Abstract
Chromosomal inversions have an important role in evolution, and an increasing number of inversion polymorphisms are being identified in the human population. The evolutionary history of these inversions and the mechanisms by which they arise are therefore of significant interest. Previously, a polymorphic inversion on human chromosome Xq28 that includes the FLNA and EMD loci was discovered and hypothesized to have been the result of nonallelic homologous recombination (NAHR) between near-identical inverted duplications flanking this region. Here, we carried out an in-depth study of the orthologous region in 27 additional eutherians and report that this inversion is not specific to humans, but has occurred independently and repeatedly at least 10 times in multiple eutherian lineages. Moreover, inverted duplications flank the FLNA–EMD region in all 16 species for which high-quality sequence assemblies are available. Based on detailed sequence analyses, we propose a model in which the observed inverted duplications originated from a common duplication event that predates the eutherian radiation. Subsequent gene conversion homogenized the duplications, thereby providing a continuous substrate for NAHR that led to the recurrent inversion of this segment of the genome. These results provide an extreme example in support of the evolutionary breakpoint reusage hypothesis and point out that some near-identical human segmental duplications may, in fact, have originated >100 million years ago.
Keywords: duplication, gene conversion, inversion polymorphism
Chromosomal rearrangements were among the first types of genetic variation to be studied and have been proposed to play an important role in genome evolution and the phenotypic differences within and between species (1, 2). The recent availability of genomic data from multiple species has led to the unexpected discovery that structural variation, including inversions, is relatively common in the human population (3, 4), as well as between closely related species, such as human and chimpanzee (5–7). Comparisons of the positions of evolutionary breakpoints in different mammalian lineages suggest that a small fraction of the mammalian genome is particularly prone to rearrangement and constitute breakpoint “hotspots” that have been reused over the course of mammalian evolution (e.g., refs. 8 and 9). Therefore, a conserved property of mammalian genomes appears to be the presence of a limited number of fragile regions that commonly mediate chromosomal rearrangements. This observation contrasts with the long-held view that evolutionary breakpoints are distributed randomly across the genome (10), and there has been considerable debate pitting the random breakage model versus the fragile breakage/breakpoint reusage hypothesis (11).
One mechanism known to mediate chromosomal rearrangements is nonallelic recombination between homologous sequences (NAHR). In humans, 5% of the genome is comprised of segmental duplications, which are typically defined as duplications >1 kb in length and >90% sequence identity, and could act as potential hotspots for genome evolution (12). NAHR between these duplications mediates both benign and pathogenic chromosomal rearrangements in the human population (13). Strikingly, not only are the locations of human segmental duplications enriched at the positions of evolutionary breakpoints that occurred in the human lineage, but the orthologous positions in other mammalian genomes are also prone to breakage (9, 14, 15). Thus, it has been postulated that segmental duplications in the human genome are indicators of conserved fragile regions found in other species (15). However, no explicit mechanism or example has been reported that could account for this association and the reusage of breakpoints during evolution. In addition, it is not clear whether the presence of the duplicated sequences precede the generation of the chromosomal rearrangements or the properties of these genomic regions account for both the tendency to break and duplicate.
Here, we report the results of a comparative genomic study focused on the segment of human chromosome Xq28 containing the FLNA and EMD loci. This region has been associated with a ≈40-kb polymorphic inversion that was originally detected at a frequency of 18% in a sample of people of European descent (16). However, whether the more common FLNA–EMD arrangement present in the reference human genome assembly or the alternative EMD–FLNA arrangement (designated as the + and − arrangements, respectively, in Fig. 1A) represents the ancestral arrangement was not established. Remarkably, we found that this locus has been the site of recurrent and independent inversions in a diverse sampling of eutherians. Furthermore, based on detailed sequence analysis, we propose a model by which the recurrent inversions were the result of NAHR between a conserved pair of inverted duplications on the X chromosome that originated before eutherian radiation >100 million years ago.
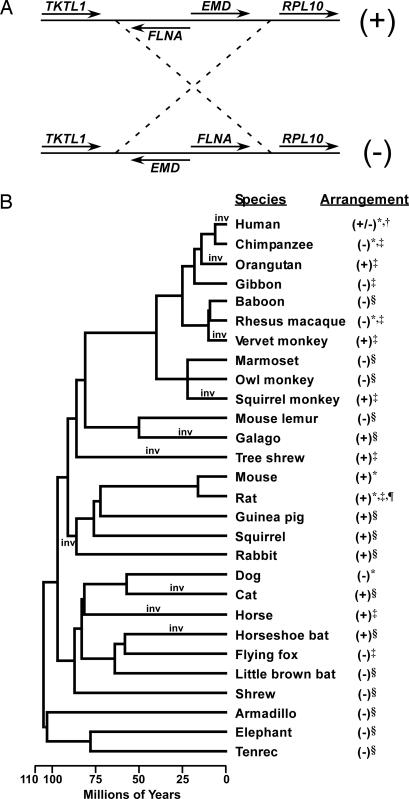
Fig. 1.
