Abstract
The evolutionary forces that establish and hone target gene networks of transcription factors are largely unknown. Transposition of retroelements may play a role, but its global importance, beyond a few well described examples for isolated genes, is not clear. We report that LTR class I endogenous retrovirus (ERV) retroelements impact considerably the transcriptional network of human tumor suppressor protein p53. A total of 1,509 of ≈319,000 human ERV LTR regions have a near-perfect p53 DNA binding site. The LTR10 and MER61 families are particularly enriched for copies with a p53 site. These ERV families are primate-specific and transposed actively near the time when the New World and Old World monkey lineages split. Other mammalian species lack these p53 response elements. Analysis of published genomewide ChIP data for p53 indicates that more than one-third of identified p53 binding sites are accounted for by ERV copies with a p53 site. ChIP and expression studies for individual genes indicate that human ERV p53 sites are likely part of the p53 transcriptional program and direct regulation of p53 target genes. These results demonstrate how retroelements can significantly shape the regulatory network of a transcription factor in a species-specific manner.
Deciphering gene regulatory networks in the postgenomic era will provide pivotal insights into genome function and human disease, but it will require a much improved knowledge of evolutionary forces that shape transcriptional networks. One key will be understanding the ≈5% of the human genome that is under purifying selection and hence likely to contain functional segments (1, 2). Two-thirds of these segments do not code for protein and likely harbor essential regulatory information. Some of these derive from transposable elements, and thus lie in the 45% of our genome once deemed “junk DNA.” Although there are examples where transposable elements played important roles in the evolution of gene regulation (3–5), and certain families have deposited putative regulatory elements that are now subjected to purifying selection (6–10), it is unclear how extensively these mobile elements have shaped gene regulatory networks.
Human endogenous retroviruses (ERVs), remnants of exogenous retroviruses that gained access to the germ line, are usually not found in gene-rich regions, consistent with an effect on gene expression that typically reduces fitness (11, 12). ERVs make up 8% of the human genome (1). Approximately 10% contain sequences that once coded for retroviral proteins flanked by two LTRs; the rest are solitary LTRs (solo LTRs). Despite the general selection against ERVs near genes, the number of examples of promoters and enhancers derived from ERVs is steadily increasing (13). The evolutionary process of “exaptation” of noncoding functional elements from viruses and transposons to benefit the host is not well understood beyond a few examples (14).
The tumor suppressor protein p53 is a sequence-specific transcription factor that responds to cellular stresses by coordinating expression of genes involved in cell-cycle arrest, senescence, and apoptosis (15). p53 regulates genes of diverse biological pathways and is considered a pleiotropic master regulator. Intense computational and experimental efforts have determined p53 DNA binding specificity, mapped many genomic binding sites, and identified numerous target genes (16–19). However, no studies have examined a relationship between p53 and transposable elements to our knowledge.
We report that human ERVs actively shape the p53 transcriptional network in a species-specific manner. p53 sites are highly enriched in LTRs of a few ERV subfamilies. These p53 site-containing LTRs are in vivo binding sites for p53 and account for >30% of p53 sites found in a genomewide ChIP analysis (16). Expression of many genes close to these LTRs is regulated by p53, based on published data and our experimental validation. These ERVs likely entered the primate ancestral genome and transposed within it ≈25 Mya to 63 Mya. Their proviruses were probably responsible for introducing a p53 site. In general, ERV insertions near genes (including those with p53 sites) were selected against (11, 12), but a significant fraction of p53 site-containing ERVs may have been exapted as regulatory sequences to expand the p53 transcriptional network. At least one ERV insertion likely reshaped the transcriptional landscape of its surrounding genomic area and was instrumental in creating a new gene that became part of the human-specific p53 regulatory network.
Results
p53 Sites Are Enriched in LTRs of Several Human ERV Subfamilies.
