Abstract
Fibrillar, or “hairy,” adhesives have evolved multiple times independently within arthropods and reptiles. These adhesives exhibit highly desirable properties for dynamic attachment, including orientation dependence, wear resistance, and self-cleaning. Our understanding of how these properties are related to their fibrillar structure is limited, although theoretical models from the literature have generated useful hypotheses. We survey the morphology of 81 species with fibrillar adhesives to test the hypothesis that packing density of contact elements should increase with body size, whereas the size of the contact elements should decrease. We test this hypothesis in a phylogenetic context to avoid treating historically related species as statistically independent data points. We find that fiber morphology is better predicted by evolutionary history and adhesive mechanism than by body size. As we attempt to identify which morphological parameters are most responsible for the performance of fibrillar adhesives, it will be important to take advantage of the natural variation in morphology and the potentially suboptimal outcomes it encompasses, rather than assuming evolution to be an inherently optimizing process.
Keywords: contact splitting, fibrillar adhesion, setae, phylogeny, independent contrasts
Geckos are best known for using their subdigital fibrillar adhesive to scale vertical and overhanging, smooth or rough surfaces with ease. Yet, “dry” fibrillar adhesives are not unique to geckos but can be found in anoles, skinks, and spiders. Some insects also bear hairy pads but augment them with “wet” sticky secretions. Although the adhesive mechanisms may differ, the similarities in their morphology have led researchers to create models in search of universal relationships between structure and function across all fibrillar adhesive organisms (1). Such relationships, if found, would do much to guide our understanding of how fibrillar adhesives function, to determine how they benefit or limit the organisms using them and to offer clear design principles for the engineers attempting fabrication of such adhesives (2–10).
Contact splitting is one important principle of fibrillar adhesives predicted by several models including fracture mechanics (11, 12) and Johnson-Kendall-Roberts theory (JKR) (13, 14). This principle suggests that a given amount of surface area detaches with higher adhesive forces (or work) if it is divided into smaller, more numerous contact elements (14). Contact splitting is, at minimum, a byproduct of fibrillar adhesives, which are by definition densely packed fine structures. Geckos and several other organisms seem to capitalize further on the advantages of contact splitting by terminally branching into many flattened tips, termed “spatulae,” of considerably smaller radius than the stalks themselves (Fig. 1). Despite the growing body of physical and mathematical literature modeling fibrillar adhesion, there have been few direct tests of the models' predictions on the biological specimens (14–22) and even fewer that attempt to describe or account for the variation in adhesive morphology seen in nature (23–25).
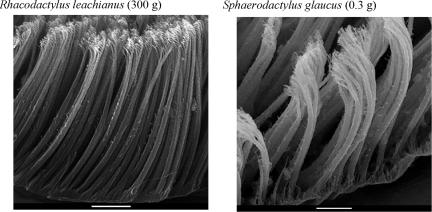
Fig. 1.
Setal morphology of two gecko species varying by three orders of magnitude in body mass but by only 2-fold in density of contact elements (spatulae). (Scale bars, 10 μm.)
Arzt et al. (24) hypothesize that organisms take advantage of contact splitting to overcome the unfavorable geometric scaling of body mass with adhesive pad area. Using JKR theory, they estimate the pull-off force (Fc) of a field of n hemispherical contact elements with diameter s in contact with a smooth surface to be
where γ is the adhesion energy per area. Contacts are square-packed, and their diameter is inversely proportional to the square root of contact density. It is assumed that adhesive pad area per unit mass decreases with size according to geometric similarity. This mass-specific decrease is predicted to drive a compensatory increase in contact density and a corresponding decrease in seta and contact diameter to support increasing body mass. Given these assumptions, Arzt et al. (24) hypothesize that spatular density should increase with body mass to the 2/3 power across all fibrillar adhesive organisms and that spatular width should decrease with body size to the −1/3 power. They survey a variety of fibrillar adhesive organisms (nine flies, five beetles, two spiders, one heteropod, three geckos, and one anole) to test their hypothesis, but did not include the effects of evolutionary history, setal branching, or adhesive mechanism (dry versus wet, or secretion-aided adhesion) in their analysis. Because the hypothesis is a logical, compelling, and important one, we decided to retest their prediction using a phylogenetic approach that considers more species over a wider range of body mass and setal morphology.