Alternative arrangements of the FLNA–EMD chromosomal segment in eutherians. (A) Schematic diagram of the alternate arrangements of the FLNA–EMD chromosomal segment, which were arbitrarily given + and − designations, as indicated. (B) Phylogenetic distribution of the FLNA–EMD chromosomal arrangements in 28 mammals. The phylogeny of the 28 species is depicted based on the branching order and dates reported in refs. 52–56. Time points of the minimum of 10 inversions required by one of the most parsimonious models are labeled on the tree (inv). The orientation of the FLNA–EMD chromosomal segment in each species was inferred from: whole-genome assemblies (*), Small et al. (16) (†), comparative mapping of paired clone-end sequence reads (‡), and targeted BAC-based assemblies (§). Genomic sequence assembly that does not include all four genes in the region is indicated by ¶.
Results
Recurrent Inversion of the FLNA–EMD Chromosomal Segment in Eutherians.
To study the evolutionary history of the FLNA–EMD inversion, we used targeted BAC-based sequencing, comparative mapping of BAC and fosmid paired-end reads, and whole-genome assemblies to determine the arrangement of this chromosomal segment in a diverse sample of 27 additional eutherians (see Methods). Because it has traditionally been considered that inversions are relatively infrequent and have a unique origin, we expected that a single orientation would be uniformly present in our sample of species. In contrast to our expectation, both the + and − chromosomal arrangements were observed (Fig. 1B). Moreover, the phylogenetic distribution of the observed arrangements suggests that multiple independent inversions have occurred since the most recent common ancestor of eutherians. Although the order of genes in this region could not be inferred from the genome assemblies of the noneutherian mammals opossum and platypus, other sequenced tetrapods, or fish because of a lack of conserved synteny (data not shown), the most parsimonious scenario that accounts for the observed variation in the arrangement of the FLNA–EMD chromosomal segment is one in which the − arrangement was present before the eutherian radiation and requires a minimum of 10 independent inversion events (Fig. 1B). These results are consistent with an inversion hotspot in this region of the eutherian X chromosome.
Inverted Duplications Flanking the FLNA–EMD Chromosomal Segment Are a Conserved Feature of Eutherian Genomes.
Small et al. (16) noted the presence of near-identical inverted duplications flanking the FLNA–EMD chromosomal segment in humans and other hominoids and proposed that the human polymorphic inversion was the result of NAHR between the duplications. To determine the degree to which inverted duplications flanking the FLNA–EMD segment are a conserved feature of eutherian X chromosomes, detailed genomic sequence annotation and comparisons were conducted in human and 15 other eutherians by using high-quality BAC-based targeted sequence assemblies of this region generated by us and sequences extracted from whole-genome assemblies [Fig. 2 and supporting information (SI) Table 1].
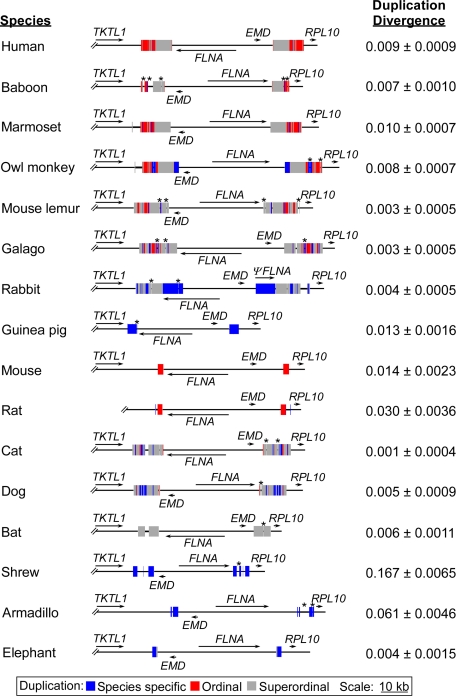
Fig. 2.
Inverted duplications flanking the FLNA–EMD chromosomal segment in eutherians. The positions and orientations of genes (arrows) and duplicated segments (filled boxes) are illustrated for each species. Species-specific refers to a sequence duplicated in a single species, ordinal refers to a sequence duplicated in more than one species from the same order, and superordinal refers to a sequence duplicated in at least two species from two distinct orders. The position of sequencing gaps within the duplications is indicated by *. The intraspecies divergence between duplications as measured by the number of substitutions per site was calculated by using the Kimura two-parameter (K2P) model (50) and is shown on the right along with the standard error.