A genomewide yeast-based screen identified certain ERV LTR elements with a p53-responsive site (J.Z. and R.K.B., unpublished data). This finding triggered a computational survey of the human genome for p53 sites in ERV LTR elements. Using RepeatMasker (44), 319,106 ERV LTR fragments were identified, accounting for 5% of the human genome and belonging to >500 families and subfamilies of LTR-containing retroelements defined in RepBase (21). Only 1,509 fragments had a near-perfect p53 site based on our stringent criteria [see Materials and Methods and supporting information (SI) Text]. Copies with a p53 site were strikingly overrepresented in the LTR10 and MER61 families of class I ERV elements, accounting for 9–53% of specific subfamilies (Table 1). Most other ERVs and repetitive element families were without p53 sites or had a frequency of p53 sites expected by chance (SI Table 4).
Table 1.
Distribution of p53 sites in ERV/LTR elements
| ERV category | Fragment no. | Copies with p53 sites | Percentage | Sites/fragment | Sites/kb |
|---|---|---|---|---|---|
| All ERVs | 319,106 | 1,509 | 0.47 | 0.0048 | 0.011 |
| LTR10B1 | 246 | 77 | 31.30 | 0.43 | 1.4 |
| LTR10C | 512 | 47 | 9.18 | 0.11 | 0.25 |
| LTR10D | 190 | 35 | 18.42 | 0.19 | 0.45 |
| LTR10E | 252 | 69 | 27.38 | 0.44 | 0.96 |
| MER61C | 294 | 157 | 53.40 | 0.53 | 1.5 |
| MER61E | 316 | 112 | 35.44 | 0.36 | 0.95 |
ERV LTRs are separated into those that contain p53 sites and those that do not, and overlap with PET3 + loci is determined for each set. Enrichment is determined as a ratio of the two overlap fractions. P value is calculated based on hypergeometric distribution. N/A, not available.
p53 Binds in Vivo to ERV LTRs with a p53 Site.
Recently, a study identified 327 high confidence binding regions for DNA damage-activated p53 (PET3+ loci) using ChIP followed by paired-end di-tags sequencing (ChIP–PET) (16). We found a 446-fold enrichment of ERV LTRs with p53 sites in these ChIP–PET-confirmed p53 binding regions (PET3+; see ref 16 for definition and statistical criteria) compared with ERV LTRs without p53 site (P < 6 × 10−198), with 10-fold or higher enrichment in each of the six LTR10 and MER61 subfamilies (Table 2). LTRs in these six subfamilies accounted for 72 of the 89 overlaps between PET3+ regions and LTRs with p53 sites. By our site definition, 250 of the 327 PET3+ loci contained a p53 site (16). Thus, the 89 ERV LTR fragments accounted for one-third of PET3+ loci with predicted p53 sites (Table 3). When we further compared the distribution of repetitive elements within PET3+ loci to their genomewide distribution, we found clear enrichment of ERV LTR copies in PET3+ loci (18.3% versus 8.7%), entirely explained by class I ERVs (14.3% versus 3%) (SI Table 5).
Table 2.
Overlap between ERV/LTR and PET3 + loci
| ERV category | ERVs without p53 sites that overlap PET3 + loci | ERVs with p53 sites that overlap PET3 + loci | Enrichment for ERVs with p53 sites (Fold) | P value |
|---|---|---|---|---|
| ERV | 42 of 317,597 (0.0132%) | 89 of 1,509 (5.9%) | 446 | 5.56E-198 |
| LTR10B1 | 0 of 169 (0%) | 12 of 77 (15.6%) | N/A | 1.08E-32 |
| LTR10C | 1 of 465 (0.22%) | 1 of 47 (2.1%) | 9.9 | 0.1045 |
| LTR10D | 0 of 155 (0%) | 11 of 35 (31.4%) | N/A | 7.83E-34 |
| LTR10E | 2 of 183 (1.1%) | 14 of 69 (20.3%) | 18.6 | 2.59E-36 |
| MER61C | 1 of 137 (0.73%) | 15 of 157 (9.6%) | 13.1 | 1.69E-35 |
| MER61E | 1 of 204 (0.49%) | 19 of 112 (17.0%) | 34.6 | 2.82E-50 |
Predicted p53 sites within repetitive sequences of the human genome. The columns contain the following: names of selected LTR families defined in RepBase (21); total number of fragments identified in the human genome; total number of fragments that contain a predicted p53 site; percentage of fragments that contain a p53 site; average number of sites per fragment; average number of sites per thousand bases. Only selected LTR10 and MER61 subfamilies are listed. For a complete table see SI Table 4.
