IAK1, caused lethality in ipl1-4 mutants but not in wild-type cells at the permissive temperature (Chan, C., unpublished observations). This strongly suggests that the observed phenotype is specific and that IAK1 is not simply causing nonspecific lethality. Some of these cytological defects are also observed in conditional ipl1 mutant cells incubated at their restrictive temperatures and are also reminiscent of the effect of the aurora mutation in Drosophila. These observations strongly suggest that IAK1 interferes with the function of Ipl1p (or a parallel pathway), thus leading to lethality in cells that already have compromised Ipl1p function (due to the ipl1-4 mutation). IAK1 could interact with Ipl1p itself, an upstream regulator of Ipl1p, a downstream effector (substrate) of Ipl1p, or a protein in a parallel pathway. Disruption of IAK1 activity in mammalian cells will probably address its role in centrosome and mitotic spindle function in mammalian cells.
The inability of IAK1 to rescue the ipl-4 mutation suggests that IAK1 may not perform the identical functions as Ipl1p. It is possible that an, as yet unidentified, relative of IAK1, fulfills the Ipl1p function or that there is no single functional homologue of Ipl1p in mammalian cells. Conceivably the function(s) carried out by Ipl1p in yeast could be carried out in mammalian cells by multiple kinases which have evolved more complex or distinct forms of regulation. Interestingly, in addition to IAK1 we have identified another mammalian kinase, related to Ipl1 and aurora, which is not expressed in NIH 3T3 cells or most adult tissues, but is expressed in germ cells (Gopalan, G., J. Centanni, and P.J. Donovan, manuscript in preparation). This kinase, which shares 52% identity at the amino acid level to IAK1, may play a similar role to IAK1 in meiotic germ cells or may function during postmeiotic germ cell differentiation. Another cell cycle–regulated kinase of unknown function, STK1, has also recently been identified and shows homology to IAK1 (see Fig. 2 B). The mouse genome therefore contains at least three members of this subfamily and, based on chromosome localization studies, may also contain another related gene (our own unpublished observations). A Xenopus cDNA sequence (Eg2) present in the GenBank database (accession number Z17207)also shows a high degree of homology to IAK1, STK1, Ipl1, and aurora (Fig. 2 B). As well as these other genes, we have identified two related genes each in human and worm (Gopalan, G., J. Schumacher, A. Golden, and P.J. Donovan, unpublished observations). These data suggest that aurora, IAK1, STK1, Eg2, and Ipl1 may be the first members of an emerging subfamily of the serine/threonine kinase superfamily that could play important roles in centrosome and mitotic spindle function during mitosis. Analysis of these related proteins in a variety of species will probably allow a more complete understanding of the function and mode of action of this protein family.
Acknowledgments
We are grateful to Ira Daar, Dineli Wickramasinghe, Leslie Lock, Andy Golden, and Peter Johnson for comments on the manuscript and for many constructive discussions. We also thank Tony Hunter for helpful suggestions and for his encouragement and Doug Ferris, Jeff Strathern, and Amar Klar for useful insights. We are extremely grateful to Jim Resau, Eric Hudson, and Richard Frederickson for their help in generating Figs. 7 and 8, Marilyn Powers for making oligonucleotides, Terry Copeland for generating the antibodies, Louise Finch for carrying out FACS® analysis, and Madeline Wilson for typing the manuscript. The sequence of IAK1 reported here is available from GenBank/EMBL/DDBJ under accession number BankIt 120295 AF007817.
Abbreviations used in this paper
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- IAK1
Ipl1- and aurora-related kinase 1
- MBP
myelin basic protein
- MPF
M-phase promoting factor
- MTOC
microtubule organizing center
- PKA
protein kinase A
- Plk1
Polo-like kinase
- ts
temperature-sensitive
- T/STK
testis-derived serine/threonine kinases
Footnotes
Please address all correspondence to Peter J. Donovan, Cell Biology of Development and Differentiation Group, ABL-Basic Research Program, NCI-FCRDC, Frederick, MD 21702-1201. Tel.: (301) 846-5176. Fax: (301) 846-6666. E-mail: donovanp@ncifcrf.gov
This research was sponsored in part by the Department of Health and Human Services under contract with Advanced Bioscience Laboratories (P.J. Donovan), and the National Institutes of Health (GM45185) and the Council for Tobacco Research (#4496) (C.S.M. Chan).