As described (16), near-identical (0.009 substitutions per site) 11.4-kb inverted duplications flank the FLNA–EMD chromosomal segment in humans (Fig. 2). Strikingly, inverted duplications were also detected at the orthologous positions in all 15 other eutherians (Fig. 2 and SI Table 1). These duplications ranged in size from ≈1.9 kb in elephant to >17 kb in rabbit and, with the exception of the rabbit duplications that contain a portion of FLNA, were devoid of known genes. As was the case in humans, the duplications flanking the FLNA–EMD chromosomal segment were near-identical (≤ 0.014 substitutions per site) in 12 of the 15 additional species analyzed (Fig. 2). In the remaining three species (rat, armadillo, and shrew) divergence between the duplications was higher and ranged from 0.03 to 0.17 substitutions per site (Fig. 2), but they did contain several tracts of perfect identity of ≥50 bp, totaling 1,422, 1,223, and 1,531 bp in rat, armadillo, and shrew, respectively. Preliminary analysis of this region in the rhesus macaque, colobus monkey, and cow, for which there are lower-quality sequence assemblies, as well as squirrel, tenrec, and little brown bat, for which high-quality assemblies recently became available, indicate that inverted duplications are also present in those species (SI Table 1). Thus, near-identical inverted duplications flanking the FLNA–EMD chromosomal segment are a conserved feature of eutherian X chromosomes.
Evolutionary Relationships Among the Duplications.
The presence of near-identical duplicated sequences at orthologous positions in a phylogenetically diverse set of species (Fig. 2) is a hallmark of sequence homogenization resulting from gene conversion, or other possible recombination processes, such as successive single cross-overs (17), between the duplications. Alternatively, this observation is also consistent with numerous recent and independent duplications of the orthologous genomic regions in multiple lineages. To establish the degree to which gene conversion and/or independent duplication events have contributed to the ubiquitous presence of inverted duplications flanking the FLNA–EMD chromosomal segment in eutherians, we examined the sequence conservation and the evolutionary relationship of the duplications across species and compared the relative genomic positions of the duplications.
Multiple sequence alignments were generated and used to classify subregions of the duplications as either species-specific, conserved within an order (ordinal), or conserved across orders (superordinal) (Fig. 2; see Methods for details and SI Table 2 for a summary of the conservation of the duplications between each pair of species). Most of the duplications were a complex mixture of two or more of the sequence classes and shared some homologous sequence with at least one other species. However, the guinea pig, shrew, and armadillo duplications were composed entirely of species-specific sequences. Within the 11 species that contained duplicated sequence classified as superordinal, we were able to detect just 102 bp duplicated in all species (SI Fig. 4A). This low degree of sequence conservation between the duplications is similar to that of the flanking intergenic regions and not unexpected for comparisons of random genomic sequence across a set of evolutionarily diverse species (18). To formally reconstruct the evolutionary relationships among the duplications, the conserved 102-bp sequence was used to generate a phylogenetic tree. As expected, the intraspecies paralogous duplications clustered most closely with one another and not with an orthologous duplication (SI Fig. 4B). An analogous result was also observed for the phylogeny of the duplicated sequence common to only mouse and rat (SI Fig. 4C). Thus, the composition of the duplications based on inferred common ancestry from sequence alignments and the reconstructed evolutionary relationships between the duplications are consistent with either a common origin followed by a high degree of gene conversion and/or independent recent duplications of the orthologous positions in multiple lineages.
To distinguish between a single origin of the duplications versus multiple independent duplication events, we used the sequence alignments of the species that shared some common duplicated sequence to compare the relative positions of the duplications in each species. The external edges of the duplications were fairly consistent between species; in four of the primates (marmoset, owl monkey, mouse lemur, and galago) and rabbit, they mapped within 30 bp of each other, and in most other species the difference in location could be explained by simple deletions (SI Fig. 5). There was more variation in the relative position of the internal edges, although the human, baboon, marmoset, cat, and dog internal edges mapped within 35 bp of one another and those of mouse lemur, galago, and bat are located a few hundred nucleotides away because of independent ≈1-kb deletions (SI Fig. 5). In addition, the species-specific duplicated regions at the internal edges in owl monkey (1.8 kb) and rabbit (7.5 kb) could be explained by an expansion of the duplications via gene conversion (see Discussion). Therefore, despite the overall low sequence conservation, the positions of the duplications suggest that they are derived from an ancestral duplication and strongly support a single common origin.
Signatures of Gene Conversion in the Duplications.
Initial sequence comparisons of the inverted duplications flanking the FLNA–EMD chromosomal segment suggested that gene conversion has likely played a role in the evolution of the duplications. We therefore performed additional intraspecies and interspecies sequence analyses to determine whether other known signatures of gene conversion could be detected within the duplications.
First, intraspecies pairwise alignments of the duplications were tested for stretches of perfect identity longer than expected by chance given the overall sequence identity between the duplications and a random distribution of nucleotide substitutions with GENECONV (19). Statistically significant identical tracts that could be the result of gene conversion were found in all 16 species analyzed (SI Table 3). Second, because the ends of duplications are less likely to be homogenized by gene conversion events, we used the same pairwise alignments to compare the frequency of single-nucleotide substitutions at the edges of the duplications versus the internal segment within each species. If gene conversion has occurred, we would expect to see elevated divergence at the edges compared with the rest of the duplication. Indeed, with the exception of rabbit and shrew, the combined divergence of the 100 bp at each edge of the duplication was significantly elevated compared with the internal segment in the remaining 14 species (SI Table 3). Finally, because there is substantial evidence that gene conversion leads to an increase in GC content resulting from a bias in mismatch repair (20), we compared the GC content of the duplicated sequences and the intergenic regions flanking each duplication. In all cases, the GC content of the duplications was significantly higher (1.3–25.6%) than that of the flanking intergenic regions (SI Table 3). Although the results of the intraspecies-based tests cannot exclude alternative mechanisms, they were consistent with homogenization of the duplications flanking the FLNA–EMD chromosomal segment by gene conversion.