Table 3.
ERVs with a p53 site enriched in PET loci
| Locus category | No. of loci | Loci with p53 sites | Loci with p53 sites in ERV |
|---|---|---|---|
| PET1 (low confidence) | 61,160 | 1,722 | 150 (8.7%) |
| PET2 (medium confidence) | 1,452 | 281 | 67 (23.8) |
| PET3+ (high confidence) | 327 | 250 | 89 (35.6) |
PET1, PET2, and PET3 + were defined by ref. 16 as potential p53 binding regions with different confidence. The number of loci in each category is listed in the second column. The third column contains the number of loci that have a predicted, near-perfect p53 site. The last column indicates the number and fraction of these predicted p53 sites residing in ERVs.
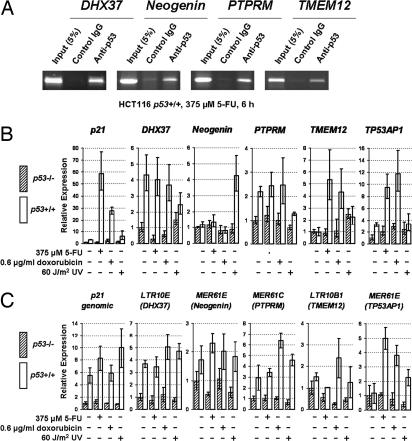
To verify these findings, we chose four genomic ERV p53 sites close to or in the introns of genes for validation. All selected loci, near DHX37, Neogenin, PTPRM, and TMEM12, showed increased p53 occupancy after DNA damage by using ChIP of p53 in HCT116 cells (Fig. 1A and SI Table 6).
Fig. 1.
Experimental validation of selected candidates. (A) ChIP of p53 with semiquantitative PCR for four LTR elements near genes, after treatment with 5-FU (375 μM for 6 h). (B) Reverse transcriptase quantitative real-time PCR for four genes close to the four LTR elements of A and TP53AP1, after treatment with 5-FU (375 μM for 24 h), doxorubicin (0.6 μg/ml for 24 h), or UV irradiation (60 J/m2). (C) p53 reporter gene assays for five firefly luciferase constructs driven by selected LTR fragments, using the same treatments as in B. Error bars in B and C represent 95% confidence interval (≈2 SDs). Relative expression levels were scaled relative to the mean of p53 (−/−) with no treatment (designated as 1). p21 served as a positive control.
ERV LTRs with a p53 Site Exhibit p53-Regulatory Potential.
We collected published data for 392 genes with p53-dependent regulation. This list was compared with the set of 440 closest genes that are no further than 1 Mb away from each of the 497 LTR10 and MER61 LTRs with a p53 site. Thirty-one of the 392 known p53 targets were among the 440 genes associated with an LTR-derived p53 site (P < 8 × 10−11) (SI Table 6).
Many of the 392 genes are direct p53 targets with a confirmed p53 binding site, but it was not previously noted that in some cases the p53 site is from an ERV LTR. For example, a p53 ChIP study of ENCODE regions confirmed a binding site close to TRIM22, which is up-regulated by p53. This binding site is provided by an LTR10D copy (22). Confirmatory ChIP for individual genomic sites of the ChIP–PET study included a site close to IFNAR. We determined that the p53 site is provided by an LTR10B1 copy (16).
We investigated the p53-dependent regulatory potential of the four p53 binding ERV LTRs we confirmed (Fig. 1A) and an additional LTR near the TP53AP1 gene (which we discuss later) by assessing expression levels of nearby genes and the enhancer capacity of these LTRs in a reporter gene assay in response to various stress treatments. We performed quantitative RT-PCR of p53+/+ and p53−/− HCT 116 cells before and after 5-fluorouracil (5-FU), doxorubicin, or UV treatments. All genes showed p53-dependent activation, albeit to varying degrees, depending on the type of DNA-damaging agent (Fig. 1B). Under similar cellular conditions, all LTR fragments tested showed clear p53-dependent enhancement of transcriptional activity in a luciferase reporter gene assay (Fig. 1C). Although other factors are likely involved in the transactivation of these target genes, these results strongly argue that these ERVs have the potential to contribute to p53-dependent expression of nearby genes.