Results and Discussion
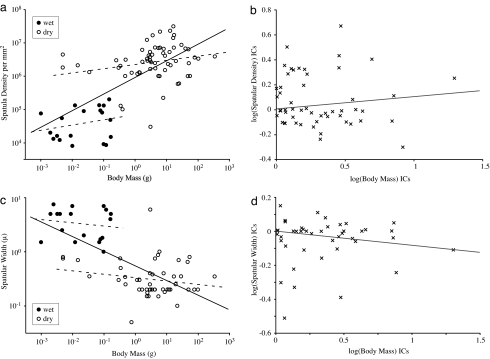
The data shown by Arzt et al. (24) in support of their scaling hypotheses assume that all taxa have evolved independently. If we make the same assumption with our expanded dataset, we would also conclude that spatular density increases significantly with body mass, although to the 0.50 power, significantly less than the predicted 2/3 [P < 0.001; r2 = 0.47; 95% confidence limits (CL) = 0.38, 0.62; Fig. 2a and Table 1]. Phylogenetically uncorrected or raw species values (SV) would suggest that spatular width varies significantly with body mass to the −0.27 power (P < 0.0001; r2 = 0.44; 95% CL: −0.35, −0.20; Fig. 2c and Table 2), not significantly different from the −1/3 prediction (P = 0.11).
Fig. 2.
Linear regression on species values compared with independent contrasts (ICs). Solid lines represent regressions through the pooled dataset. Dashed lines represent partitioned (wet/dry) data. (a) Log–log plot of spatular density versus body mass for 81 taxa. (b) Phylogenetically corrected values for spatular density versus body mass. (c) Log–log plot of spatular width (μm) versus body mass (g) for 67 taxa. (d) Phylogenetically corrected values for spatular width versus body mass.
Table 1.
Regression results for spatula-density versus body-mass analyses
| Partition | Scaling exponents |
Comparison of analyses | |
|---|---|---|---|
| Raw species values | Phylogenetically corrected (ICs) | ||
| Dry | 0.13 (ns); 95% CL (−0.020, 0.27); P = 0.08; df = 61 | 0.064 (ns); 95% CL (−0.13, 0.26); P = 0.50; df = 37 | ns |
| Wet | 0.16 (ns); 95% CL (−0.16, 0.47); P = 0.30; df = 18 | 0.093 (ns); 95% CL (−0.10, 0.29); P = 0.32; df = 15 | ns |
| Branched | 0.22 (+); 95% CL (0.052, 0.38); P = 0.014; df = 43 | −0.030 (ns); 95% CL (−0.25, 0.19); P = 0.78; df = 22 | * |
| Unbranched | 0.29 (+); 95% CL (0.089, 0.43); P = 0.006; df = 36 | 0.12 (ns); 95% CL (−0.024, 0.26); P = 0.10; df = 31 | * |
| Pooled | 0.50 (+); 95% CL (0.38, 0.62); P < 0.001; df = 80 | 0.10 (ns); 95% CL (−0.036, 0.24); P = 0.14; df = 53 | * |
Each analysis was conducted on species values and ICs by using pooled and partitioned datasets (dry/wet, branched/unbranched). P values indicate whether the exponent is significantly different from zero. The rightmost column indicates where results from phylogenetic analyses were significantly different from nonphylogenetic analyses. +, significant and positive relationship; *, analyses yielded significantly different conclusions; ns, no significant relationship or no significant difference between analyses.
Table 2.
Regression results for spatula-width versus body-mass analyses
| Partition | Scaling exponents |
Comparison of analyses | |
|---|---|---|---|
| Raw species values | Phylogenetically corrected (ICs) | ||
| Dry | −0.064 (ns); 95% CL (−0.16, 0.031); P = 0.19; df = 48 | −0.075 (ns); 95% CL (−0.21, 0.058); P = 0.26; df = 28 | ns |
| Wet | −0.060 (ns); 95% CL (−0.26, 0.14); P = 0.54; df = 17 | −0.031 (ns); 95% CL (−0.15, 0.087); P = 0.59; df = 14 | ns |
| Branched | 0.040 (ns); 95% CL (−0.045, 0.13); P = 0.32; df = 33 | 0.040 (ns); 95% CL (−0.11, 0.19); P = 0.59; df = 15 | ns |
| Unbranched | −0.17 (−); 95% CL (−0.27, −0.062); P = 0.003; df = 32 | −0.11 (ns); 95% CL (−0.22, 0.007); P = 0.065; df = 27 | * |
| Pooled | −0.27 (−); 95% CL (−0.35, −0.20); P < 0.001; df = 66 | −0.086 (ns); 95% CL (−0.18, 0.007); P = 0.068; df = 42 | * |
For an explanation, see the legend of Table 1. +, significant and positive relationship; −, significant and negative relationship; *, analyses yielded significantly different conclusions; ns, no significant relationship or no significant difference between analyses.