As a complement to the above tests, we used interspecies sequence comparisons of the duplications and flanking regions between three pairs of closely related species to look for signatures of gene conversion (SI Table 4). We detected discrete regions at the edges of the duplications that were more similar to the orthologous region from the other species in each pair than to the intraspecies paralogous sequence. Specifically, in the human–baboon alignment, we detected tracts of 82 and 76 bp at the outer and inner ends of the duplications, respectively, that include several intraspecies paralogous sequence variants that are shared between the orthologous duplications (SI Table 4 and SI Fig. 6). A similar pattern was observed in a 3,092-bp tract (including a 2.7-kb deletion) and a 79-bp tract at the outer edge of the marmoset–owl monkey and mouse–rat duplication alignments, respectively. One interpretation of these observations is that these regions escaped the recent homogenization effect of gene conversion and represent the divergence accumulated between duplications in the common ancestor of each species pair. Such a model is consistent with a common origin for the duplications in each of the three pairs of species, followed by gene conversion in both the primate and rodent lineages, and further supports the assertion that gene conversion is actively homogenizing the paralogous duplications within each species.
Discussion
The ever-expanding list of sequenced mammalian genomes and human variation data is providing novel and refined views of the differences between genomes and the molecular mechanisms by which those differences arise. Here, we have described the results of our study focused on a segment of the eutherian X chromosome that displays two highly unusual properties: recurrent inversions and the ubiquitous presence of near-identical duplications in a diverse sampling of eutherians. As such, this locus provides a unique perspective on how mammalian genomes can evolve.
Chromosomal rearrangements, including inversions, are generally thought to occur infrequently. Moreover, if chromosomal breakpoints occur at random in the genome, the chance of the “same” inversion arising independently and repeatedly at the same locus in a broad sampling of taxa should be vanishingly small. In contrast to the above expectation, we were able to infer that a minimum of 10 inversions have flipped the arrangement of the FLNA–EMD chromosomal segment since the eutherian radiation some ≈100 million years ago. Considering the estimated rate for the occurrence of evolutionary breakpoints in mammalian lineages of ≈0.11–2.25 breakpoints per genome every million years (9) and the ≈1,642 million years of evolution represented in our phylogeny (Fig. 1), only 0.005–0.11 breakpoints would be expected to map within a genomic interval the size of the FLNA-EMD segment and flanking regions (≈75 kb). The 20 breakpoints we observed corresponding to a minimum of 10 inversions are therefore at least ≈180-fold higher than expected. Thus, even by this conservative estimate of the number of independent inversions (see below), the FLNA–EMD chromosomal segment is clearly an inversion hotspot and supports the breakpoint reusage hypothesis (8).
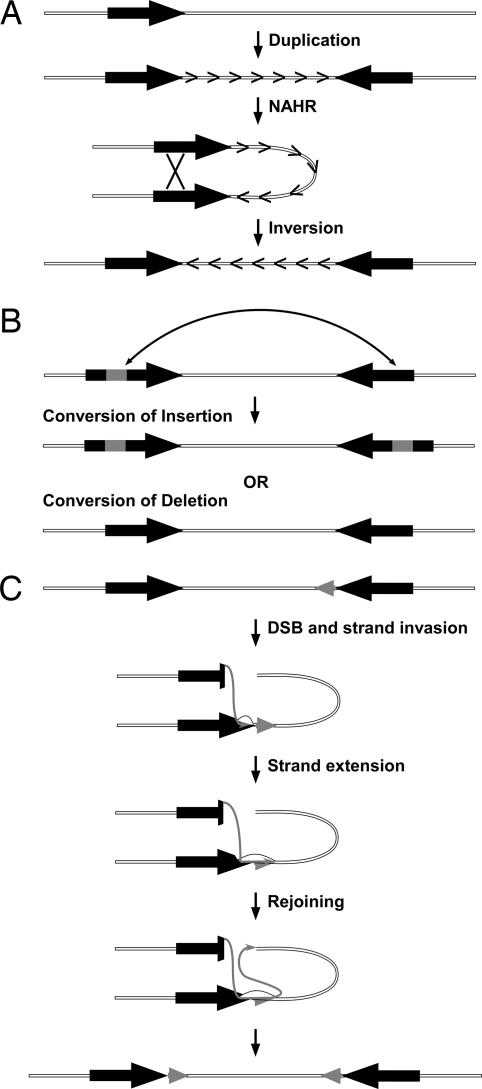
Although remarkable, the two unique properties of the FLNA–EMD region, i.e., recurrent inversions and the ubiquitous presence of near-identical duplications in a diverse sampling of eutherians, can be explained by a single molecular mechanism: NAHR between inverted repeats. As with any other type of recombination event, NAHR can be resolved through gene conversion or through crossing-over, which will result, respectively, in high identity between the duplications or inversion of the segment between them. Therefore, to account for the evolution of the FLNA–EMD region, we propose the following model (Fig. 3). First, before the most recent common ancestor of eutherians, an ancestral duplication event occurred (Fig. 3A). Subsequently, over the past ≈100 million years, the duplications were continuously homogenized by gene conversion and the near-identical tracts of sequence acted as substrate for additional NAHR and recurrent inversions. However, periodically a fraction of the ancestral duplications accumulated enough sequence differences between paralogs to escape conversion, most notably exemplified in the shrew lineage, thus likely limiting their potential to act as substrates for future inversions by NAHR. In addition, we hypothesize that the process of gene conversion has included not only the conversion of single-nucleotide variants, but also of insertions and deletions (Fig. 3B). Such a mechanism is consistent with studies from various species showing the conversion of insertions and deletions of various sizes (21–24) and would explain the presence of species- and ordinal-specific sequences embedded within the duplications. Finally, the expansion of the duplications in some species could be explained by so-called long tract gene conversion events (25), in which strand extension proceeds through the homologous duplicated sequence and into the flanking “unique” region, ultimately resulting in a newly duplicated sequence (Fig. 3C). Conversely, simple deletions at the edges of the duplications could have progressively contracted the duplicated regions in other species.