Evolutionary History of ERVs with p53 Sites.
LTR10 families are related to provirus HERVIP10, and MER61 families are related to provirus HUERS-P3B (23). We used three independent methods to estimate the age of these ERV elements.
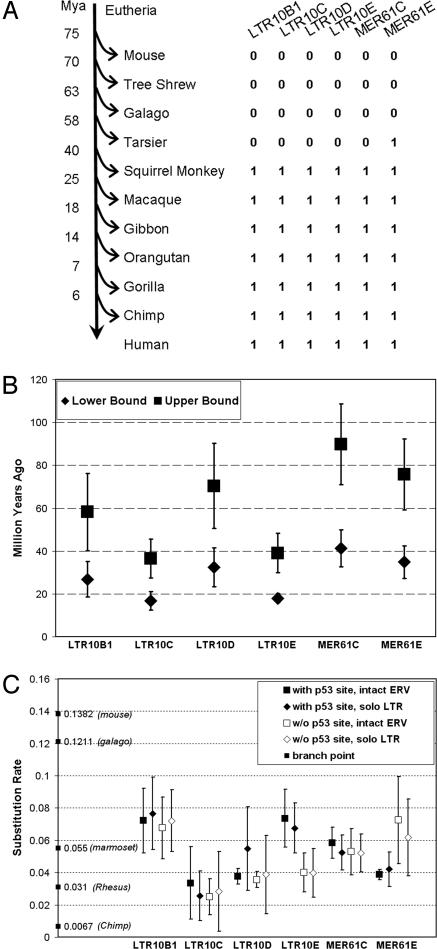
First, we looked for the presence of sequences related to LTR10 and MER61 families in extant species by searching all nucleotide sequence in National Center for Biotechnology Information databases, including trace archives. Copies of LTR10 and MER61 families were identifiable in both branches of anthropoids, i.e., New World monkeys (including squirrel monkey and marmoset) and Catarrhini (Old World monkeys, e.g., rhesus monkey, and apes, e.g., human) but not in Strepsirrhini prosimians, such as lemurs and galagos, and not in tree shrews (Fig. 2A). The tarsier is thought to be more closely related to the anthropoids than are the Strepsirrhini prosimians, forming a clade with anthropoids called Haplorrhini (24). By searching 8.7 Gb of sequence from the trace archive, about three times coverage of the tarsier genome, we found further evidence for this phylogenetic relationship in 790 matches to the anthropoid MER61E element. However, we cannot confidently identify a tarsier MER61E element that is in the orthologous location in an anthropoid, so we cannot rule out the possibility that these matches are caused by an independent endogenization of a similar, but not identical, retrovirus. None of the five other p53 site-containing LTR subfamilies are found in tarsier. Thus, at least five of these six ERV families did not show activity in Haplorrhini before the split between anthropoid and tarsier, but do show wide activity in anthropoids. This finding places the time of original activity for these ERVs roughly between 40 Mya and 63 Mya. The spreading of these elements was likely attenuated 25 Mya when Old World monkeys diverged from apes. This hypothesis is based on the fact that human and rhesus share the majority, but not all, of these LTRs at orthologous sites (some differences possibly caused by lineage-specific losses), and human and chimpanzee share essentially all. These estimations are similar to results based on analyzing coding sequence divergence in the respective ERVs (23, 25).
Fig. 2.
Estimated age of p53 site-containing ERV families. Insertion time of individual families of ERV LTRs is estimated by three methods. (A) Age of each LTR family, estimated by examining the presence of sequences in different species. 0 indicates no homologous hit, and 1 indicates that there are homologous hits. (B) Ages of near-complete ERVs, estimated by comparing their 5′ and 3′ LTR sequence divergence. Upper and lower bound were calculated by using a mutation rate of 2.3 × 10−9 and 5 × 10−9 substitutions per site per year, respectively. (C) Substitution rate, calculated by comparing individual fragments to the consensus by using the Jukes–Cantor formula (26). Fragments were grouped based on the presence of a p53 site and if they are solo LTRs, then the average and SD were calculated for each group. Branch length is taken from that of ref. 27.