After correcting for the fact that some species are more closely related phylogenetically than others, we found that no parameter describing setal morphology scaled with body size to an exponent significantly different from zero (spatula density, P = 0.16, Fig. 2b; spatula width, P = 0.072, Fig. 2d; seta length, P = 0.63; seta width, P = 0.30). Furthermore, neither phylogenetically corrected nor uncorrected values for spatula density and spatula width scaled with body mass to an exponent significantly different from zero when partitioned into wet (SV: density, P = 0.30; width, P = 0.54) and dry adhesive organisms (SV: density, P = 0.086; width, P = 0.19; Fig. 2 a and c and Tables 1 and 2). Our expanded dataset revealed that within groups of related taxa, there were no strong trends relating spatular density or width with body mass (Figs. 1 and 3). By partitioning corrected values according to setal branching structure, we found that the assumed inverse relationship between spatular density and spatula width was only present across unbranched setae (95% CL: −0.78, −0.38; P < 0.001). The spatular density of branched setae did not scale predictably with spatula size (95% CL: −0.24, 0.50; P = 0.45) but was in general much greater than that of unbranched setae.
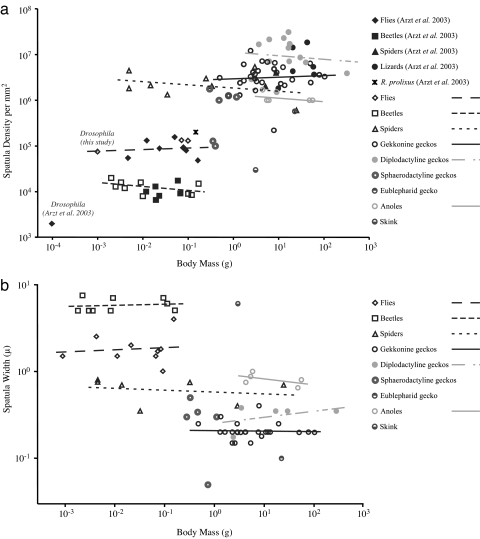
Fig. 3.
Phylogenetically uncorrected species values for spatular density (a) and spatula width (b) reveal no clear trends within groups of related taxa.
The hypothesis that larger animals should benefit from higher spatular density is the most reasonable null model. In fact, available data appear to support the notion. Among fibrillar adhesive organisms, certain geckos have attained their relatively large size because they inherited from their ancestors especially small, densely packed spatulae. Both the largest living and extinct geckos (Rhacodactylus leachianus and Hoplodactylus delcourti; ref. 26) belong to a clade of geckos whose current members possess the highest spatular densities of all organisms with setae. However, miniaturized geckos (various Sphaerodactylus spp., at <1 g, rank among the smallest of the tetrapods; ref. 27) share the same spatular morphology and density as many of their much larger relatives (Fig. 1). To invoke a scaling law, body mass must predominately determine adhesive morphology, and morphology must not be constrained by ancestry. It appears that natural selection has driven gekkonid body size in divergent directions under divergent pressures, rather than larger and larger as their spatular morphology alone might allow. This result yields new predictions. For instance, we can hypothesize that the smaller organisms within each group possess an advantage. Animals with lower body mass may either enjoy a higher safety factor or require less total pad area per body mass than their larger relatives.
Our results question the assumption that natural selection has acted to increase spatular density solely due to increased static normal forces during station-keeping. Considering that even large Tokay geckos hang easily by a single toe (20) and that the shear force generated at that toe fails under a load 10–20 times their body weight (14), it is unlikely that the adhesive performance of most organisms is limited by the static support of their body mass. Geckos running up a vertical surface at nearly one meter per second attach their adhesive pads in 5 ± 2 ms and detach them in 15 ± 4 ms (28). Peak shear forces exceed twice body weight at midstance. Adhesive function depends on how effectively single setae, arrays, toes and feet all function synergistically during dynamic attachment and detachment, not just the density and size of contact elements. Additional variation is likely present for species that live on different substrates, because attachment to rougher surfaces may depend on, among other things, seta length, angle, and curvature, as well as effective seta, array, and toe compliance (12, 21, 29–32). The setae of arthropods are made of cuticle, which can vary in stiffness from 1kPa to tens of GPa (33), whereas reptile setae are likely confined to a narrow range of high elastic moduli (25). Furthermore, most animals do not use fibrillar adhesives exclusively but are aided by alternative attachment mechanisms such as claws. Finally, each taxon must grow through a range of body sizes during its lifespan, with undetermined constraints on its adhesive pad morphology as it develops. Although it is not unreasonable to choose body mass as a proxy for adhesive force, direct simultaneous measurements of shear and normal forces will contribute far more to our eventual understanding of fibrillar adhesive mechanics.