Fig. 3.
Model for the evolution of the FLNA–EMD region. (A) Generation of an inverted duplication (filled arrows) in a common ancestor of eutherians and continuous NAHR between the duplications led to the homogenization of the duplications by gene conversion and the recurrent inversion of the locus. (B) Remodeling of the ancient duplications by the conversion of insertions (gray boxes) and deletions. (C) Long-tract gene conversion (25) resulted in the expansion of the duplications (gray arrows). DSB, double-strand break.
NAHR between inverted repeats is a commonly accepted molecular basis for inversions (13) and is known to give rise to recurrent inversions in somatic cells (26) and in the germ line (27). However, we are aware of just one other example by which NAHR between near-identical segmental duplications at orthologous positions in highly divergent species has been directly implicated in mediating convergent inversions. Specifically, Lozier et al. (28) observed that, as is the case in some human patients, factor VIII deficiency in dogs is caused by a chromosomal inversion most likely caused by NAHR between duplicated sequences analogous to those found in humans. Strikingly, this locus also maps to Xq28 and is located just ≈1 Mb telomeric of the FLNA–EMD region. Thus, while presumably rare, NAHR between near-identical segmental duplications at orthologous positions can lead to the clustering of evolutionary breakpoints. One interesting prediction of this model is that, as in humans, inversion polymorphism of the FLNA–EMD region may be a genetic variant common to most, if not all, eutherians. Therefore, prevalent intraspecies variation is an alternative explanation for the pattern of chromosomal arrangements seen in Fig. 1B.
With regard to the evolution of the duplications, it has been argued that gene conversion is not prevalent enough to obscure the true age of most duplications in the human genome, and near-identical duplications are likely to have arisen in the very recent past and be species- or clade-specific (29, 30). In contrast to this assertion, our detailed sequence analysis of the duplications flanking the FLNA–EMD region is consistent with the action of gene conversion and suggests a single origin for the duplications common to all eutherians, thereby dating the duplication to a time point >100 million years ago. Gene conversion is a dominant force dictating the evolution of segmental duplications on the human Y chromosome (31) and is common between autosomal duplications as well (32). In fact, previous work in worms (33) and mammals (34) has already demonstrated that homogenization of duplicated sequences by gene conversion can be sustained over long evolutionary timeframes on the order of ≈100 million to 200 million years. However, although there are other cases of near-identical human duplications that are older than expected based on the divergence estimates (24, 35), to our knowledge, the duplications flanking the FLNA–EMD region represent the most extreme example of such a finding, both in terms of the estimated age of the duplications and the phylogenetic breadth of conservation.
Although models other than those proposed above for the history of the FLNA–EMD region may certainly also be possible, perhaps the most intriguing question has nothing to do with how and when the inversions and segmental duplications occurred, but rather with the pervasive conservation of near-identical duplications at this locus. Because there is no evidence to suggest that the duplications encode genes, save for the 3′ end of the FLNA gene in rabbit, one possibility is that the duplications themselves are entirely dispensable, but just happen to be located in an optimal genomic environment, i.e., spacing, orientation, and chromosome location, for gene conversion. It is therefore tempting to speculate that hemizogosity of the X chromosome in the male germ line has led to an elevation of intrachromosomal NAHR, as has been observed for the duplications that mediate the inversion that causes factor VIII deficiency (27). Indeed, gene conversion is thought to be a key mechanism for the long-term survival of genes on the Y chromosome (36), and both the human X and Y chromosomes have a disproportionately high percentage of intrachromosomal duplications with >99% identity (30, 31) and inverted repeats (37). A second possibility is that it is not the retention of the duplications that is being selected for, but the ability to alter the orientation of the inverted region, which may have some unknown beneficial consequence. For example, a series of newly described 5′ extended transcripts (38) that map both within and outside of the inverted region, and thus are predicted to be arrangement-specific, have been associated with FLNA, which encodes an actin binding protein involved in cytoskeletal assembly (39), EMD, a nuclear envelope protein mutated in X-linked Emery-Dreifuss muscular dystrophy (40), and RPL10, a component of the 60s ribosomal subunit (41). Lastly, it is also possible that the duplications are being actively conserved because they are functionally important, and as a pair might act in concert to modulate local chromatin structure and/or contribute to the regulation of neighboring genes via the formation of a cruciform structure (36, 37).