Second, ≈7% of LTRs in each family are linked to an almost intact ERV internal structure with both flanking LTRs. Knowing that the 5′ and 3′ LTR were identical at the time of insertion into the ancestral genome, and assuming that ERVs accumulate mutations at a rate of 2.3 to 5 × 10−9 substitutions per site per year (25), we estimated that LTR10 and MER61 LTRs were transposed ≈40 Mya (Fig. 2B), perhaps just before the split of anthropoids into New World monkeys and Catarrhini.
Lastly, assuming that the consensus sequence is a good replica of what was originally inserted for each copy and using the Juke–Cantor formula (26), we calculated a substitution rate for each individual LTR (Fig. 2C). Using branch lengths calculated as in ref. 27, we estimated that most were transposed in the same time window as above, after the split with prosimians and before the split with New World monkeys. Combining the above analyses, we estimate that ERVs with p53 sites were spreading in the anthropoid lineage in a time window of ≈25 Mya to 63 Mya.
The p53 Sites Were Likely Present in Progenitor LTRs.
LTR elements of LTR10 and MER61 families became fixed in the primate lineage long ago. One of two main scenarios likely explains p53 sites in a subset of elements in each subfamily. (i) The p53 site was present in the LTRs of the founder retroviruses or proviruses, and some copies lost the p53 site over time. (ii) The founder provirus had no p53 site, and a later copy acquired a p53 site through mutations, started to propagate, and created a subset of ERV insertions with a p53 site.
Our analysis is most consistent with the first model. First, it is unlikely that individual LTRs evolved a p53 site independently as the phylogenetically reconstructed LTR consensus of each of the six subfamilies clearly contains a p53 site. Second, for each of the six subfamilies, sequence comparison shows that LTRs with p53 site are no more similar to one another than to LTRs without a p53 site, arguing against a single ancestor for LTRs with a p53 site that is distinct from all other LTRs. Third, the estimated age of individual LTRs with a p53 site has a wide range and is slightly biased toward older age compared with LTRs without a p53 site, except for MER61E (Fig. 2C), indicating that p53 site-containing ERVs occurred relatively early in the evolution of these ERV families.
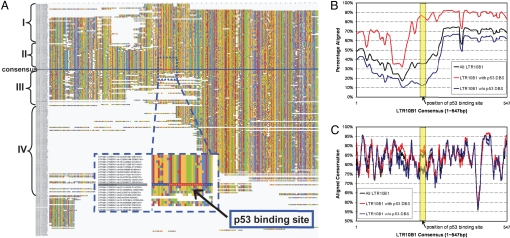
To further support the hypothesis that the p53 site was present in the progenitor LTR, we aligned all genomic copies of each LTR family and found that many copies without a p53 site likely lost it because of deletions. For example, we aligned 246 human LTR10B1 copies and clustered them based on the presence of a p53 site and percentage identity with the consensus (Fig. 3A). The horizontal blue line in Fig. 3A represents the consensus LTR10B1, and the red stretch identifies the reconstructed ancestral p53 site. The reconstructed site (TGACATGCCCAGACATGCCT) is an almost perfect p53 site. Only the first position “T” is different from the “R (purine)” in the canonical consensus (28), the influence of which is not likely significant (18). Sequences above the blue line in Fig. 3A have a p53 site, whereas sequences below do not. More than 10% of LTR10B1 fragments are almost free of large deletions or insertions and contain a p53 site (Fig. 3A, cluster II). Another 20% with a p53 site display deletions at the 5′ end that stop right before the p53 site (Fig. 3A, cluster I). LTRs without a p53 site either have a small deletion around the p53 site (Fig. 3A, cluster III), or a large deletion from the 5′ end to just 3′ of the p53 site (Fig. 3A, cluster IV). This trend is summarized in Fig. 3B by plotting the coverage of the LTR10B1 consensus by all genomic fragments. The sequence identity of the aligned bases across the length of LTR10B1 is ≈85% (Fig. 3C). The percentage identity within the p53 site is no better than the background because the binding site allows similar levels of degeneracy. The preponderance of large deletions in LTR10B1 elements without a p53 site is consistent with the model in which the progenitor sequence had an intact p53 site.