Although variation in setal morphology cannot be predicted by body mass, we can identify some trends by taking into account the history, presence or absence of setal branching, and adhesive mechanism of each taxon. Hierarchical structure (branching) has been predicted to confer multiple advantages, including decreased self-adhesion (1, 34), orientation dependence (35) and increased tolerance to rough surfaces (36, 37). Our results indicated that branching allows higher packing density of smaller spatulae compared with unbranched setae. Animals using dry adhesives possessed spatular densities one to three orders of magnitude greater than those with wet adhesives (Fig. 2a), and much smaller spatula size (Fig. 2c). Beetles and flies surveyed in our study possess spatular tips 1–10 μm wide, whereas geckos possess spatulae ≈200 nm in width, and spider and anole spatulae fall between 350 nm to 1 μm. Federle (1) has suggested that secretions compensate for larger spatulae in insects, specifically on substrates with finer roughness scales than the width of the spatula, where the fluid can bridge small scale gaps. Because secretions predate hairy adhesives in insects, and because many insects adhere without hairs at all, it is possible that selection pressure driving a decrease in spatula size was weaker than selection pressure driving the evolution of dry adhesive structures found in spiders and reptiles. Qian and Gao (38) have hypothesized that decreasing spatula width further than the micrometer scale would actually hinder adhesion in secretion-aided systems. However, their prediction only addresses perpendicular (pulloff) forces. Animals climbing vertically load their adhesives primarily in the shear direction (15, 39), for which few, if any, models currently exist to generate scaling predictions in either dry or wet fibrillar adhesives.
Although several independent hypotheses lead to a contact splitting prediction, we question the assumption that biological data must necessarily confirm it, or any other still untested models of fibrillar adhesion (1). This outcome should serve as a caveat to scientists working with biological datasets, or engineers who would consider organisms as biological “prototypes”. Because of varying degrees of historical relatedness among the subjects of a comparative study, species values cannot be treated as independent data points in statistical analyses. Such assumptions often lead to statistically significant but invalid results (40–43). Investigators must be aware of the developmental and historical constraints on mechanical “design” in animals before they assume evolution to be an optimizing process (44, 45). Synthetic versions of fibrillar adhesives with human-manipulated material properties and morphology could serve as physical models in reconstructing the evolution of these unique structures.
Materials and Methods
Scanning Electron Microscopy.
Gecko setae were harvested from either live or ethanol-preserved museum specimens of 31 gecko species and four anoles [supporting information (SI) Table 3]. Samples from live animals were acquired in accordance with University of California, Berkeley, animal use protocol no. R137. Setae were mounted on stubs using double-sided carbon conductive tape (Ted Pella, Redding, CA) before sputter-coating on two sides with a platinum/palladium alloy (Hummer VI; Technics, Springfield, VA). Samples were viewed with a scanning electron microscope (Amray 1810; Amray, Bedford, MA) at Lewis and Clark College (Portland, OR).
Literature Data.
Characteristic adult live body mass was determined from the literature when possible but was interpolated from body length when necessary. For geckos, we used a scaling relationship based on 16 species of known mass and length (mass = 3.1 × 10−6 × length3.4; r2 = 0.97). Among beetles, we used the scaling relationship from Oertli (46). Additional values for spatula density and spatula size were gathered or estimated from the literature (refs. 16, 23, 24, and 47–88; N. E. Stork, unpublished data; SI Table 3). We included data from species in Arzt et al. (24), but those data were based on our own observations or alternate sources.
Phylogenetic and Statistical Analyses.
We calculated independent contrasts (40) from log-transformed data using CAIC version 2.6.9 (89), which outputs ordinary least squares (OLS) regression statistics (slope, r2, P value) and notifies the user of outliers, as well as any statistical or evolutionary assumptions violated by the data. We calculated regressions on phylogenetically uncorrected species values for comparison. All 81 taxa were included in the spatular density versus body mass analysis. Sixty-seven taxa were available for the spatular width versus body mass scaling analysis. The phylogenetic hypothesis used to generate independent contrasts is a conservative (consensus) composite tree assembled from phylogenies published for the constituent groups (geckos, refs. 90–98; Anolis, ref. 99; squamates, refs. 100 and 101; spiders, refs. 102 and 103; hexapods, ref. 104; beetles, based on refs. 105 and 106; flies, refs. 107 and 108; SI Fig. 4). Where no phylogenetic information was available for a species, the current accepted taxonomic relationship was used. Because statistically supported branch lengths were lacking, we compared results across four trees with the same topology but with widely varying branch lengths transformations (Equal, Grafen, Nee, and Pagel; constructed with Mesquite version 1.12, ref. 109). Because there were no significant differences across branch-length assumptions for any analysis, all plots and statistics representing phylogenetic analyses assume equal branch lengths for simplicity.