In conclusion, our results have revealed an extreme example of breakpoint reusage and long-term gene conversion on the eutherian X chromosome and led us to propose an explicit model to account for both of those unique genomic properties. Future efforts are needed to determine whether the evolution of this locus is a genomic oddity or is, in fact, representative of a discrete fraction of other evolutionary breakpoints and/or “recent” segmental duplications.
Methods
BAC-Based Mapping, Sequencing, and Assembly.
Targeted BAC-based mapping, sequencing, and ordered and oriented assembly of the orthologous genomic segments in baboon (Papio anubis), marmoset (Callithrix jacchus), owl monkey (Aotus nancymaae), dusky titi (Calicebus moloch), galago (Otolemur garnettii), mouse lemur (Microcebus murinus), rabbit (Oryctolagus cuniculus), guinea pig (Cavia porcellus), squirrel (Spermophilus tridecemlineatus), cat (Felis catus), horseshoe bat (Rhinolophus ferrumequinum), little brown bat (Myotis lucifugus), shrew (Sorex araneus), armadillo (Dasypus novemcinctus), tenrec (Echinops telfairi), and elephant (Loxodonta africana) were done by the National Institutes of Health Intramural Sequencing Center Comparative Sequencing Program as described (42, 43) as part of the ENCODE project (44). An unordered and oriented BAC-based assembly was also generated for the colobus monkey (Colobus guereza). Additional genomic sequences corresponding to the TKTL1–RPL10 segment were extracted from the whole-genome assemblies of human (Homo sapiens, hg17), chimpanzee (Pan troglodytes, panTro1), rhesus macaque (Macaca mulatta, rheMac2), dog (Canis familiaris, canFam2), cow (Bos taurus, bosTau2), mouse (Mus musculus, mm8), and rat (Rattus norvegicus, rn4). A summary of the genomic data used in this study is provided in SI Table 1, and the evidence supporting the sequence assemblies is listed in SI Table 5.
Comparative Mapping of BAC and Fosmid Paired-End Reads.
Available BAC and fosmid paired-end reads from eutherians were used to infer the arrangement of the FLNA–EMD region in a greater sampling of species, and in some cases to confirm the arrangement of the FLNA–EMD chromosomal segment present in the whole-genome assemblies. Fosmid and BAC-end reads were imported from the National Center for Biotechnology Information and compared by BLAST (45) and/or BLAT (46) searches to the finished human or mouse genomic sequence. The orientation of the FLNA–EMD segment was then inferred in chimpanzee, orangutan (Pongo pygmaeus), gibbon (Nomascus leucogenys), rhesus macaque, vervet monkey (Chlorocebus aethiops), squirrel monkey (Saimiri boliviensis), tree shrew (Tupaia belangeri), rat (Rattus norvegicus), horse (Equus caballus), and flying fox (Pteropus vampyrus) by identifying paired-end reads in which one end-read fell between the duplications flanking the FLNA–EMD loci, and the mate-pair mapped within a distance of 27–78 kb for fosmids and 135–318 kb for BAC clones. The relative orientation of the paired-end reads to the human or mouse genomes were then used as the basis to infer the arrangement of the FLNA–EMD segment in each species. A summary of the paired-end read mapping results are provided in SI Table 6.
Multiple Sequence Alignment and Sequence Analysis.
Gene annotation of the region of interest was either lifted from the whole-genome assemblies or generated locally with a combination of interspecies cDNA–genomic and genomic–genomic alignments. Genomic alignments between and within species were generated with the BLASTZ suite of alignment programs, including TBA and MultiPipMaker (47, 48). The location of the duplications within each species was inferred from sequence alignments of each species to itself, and the edges of the duplications were further refined by comparison with those of other species by using multiple sequence alignments. Pairs of duplications or flanking sequences were aligned with MUSCLE (49) by using default parameters, and alignments were manually inspected to check for potential errors. Sequence divergence was calculated by the Kimura two-parameter (K2P) model (50) and the standard error was estimated with 1,000 bootstrap replicates by using MEGA (51). Phylogenetic trees of the duplicated sequence were constructed by using the neighbor-joining method and 100 bootstrap replicates as implemented in PAUP*. Gene-conversion tracts within the pairwise alignments of the duplications of each species were estimated with GENECONV (19) by detecting stretches of perfect identity longer than expected by chance using the “include monomorphic sites” option.