Fig. 3.
Evolutionary pattern of LTR10B1 genomic copies. Genomic copies of LTR10B1 are aligned to the consensus sequence. (A) Multiple sequence alignments, clustered based on existence of a p53 site and the percentage identity to the consensus. The blue line in the middle is the consensus sequence, and the red stretch indicates the position of the reconstructed p53 site. Every copy above the blue line contains a predicted p53 site. Sequences that are placed closer to the consensus have higher sequence similarity. The nucleotides are color-coded. A, green; C, yellow; G, red; T, blue. The image was created with Jalview (20). (B) Frequency of coverage of the consensus sequence by the genomic copies. (C) Average percentage identity of each base in the consensus sequence that is aligned to multiple genomic copies.
Impact of ERVs with p53 Sites on the Dynamics of Genome Regulation.
ERV insertions tend to be distant from genes and those fixed in introns are preferentially oriented antisense to the enclosing gene (11, 12). We observed the same for LTR10 and MER61 families. Compared with LTRs without a p53 site, LTRs with a p53 site are even less frequently found close to genes and show stronger strand bias, suggesting overall selection against insertion that could influence gene expression inappropriately (SI Text and SI Fig. 4). However, our data indicate that in a subset of LTRs, which have a p53 site positioned correctly to affect gene regulation, the site is likely to be bound by p53 and affect regulation of the nearby gene. For example, for 21 of 31 known p53 target genes associated with an ERV-derived p53 site, the ERV p53 site is the closest one to the gene. Similarly, for those genes identified by us as being close to an LTR with a p53 site, the LTR p53 site is also the closest one to the gene in the majority of cases (319/440, or 72%). Nevertheless, there may exist additional p53 sites that are functional between the ERV p53 site and the candidate gene, or in the vicinity, especially when the ERV is relatively faraway from the gene. The ERV p53 sites may act as an enhancer or work in concert with other sites to convey regulation, the elucidation of which awaits further investigation.
Given the importance of p53 regulation in all mammals, we expect that the already established and fine-tuned parts of the p53 regulatory network were not substantially disturbed when ERVs with a p53 site populated the genome ≈40 Mya. More likely, the introduction of ERVs resulted in new lineage-specific subnetworks of p53 regulation (29). Indeed, when we analyzed Gene Ontology annotation of potential new p53 target genes, we found that genes related to well known and conserved p53 functions, such as response to DNA damage, are completely absent from our list of genes close to ERVs with a p53 site (P < 6 × 10−13). In contrast, cell adhesion-related genes are enriched (30/413, P < 7 × 10−8). This finding gives indirect evidence that p53 adopted a new role of regulating cell adhesion-related processes in the primate lineage because of ERV-mediated depositing of p53 sites near genes linked to this process. Interestingly, cell adhesion genes are known to be enriched for exapted elements and to evolve rapidly in the human lineage (6, 9, 30).
Our model predicts that insertion of an ERV with a p53 site has the potential to significantly change the dynamics of regional transcriptional activity. In one case, such ERV activities may even have led to the creation of a new gene, TP53AP1 (see SI Fig. 5A for details of genomic locus). TP53AP1 is known to be up-regulated by p53 in human cells (31, 32) (Fig. 1 B and C), but its exact function remains unknown. p53 binds to the second intron of TP53AP1 (16), and the p53 site is within the MER61E LTR element of a near-intact HUER-P3B ERV copy that inserted ≈40 Mya. It is shared among human, chimp, and rhesus, but not outgroup species such as galago and treeshrew.