Supplementary Material
Acknowledgments
We thank Kellar Autumn, Chris Conroy, Jens Vindum, Walter Federle, Ed Florence, Ted Garland, Sheila Patek, Shai Revzen, and Nigel Stork. Funding was provided by National Science Foundation Nanoscale Interdisciplinary Research Teams Grant 0304730, Defense Advanced Research Projects Agency/Space and Naval Warfare Systems Command Grant N66001-03-C-8045, the American Association of University Women, the National Science Foundation Graduate Research Fellowship Program, and the University of California, Berkeley, Integrative Biology Department.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707591104/DC1.
References
- 1.Federle W. J Exp Biol. 2006;209:2611–2621. doi: 10.1242/jeb.02323. [DOI] [PubMed] [Google Scholar]
- 2.Sitti M, Fearing R. Proceedings of the Second IEEE Conference on Nanotechnology; Piscataway, NJ: IEEE; 2002. pp. 137–140. [Google Scholar]
- 3.Geim AK, Dubonos SV, Grigorieva IV, Novoselov KS, Zhukov AA, Shapoval SY. Nat Mater. 2003;2:461–463. doi: 10.1038/nmat917. [DOI] [PubMed] [Google Scholar]
- 4.Majidi C, Groff R, Fearing RS. Proceedings of the ASME International Mechanical Engineering Congress and Exposition; New York: Am Soc Mechanical Engineers; 2004. p. IMECE2004-62142. [Google Scholar]
- 5.Daltorio KA, Gorb S, Peressadko A, Horchler AD, Ritzmann RE, Quinn RD. In: Tokhi MO, Virk GS, Hassain MA, editors. Climbing and Walking Robots: Proceedings of the Eighth International Conference on Climbing and Walking Robots and the Support Technologies for Mobile Machines; Berlin: Springer; 2005. pp. 131–138. [Google Scholar]
- 6.Northen MT, Turner KL. Nanotechnology. 2005;16:1159–1166. [Google Scholar]
- 7.Yurdumakan B, Raravikar NR, Ajayan PM, Dhinojwala A. Chem Commun. 2005:3799–3801. doi: 10.1039/b506047h. [DOI] [PubMed] [Google Scholar]
- 8.Majidi C, Groff RE, Maeno Y, Schubert B, Baek S, Bush B, Maboudian R, Gravish N, Wilkinson M, Autumn K, Fearing RS. Phys Rev Lett. 2006;97 doi: 10.1103/PhysRevLett.97.076103. 076103. [DOI] [PubMed] [Google Scholar]
- 9.del Campo A, Arzt E. Macromol Biosci. 2007;7:118–127. doi: 10.1002/mabi.200600214. [DOI] [PubMed] [Google Scholar]
- 10.Gorb S, Varenberg M, Peressadko A, Tuma J. J R Soc Interface. 2007;4:271–275. doi: 10.1098/rsif.2006.0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffith AA. Philos Trans R Soc Lond Ser A. 1921;221:163–198. [Google Scholar]
- 12.Gao H, Yao H. Proc Natl Acad Sci USA. 2004;101:7851–7856. doi: 10.1073/pnas.0400757101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson KL, Kendall K, Roberts AD. Proc R Soc Lond Ser A. 1971;324:301–313. [Google Scholar]
- 14.Autumn K, Sitti M, Liang YA, Peattie AM, Hansen WR, Sponberg S, Kenny TW, Fearing R, Israelachvili JN, Full RJ. Proc Natl Acad Sci USA. 2002;99:12252–12256. doi: 10.1073/pnas.192252799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Autumn K, Liang YA, Hsieh ST, Zesch W, Chan W-P, Kenny WT, Fearing R, Full RJ. Nature. 2000;405:681–685. doi: 10.1038/35015073. [DOI] [PubMed] [Google Scholar]
- 16.Kesel A, Seidl T. J Exp Biol. 2003;206:2733–2738. doi: 10.1242/jeb.00478. [DOI] [PubMed] [Google Scholar]