Supplementary Material
Acknowledgments
We thank L. Armengol, X. Estivill, K. Garber, J. R. González, L. McGraw, J. M. Ranz, A. Ruiz, S. Yi, and Y. Tao for helpful comments and discussion. This research was supported in part by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health. M.C. was supported by the Ramón y Cajal Program (Ministerio de Educación y Ciencia, Spain).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AC135553.3, AC149172.3, AC158724.2, AC166863.2, AC174861.2, AC174925.2, AC182364.2, AC186089.3, AC186101.2, AC186105.2, AC187641.3, AC187677.2, AC190013.1, AC190442.2, AC192052.2, AC196879.3, AC196919.2, AC199692.2, AC202232.2, and AC206365.2).
This article contains supporting information online at www.pnas.org/cgi/content/full/0706604104/DC1.
References
- 1.Sturtevant AH. Carnegie Inst Washington Publ. 1919;278:305–341. [Google Scholar]
- 2.Dobzhansky T. Genetics of the Evolutionary Process. New York: Columbia Univ Press; 1970. [Google Scholar]
- 3.Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D, et al. Nat Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 4.Khaja R, Zhang J, MacDonald JR, He Y, Joseph-George AM, Wei J, Rafiq MA, Qian C, Shago M, Pantano L, et al. Nat Genet. 2006;38:1413–1418. doi: 10.1038/ng1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newman TL, Tuzun E, Morrison VA, Hayden KE, Ventura M, McGrath SD, Rocchi M, Eichler EE. Genome Res. 2005;15:1344–1356. doi: 10.1101/gr.4338005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feuk L, MacDonald JR, Tang T, Carson AR, Li M, Rao G, Khaja R, Scherer SW. PLoS Genet. 2005;1:e56. doi: 10.1371/journal.pgen.0010056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaisson MJ, Raphael BJ, Pevzner PA. Proc Natl Acad Sci USA. 2006;103:19824–19829. doi: 10.1073/pnas.0603984103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pevzner P, Tesler G. Proc Natl Acad Sci USA. 2003;100:7672–7677. doi: 10.1073/pnas.1330369100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy WJ, Larkin DM, Everts-van der Wind A, Bourque G, Tesler G, Auvil L, Beever JE, Chowdhary BP, Galibert F, Gatzke L, et al. Science. 2005;309:613–617. doi: 10.1126/science.1111387. [DOI] [PubMed] [Google Scholar]
- 10.Nadeau JH, Taylor BA. Proc Natl Acad Sci USA. 1984;81:814–818. doi: 10.1073/pnas.81.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng Q, Pevzner PA, Tesler G. PLoS Comput Biol. 2006;2:e14. doi: 10.1371/journal.pcbi.0020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bailey JA, Eichler EE. Nat Rev Genet. 2006;7:552–564. doi: 10.1038/nrg1895. [DOI] [PubMed] [Google Scholar]
- 13.Stankiewicz P, Lupski JR. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 14.Armengol L, Pujana MA, Cheung J, Scherer SW, Estivill X. Hum Mol Genet. 2003;12:2201–2208. doi: 10.1093/hmg/ddg223. [DOI] [PubMed] [Google Scholar]
- 15.Bailey JA, Baertsch R, Kent WJ, Haussler D, Eichler EE. Genome Biol. 2004;5:R23. doi: 10.1186/gb-2004-5-4-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Small K, Iber J, Warren ST. Nat Genet. 1997;16:96–99. doi: 10.1038/ng0597-96. [DOI] [PubMed] [Google Scholar]
- 17.Hurles ME, Willey D, Matthews L, Hussain SS. Genome Biol. 2004;5:R55. doi: 10.1186/gb-2004-5-8-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Margulies EH, Chen CW, Green ED. Trends Genet. 2006;22:187–193. doi: 10.1016/j.tig.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 19.Sawyer S. Mol Biol Evol. 1989;6:526–538. doi: 10.1093/oxfordjournals.molbev.a040567. [DOI] [PubMed] [Google Scholar]
- 20.Galtier N, Piganeau G, Mouchiroud D, Duret L. Genetics. 2001;159:907–911. doi: 10.1093/genetics/159.2.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Radding CM. Cold Spring Harb Symp Quant Biol. 1979;43:1315–1316. doi: 10.1101/sqb.1979.043.01.150. [DOI] [PubMed] [Google Scholar]
- 22.Nassif N, Penney J, Pal S, Engels WR, Gloor GB. Mol Cell Biol. 1994;14:1613–1625. doi: 10.1128/mcb.14.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang H, Hasty P, Bradley A. Mol Cell Biol. 1994;14:2404–2410. doi: 10.1128/mcb.14.4.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boissinot S, Tan Y, Shyue SK, Schneider H, Sampaio I, Neiswanger K, Hewett-Emmett D, Li WH. Proc Natl Acad Sci USA. 1998;95:13749–13754. doi: 10.1073/pnas.95.23.13749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richardson C, Moynahan ME, Jasin M. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flores M, Morales L, Gonzaga-Jauregui C, Dominguez-Vidana R, Zepeda C, Yanez O, Gutierrez M, Lemus T, Valle D, Avila MC, et al. Proc Natl Acad Sci USA. 2007;104:6099–6106. doi: 10.1073/pnas.0701631104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rossiter JP, Young M, Kimberland ML, Hutter P, Ketterling RP, Gitschier J, Horst J, Morris MA, Schaid DJ, de Moerloose P, et al. Hum Mol Genet. 1994;3:1035–1039. doi: 10.1093/hmg/3.7.1035. [DOI] [PubMed] [Google Scholar]