References
- Altschul, S.F., W. Gish, W. Miller, E.W. Myers, and D.J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol., 215:403–410. [DOI] [PubMed]
- Bailly E, Doree M, Nurse P, Bornens M. p34cdc2is located in both nucleus and cytoplasm: part is centrosomally associated at G2/M and enters vesicles at anaphase. EMBO (Eur Mol Biol Organ) J. 1989;8:3985–3995. doi: 10.1002/j.1460-2075.1989.tb08581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangy A, Lane HA, d'Herin P, Harper M, Kress M, Nigg EA. Phosphorylation by p34cdc2regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 1995;83:1159–1169. doi: 10.1016/0092-8674(95)90142-6. [DOI] [PubMed] [Google Scholar]
- Brockman J, Gross SD, Sussman MR, Anderson RE. Cell cycle dependent localization of casein kinase I to mitotic spindles. Proc Natl Acad Sci USA. 1992;89:9454–9458. doi: 10.1073/pnas.89.20.9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne CL, Lockwood AH, Su JL, Steiner AL. Immunofluorescent localization of cyclic nucleotide-dependent protein kinases on the mitotic apparatus of cultured cells. J Cell Biol. 1980;87:336–345. doi: 10.1083/jcb.87.2.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SM, Botstein D. Isolation and characterisation of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics. 1993;135:677–691. doi: 10.1093/genetics/135.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Brabander, M., G. Genens, R. Nuydens, R. Willebrords, and J. De Mey. 1980. The microtubule nucleating activity of kinetochores and centrosomes in living PTK2-cells. In Microtubules and Microtubule Inhibitors. M. De Brabander and J. De Mey, editors. Elsevier, North Holland, Amsterdam. 255–268.
- Francisco L, Chan CSM. Regulation of yeast chromosome segregation by Ipl1 protein kinase and type 1 protein phosphatase. Cell Mol Biol Res. 1994;40:207–213. [PubMed] [Google Scholar]
- Francisco L, Wang W, Chan CSM. Type 1 phosphatase acts in opposition to Ipl1 protein kinase in regulating yeast chromosome segregation. Mol Cell Biol. 1994;14:4731–4740. doi: 10.1128/mcb.14.7.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs CS, Zoller MJ. Rational scanning mutagenesis of a protein kinase identifies functional regions involved in catalysis and substrate interactions. J Biol Chem. 1991;266:8923–8931. [PubMed] [Google Scholar]
- Glover DM, Leibowitz MH, McLean DA, Parry H. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell. 1995;81:95–105. doi: 10.1016/0092-8674(95)90374-7. [DOI] [PubMed] [Google Scholar]
- Golsteyn R, Mundt KE, Fry AM, Nigg EA. Cell cycle regulation of the activity and subcellular localization of PLK1, a human protein kinase implicated in mitotic spindle function. J Cell Biol. 1995;129:1617–1628. doi: 10.1083/jcb.129.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science (Wash DC) 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- Houliston E, Le Guellec R, Kress M, Philippe M, Le Guellec K. The kinesin-related protein Eg5 associates with both interphase and spindle microtubules during Xenopusearly development. Dev Biol. 1994;164:147–159. doi: 10.1006/dbio.1994.1187. [DOI] [PubMed] [Google Scholar]
- Jinno S, Suto K, Nagata A, Igarashi M, Kanaoka Y, Nojima H, Okayama H. cdc25A is a novel phosphatase functioning early in the cell cycle. EMBO (Eur Mol Biol Organ) J. 1994;13:1549–1556. doi: 10.1002/j.1460-2075.1994.tb06417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AW, Kolodner RD. Strand exchange protein 1 from Saccharomyces cerevisiae. . J Biol Chem. 1991;266:14046–14054. [PubMed] [Google Scholar]