- 17.Sun W, Neuzil P, Kustandi TS, Oh S, Samper VD. Biophys J. 2005;89:L14–L17. doi: 10.1529/biophysj.105.065268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huber G, Mantz H, Spolenak R, Mecke K, Jacobs K, Gorb SN, Arzt E. Proc Natl Acad Sci USA. 2005;102:16293–16296. doi: 10.1073/pnas.0506328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen WR, Autumn K. Proc Natl Acad Sci USA. 2005;102:385–389. doi: 10.1073/pnas.0408304102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Autumn K, Dittmore A, Santos D, Spenko M, Cutkosky M. J Exp Biol. 2006;209:3569–3579. doi: 10.1242/jeb.02486. [DOI] [PubMed] [Google Scholar]
- 21.Autumn K, Majidi C, Groff RE, Dittmore A, Fearing R. J Exp Biol. 2006;209:3558–3568. doi: 10.1242/jeb.02469. [DOI] [PubMed] [Google Scholar]
- 22.Niederegger S, Gorb SN. J Comp Physiol A. 2006;192:1223–1232. doi: 10.1007/s00359-006-0157-y. [DOI] [PubMed] [Google Scholar]
- 23.Irschick DJ, Austin CC, Petren K, Fisher R, Losos JB, Ellers O. Biol J Linn Soc. 1996;59:21–35. [Google Scholar]
- 24.Arzt E, Gorb S, Spolenak R. Proc Natl Acad Sci USA. 2003;100:10603–10606. doi: 10.1073/pnas.1534701100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peattie AM, Majidi C, Corder AB, Full RJ. J R Soc Interface. 2007;4:1071–1076. doi: 10.1098/rsif.2007.0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Russell AP, Bauer AM. Bull Chic Herp Soc. 1991;26:26–30. [Google Scholar]
- 27.Hedges SB, Thomas R. Caribb J Sci. 2001;37:168–173. [Google Scholar]
- 28.Autumn K, Hsieh ST, Dudek DM, Chen J, Chitaphan C, Full RJ. J Exp Biol. 2006;209:260–272. doi: 10.1242/jeb.01980. [DOI] [PubMed] [Google Scholar]
- 29.Sitti M, Fearing RS. J Adhes Sci Technol. 2003;17:1055–1073. [Google Scholar]
- 30.Campolo D, Jones S, Fearing RS. Proceedings of the Third IEEE Conference on Nanotechnology; Piscataway, NJ: IEEE; 2003. pp. 856–859. [Google Scholar]
- 31.Hui CY, Glassmaker NJ, Jagota A. J Adhes. 2005;81:699–721. [Google Scholar]
- 32.Persson BNJ. J Chem Phys. 2003;118:7614–7621. [Google Scholar]
- 33.Vincent JFV. Composites A. 2002;33:1311–1315. [Google Scholar]
- 34.Stork NE. J Nat Hist. 1983;17:829–835. [Google Scholar]
- 35.Yao H, Gao H. J Mech Phys Solids. 2006;54:1120–1146. [Google Scholar]
- 36.Bhushan B, Peressadko AG, Kim TW. J Adhes Sci Technol. 2006;20:1475–1491. [Google Scholar]
- 37.Kim TW, Bhushan B. J Adhes Sci Technol. 2007;21:1–20. [Google Scholar]
- 38.Qian J, Gao H. Acta Biomater. 2006;2:51–58. doi: 10.1016/j.actbio.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 39.Goldman DI, Chen TS, Dudek DM, Full RJ. J Exp Biol. 2006;209:2990–3000. doi: 10.1242/jeb.02322. [DOI] [PubMed] [Google Scholar]
- 40.Felsenstein J. Am Nat. 1985;125:1–15. [Google Scholar]
- 41.Harvey PH, Pagel MD. The Comparative Method in Evolutionary Biology. Oxford: Oxford Univ Press; 1991. [Google Scholar]
- 42.Nunn CL, Barton RA. Am Nat. 2000;156:519–533. doi: 10.1086/303405. [DOI] [PubMed] [Google Scholar]
- 43.Garland T, Bennett AF, Rezende EL. J Exp Biol. 2005;208:3015–3035. doi: 10.1242/jeb.01745. [DOI] [PubMed] [Google Scholar]
- 44.Lauder GV. J Theor Biol. 1982;97:57–67. doi: 10.1016/0022-5193(82)90276-4. [DOI] [PubMed] [Google Scholar]