- 28.Lozier JN, Dutra A, Pak E, Zhou N, Zheng Z, Nichols TC, Bellinger DA, Read M, Morgan RA. Proc Natl Acad Sci USA. 2002;99:12991–12996. doi: 10.1073/pnas.192219599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng Z, Ventura M, She X, Khaitovich P, Graves T, Osoegawa K, Church D, DeJong P, Wilson RK, Paabo S, et al. Nature. 2005;437:88–93. doi: 10.1038/nature04000. [DOI] [PubMed] [Google Scholar]
- 30.She X, Liu G, Ventura M, Zhao S, Misceo D, Roberto R, Cardone MF, Rocchi M, Green ED, Archidiacano N, et al. Genome Res. 2006;16:576–583. doi: 10.1101/gr.4949406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rozen S, Skaletsky H, Marszalek JD, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Page DC. Nature. 2003;423:873–876. doi: 10.1038/nature01723. [DOI] [PubMed] [Google Scholar]
- 32.Jackson MS, Oliver K, Loveland J, Humphray S, Dunham I, Rocchi M, Viggiano L, Park JP, Hurles ME, Santibanez-Koref M. Am J Hum Genet. 2005;77:824–840. doi: 10.1086/497704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas JH. Genetics. 2006;172:2269–2281. doi: 10.1534/genetics.105.052746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winter EE, Ponting CP. BMC Evol Biol. 2005;5:54. doi: 10.1186/1471-2148-5-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bagnall RD, Ayres KL, Green PM, Giannelli F. Genome Res. 2005;15:214–223. doi: 10.1101/gr.2946205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, et al. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 37.Warburton PE, Giordano J, Cheung F, Gelfand Y, Benson G. Genome Res. 2004;14:1861–1869. doi: 10.1101/gr.2542904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Denoeud F, Kapranov P, Ucla C, Frankish A, Castelo R, Drenkow J, Lagarde J, Alioto T, Manzano C, Chrast J, et al. Genome Res. 2007;17:746–759. doi: 10.1101/gr.5660607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vadlamudi RK, Li F, Adam L, Nguyen D, Ohta Y, Stossel TP, Kumar R. Nat Cell Biol. 2002;4:681–690. doi: 10.1038/ncb838. [DOI] [PubMed] [Google Scholar]
- 40.Holaska JM, Wilson KL. Anat Rec. 2006;288:676–680. doi: 10.1002/ar.a.20334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dick FA, Karamanou S, Trumpower BL. J Biol Chem. 1997;272:13372–13379. doi: 10.1074/jbc.272.20.13372. [DOI] [PubMed] [Google Scholar]
- 42.Thomas JW, Touchman JW, Blakesley RW, Bouffard GG, Beckstrom-Sternberg SM, Margulies EH, Blanchette M, Siepel AC, Thomas PJ, McDowell JC, et al. Nature. 2003;424:788–793. doi: 10.1038/nature01858. [DOI] [PubMed] [Google Scholar]
- 43.Blakesley RW, Hansen NF, Mullikin JC, Thomas PJ, McDowell JC, Maskeri B, Young AC, Benjamin B, Brooks SY, Coleman BI, et al. Genome Res. 2004;14:2235–2244. doi: 10.1101/gr.2648404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.The Encode Project Consortium. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kent WJ. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, et al. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwartz S, Elnitski L, Li M, Weirauch M, Riemer C, Smit A, Green ED, Hardison RC, Miller W. Nucleic Acids Res. 2003;31:3518–3524. doi: 10.1093/nar/gkg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edgar RC. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kimura M. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 51.Kumar S, Tamura K, Nei M. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 52.Murphy WJ, Eizirik E, O'Brien SJ, Madsen O, Scally M, Douady CJ, Teeling E, Ryder OA, Stanhope MJ, de Jong WW, et al. Science. 2001;294:2348–2351. doi: 10.1126/science.1067179. [DOI] [PubMed] [Google Scholar]
- 53.Goodman M. Am J Hum Genet. 1999;64:31–39. doi: 10.1086/302218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Teeling EC, Springer MS, Madsen O, Bates P, O'Brien SJ, Murphy WJ. Science. 2005;307:580–584. doi: 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- 55.Murphy WJ, Pringle TH, Crider TA, Springer MS, Miller W. Genome Res. 2007;17:413–421. doi: 10.1101/gr.5918807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nishihara H, Hasegawa M, Okada N. Proc Natl Acad Sci USA. 2006;103:9929–9934. doi: 10.1073/pnas.0603797103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.