- Kalt A, Schliwa M. Molecular components of the centrosome. Trends Cell Biol. 1993;3:118–127. doi: 10.1016/0962-8924(93)90174-y. [DOI] [PubMed] [Google Scholar]
- Krek W, Maridor G, Nigg EA. Casein kinase II is a predominantly nuclear enzyme. J Cell Biol. 1992;116:43–55. doi: 10.1083/jcb.116.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake RJ, Jelinek W. Cell-cycle and terminal differentiation-associated-regulation of the mouse mRNA encoding a conserved mitotic protein kinase. Mol Cell Biol. 1993;13:7793–7801. doi: 10.1128/mcb.13.12.7793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar JB, Russell P. The cdc25 M-phase inducer: an unconventional protein phosphatase. Cell. 1992;68:407–410. doi: 10.1016/0092-8674(92)90177-e. [DOI] [PubMed] [Google Scholar]
- Nigg EA, Schaefer G, Hilz H, Eppenberger HM. Cyclic AMP dependent protein kinase type II is associated with the Golgi complex and with centrosomes. Cell. 1985;41:1039–1051. doi: 10.1016/s0092-8674(85)80084-2. [DOI] [PubMed] [Google Scholar]
- Niwa H, Abe K, Kunisada T, Yamamura K. Cell-cycle-dependent expression of the STK1gene encoding a novel murine putative protein kinase. Gene. 1996;169:197–201. doi: 10.1016/0378-1119(95)00809-8. [DOI] [PubMed] [Google Scholar]
- Ohta Y, Ohba T, Miyamoto E. Ca2+/calmodulin-dependent protein kinase II: localization in the interphase nucleus and the mitotic apparatus of mammalian cells. Proc Natl Acad Sci USA. 1990;87:5341–5345. doi: 10.1073/pnas.87.14.5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris J, Leplatois P, Nurse P. Study of the higher eukaryotic gene function CDK2 using fission yeast. J Cell Sci. 1994;107:615–623. doi: 10.1242/jcs.107.3.615. [DOI] [PubMed] [Google Scholar]
- Pearson RB, Kemp BE. Protein kinase phosphorylation site sequences and consensus specificity motifs: tabulation. Methods Enzymol. 1991;200:62–81. doi: 10.1016/0076-6879(91)00127-i. [DOI] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle JR, Preston RA, Adams A, Stearns T, Drubin D, Haarer BK, Jones E. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;11:237–243. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- Rose, M.D., F. Winston, and P. Heiter. 1990. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. 2nd Ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Stoker AW. Cloning of PCR products after defined cohesive termini are created with T4 DNA polymerase. Nucleic Acids Res. 1990;18:4290–4290. doi: 10.1093/nar/18.14.4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsfarty I, Resau JH, Rulong S, Keydar I, Faletto DL, Vande GF, Woude The met proto-oncogene receptor and lumen formation. Science (Wash DC) 1992;257:1258–1261. doi: 10.1126/science.1387731. [DOI] [PubMed] [Google Scholar]
- Vandre DD, Davis FM, Rao PN, Borisy GG. Phosphoproteins are components of mitotic microtubule organization centres. Proc Natl Acad Sci USA. 1984;81:4439–4443. doi: 10.1073/pnas.81.14.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandre DD, Davis FM, Rao PN, Borisy GG. Distribution of cytoskeletal proteins sharing a conserved phosphorylated epitope. Eur J Cell Biol. 1986;41:72–81. [PubMed] [Google Scholar]
- Verde F, Labbe JC, Doree M, Karsenti E. Regulation of microtubule dynamics by cdc2 kinase in cell-free extracts of Xenopuseggs. Nature (Lond) 1990;343:233–238. doi: 10.1038/343233a0. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Mitchison TJ. Kinesin-related proteins at mitotic spindle poles: function and regulation. Cell. 1996;85:943–946. doi: 10.1016/s0092-8674(00)81295-7. [DOI] [PubMed] [Google Scholar]