- 45.Autumn K, Ryan MJ, Wake DB. Q Rev Biol. 2002;77:383–408. doi: 10.1086/344413. [DOI] [PubMed] [Google Scholar]
- 46.Oertli JJ. Mitt Schweiz Entomol Ges. 1991;64:139–154. [Google Scholar]
- 47.Anderson JF. J Arachnol. 1996;24:129–134. [Google Scholar]
- 48.Bauer AM. J Morphol. 1998;235:41–58. doi: 10.1002/(SICI)1097-4687(199801)235:1<41::AID-JMOR4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 49.Bauer AM, Sadlier RA. The Herpetofauna of New Caledonia. New York: Society for the Study of Amphibians and Reptiles; 2000. [Google Scholar]
- 50.Brady AR, McKinley KS. J Arachnol. 1994;22:138–160. [Google Scholar]
- 51.Bustard HR. Proc K Ned Akad Wet C. 1969;72:451–466. [Google Scholar]
- 52.Butler MA, Losos JB. Ecol Monogr. 2002;72:541–559. [Google Scholar]
- 53.Cree A. N Z J Ecol. 1994;21:351–372. [Google Scholar]
- 54.Decae A. Newsl Br Arachnol Soc. 1986;46:3–4. [Google Scholar]
- 55.Elstrott J, Irschick DJ. Biol J Linn Soc. 2004;83:389–398. [Google Scholar]
- 56.Feder ME, Feder JH. Copeia. 1981;1981:204–209. [Google Scholar]
- 57.Foelix RF, Chu-Wang IW. Proceedings of the Sixth International Arachnology Congress; Amsterdam: Nederlandse Entomologische Vereniging; 1974. pp. 156–157. [Google Scholar]
- 58.Foelix R, Jackson RR, Henksmeyer A, Hallas S. Rev Arachnol. 1984;5:329–334. [Google Scholar]
- 59.Gasc JP, Renous S. C R Hebd Seances Acad Sci. 1980;290:675–678. [Google Scholar]
- 60.Gasc JP, Renous S, Diop A. C R Seances Acad Sci III. 1982;294:169–174. [Google Scholar]
- 61.Gill B, Whitaker T. New Zealand Frogs and Reptiles. Auckland: David Bateman; 1996. [Google Scholar]
- 62.Gorb SN. Proc R Soc London B. 1998;265:747–752. [Google Scholar]
- 63.Gorb S, Gorb E, Kastner V. J Exp Biol. 2001;204:1421–1431. doi: 10.1242/jeb.204.8.1421. [DOI] [PubMed] [Google Scholar]
- 64.Hare KM, Cree A. N Z J Ecol. 2005;29:137–142. [Google Scholar]
- 65.Hill DE. Zool J Linn Soc. 1977;60:319–338. [Google Scholar]
- 66.Homann H. Naturwissenschaften. 1957;44:318. [Google Scholar]
- 67.Lehmann F-O, Dickinson MH. J Exp Biol. 1997;200:1133–1143. doi: 10.1242/jeb.200.7.1133. [DOI] [PubMed] [Google Scholar]
- 68.Marcellini DL, Keefer TE. Herpetologica. 1976;32:362–366. [Google Scholar]
- 69.Murphey RK, Possidente D, Pollack G, Merritt DJ. J Comp Neurol. 1989;290:185–200. doi: 10.1002/cne.902900203. [DOI] [PubMed] [Google Scholar]
- 70.Niederegger S, Gorb SN, Jiao Y. J Comp Physiol A. 2002;187:961–970. doi: 10.1007/s00359-001-0265-7. [DOI] [PubMed] [Google Scholar]
- 71.Norberg RA. Oikos. 1978;31:222–229. [Google Scholar]
- 72.Röll B. J Zool. 1995;235:289–300. [Google Scholar]
- 73.Rooij Nd. The reptiles of the Indo-Australian Archipelago. Leiden, The Netherlands: EJ Brill; 1915. [Google Scholar]
- 74.Roscoe DT, Walker G. Bull Br Arachnol Soc. 1991;8:224–226. [Google Scholar]
- 75.Rovner JS. J Arachnol. 1980;8:201–215. [Google Scholar]
- 76.Ruibal R, Ernst V. J Morphol. 1965;117:271–294. doi: 10.1002/jmor.1051170302. [DOI] [PubMed] [Google Scholar]
- 77.Schleich HH, Kästle W. Amphibia-Reptilia. 1986;7:141–166. [Google Scholar]
- 78.Schleich HH. Spixiana. 1987;78(Suppl):4–74. [Google Scholar]
- 79.Schmitz A, Perry SF. J Exp Biol. 2001;204:4321–4334. doi: 10.1242/jeb.204.24.4321. [DOI] [PubMed] [Google Scholar]
- 80.Stork NE. Zool J Linn Soc. 1980;68:173–306. [Google Scholar]
- 81.Szczerbak NN. Guide to the Reptiles of the Eastern Palearctic. Malabar, FL: Krieger Publishing; 2003. [Google Scholar]
- 82.Tang YZ, Zhuang LZ, Wang ZW. Copeia. 2001;2001:248–253. [Google Scholar]
- 83.Tu M, Dickinson M. J Exp Biol. 1994;192:207–224. doi: 10.1242/jeb.192.1.207. [DOI] [PubMed] [Google Scholar]
- 84.Vitt LJ, Zani PA. J Trop Ecol. 1998;14:63–86. [Google Scholar]
- 85.Walker G, Yule AB, Ratcliffe J. J Zool (Lond) 1985;205:297–307. [Google Scholar]
- 86.Williams EE, Peterson JA. Science. 1982;215:1509–1511. doi: 10.1126/science.215.4539.1509. [DOI] [PubMed] [Google Scholar]
- 87.Wilson SK, Swan G. Reptiles of Australia. Princeton: Princeton Univ Press; 2003. [Google Scholar]
- 88.Zaaf A, Van Damme R. Zoomorphology. 2001;121:45–53. [Google Scholar]
- 89.Purvis A, Rambaut A. Comput Appl Biosci. 1995;11:247–251. doi: 10.1093/bioinformatics/11.3.247. [DOI] [PubMed] [Google Scholar]
- 90.Kluge AG. Misc Publ Mus Zool Univ Mich. 1987;173:1–54. [Google Scholar]
- 91.Bauer AM. Bonn Zool Monogr. 1990;30:1–220. [Google Scholar]
- 92.Kluge AG. Am Mus Novit. 1995;3139:1–23. [Google Scholar]
- 93.Good DA, Bauer AM, Sadlier RA. Aust J Zool. 1997;45:317–330. [Google Scholar]
- 94.Chambers GK, Boon WM, Buckley TR, Hitchmough RA. Aust J Bot. 2001;49:377–387. [Google Scholar]
- 95.Carranza S, Arnold EN, Mateo JA, Geniez P. Mol Phylogenet Evol. 2002;23:244–256. doi: 10.1016/S1055-7903(02)00024-6. [DOI] [PubMed] [Google Scholar]
- 96.Lamb T, Bauer AM. Copeia. 2002;2002:586–596. [Google Scholar]
- 97.Han D, Zhou K, Bauer AM. Biol J Linn Soc. 2004;83:353–368. [Google Scholar]
- 98.Melville J, Schulte JA, Larson A. Biol J Linn Soc. 2004;82:123–138. [Google Scholar]
- 99.Poe S. Herpetol Monogr. 2004;18:37–89. [Google Scholar]
- 100.Townsend TM, Larson A, Louis E, Macey JR. Syst Biol. 2004;53:735–757. doi: 10.1080/10635150490522340. [DOI] [PubMed] [Google Scholar]
- 101.Zhou K, Li H, Han D, Bauer AM, Feng J. Mol Phylogenet Evol. 2006;40:887. doi: 10.1016/j.ympev.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 102.Coddington JA, Levi HW. Annu Rev Ecol Syst. 1991;22:565–592. [Google Scholar]
- 103.Maddison WP, Hedin MC. Invertebr Syst. 2003;17:529–549. [Google Scholar]
- 104.Beutel RG, Gorb SN. J Zool Syst Evol Res. 2001;39:177–207. [Google Scholar]
- 105.Howland DE, Hewitt GM. Insect Mol Biol. 1995;4:203–215. doi: 10.1111/j.1365-2583.1995.tb00026.x. [DOI] [PubMed] [Google Scholar]
- 106.Farrell BD, Sequeira AS. Evolution (Lawrence, Kans) 2004;58:1984–2001. doi: 10.1111/j.0014-3820.2004.tb00484.x. [DOI] [PubMed] [Google Scholar]
- 107.Yeates DK, Wiegmann BM. Annu Rev Entomol. 1999;44:397–428. doi: 10.1146/annurev.ento.44.1.397. [DOI] [PubMed] [Google Scholar]
- 108.Ståhls G, Hippa H, Rotheray G, Muona J, Gilbert F. Syst Entomol. 2003;28:433–450. [Google Scholar]
- 109.Maddison WP, Maddison DR. Mesquite: A Modular System for Evolutionary Analysis, Version 1.12. 2005 Available at http://mesquiteproject.org.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.