The predicted protein sequence of TP53AP1 is not homologous to any other protein outside of the primate lineage, and the gene structure suggests it to be a target of nonsense-mediated decay (33). Thus, it is possible that the gene is not translated, but rather functions through its RNA product. We compared the genomic regions in chimp, rhesus, galago, treeshrew, mouse, and rat that are orthologous to the predicted human ORF (SI Fig. 5C). The nucleotide sequences of these species share modest similarity, but their coding potential is quite different. The sequence of rodents allows for no ORF. Although human and chimp genomic sequences are 100% identical, the rhesus sequence contains 14 nucleotide substitutions in the presumed ORF region with 11 resulting in amino acid changes and 1 in a premature stop codon (SI Fig. 5C). Assuming that TP53AP1 is indeed a protein-coding gene, then this gene must have experienced very strong positive selection in the ape lineage to form the novel protein that exists in apes today. The simpler explanation may be that the transcript is actually not protein coding.
Regardless of whether TP53AP1 is translated or not, there is a large difference in transcript distribution in this area. Human TP53AP1 is very close to the divergently transcribed CROT. Despite a gene structure suggestive of nonsense mediated decay, in humans GenBank mRNA evidence for TP53AP1 is as abundant as for CROT. For the orthologous mouse region, ample transcripts exist for CROT, but none for the corresponding region of TP53AP1 (SI Fig. 5 A and B). This finding suggests that insertion of a human ERV with a p53 site correlates significantly with reshaping of the transcriptional landscape in its vicinity, creating a transcript that is now part of the p53 regulatory network.
Discussion
When Barbara McClintock (34) first discovered transposable elements in maize >50 years ago, she called them “controlling elements” because they altered gene expression. Roy Britten and Eric Davidson (35) then proposed that coordinated regulatory systems in animal genomes are encoded by networks of repetitive sequence relationships and that this presents an attractive evolutionary scenario for gene regulatory networks. This idea is now supported by the discovery of numerous promoters and enhancers born by exaptation (13). Recent comparative genomics studies have shown that a substantial proportion of constrained nonexonic elements unique to mammals arose from mobile elements, pointing to transposons as a major creative force in the evolution of mammalian gene regulation (9, 10).
Our study brings p53, a pleiotropic transcription factor and one of the most important master regulators, into this paradigm. We discovered a unique distribution pattern of p53 sites within repetitive sequences of the human genome, and several ERV families emerged as being substantially enriched for p53 sites in their LTRs. Whole-genome ChIP data (16) revealed that p53 occupies such LTR p53 sites in vivo, and our targeted ChIP analysis for four LTR p53 sites confirmed this assessment.
Our data indicate that LTRs with a p53 site contain strong p53-dependent regulatory potential. Our five chosen LTRs all exhibited p53-dependent enhancer activity in reporter gene assay, and expression of genes near these loci correlated with p53 activity. The p53 effect depended on the type of DNA-damaging agent, consistent with a crucial contribution of damage-specific cofactors to p53 activity. The impact of ERV p53 sites on gene expression is likely to be modulated by cofactors, long-range interactions, and local chromatin structure, as well as additional genomic p53 sites. The details of such impact await further, more systematic investigation.
Our evolutionary analysis determined that ERVs with a p53 site populated the ancestral primate genomes ≈40 Mya. Insertions and deletions of these elements created a turnover of p53 sites adjacent to many genes. In this manner, the spread of ERVs may have accelerated the evolution of the host genome (36). Specifically, by depositing p53 sites throughout the genome, ERVs may have recruited and even created new target genes to be part of the primate-specific p53 regulatory network. Other sites not directly driving expression of nearby genes may also play an important role in the p53 network by sequestering activated p53 en masse, thereby titering the amount of active p53 needed for the appropriate response to a cellular stress.
In addition to its effect on the host, the presence of a p53 site may have benefited the retrovirus. Retroviral endogenization is a parasitic process that allows retroviruses to survive and pass on their own genetic material. A p53 site in their LTRs may have given retroviruses the advantage of quick transcription to exit a cell under stress, a condition known to activate ERVs in some species (37). Thus, interaction between what was once foreign genetic information and the host regulatory systems could have impacted evolution in multiple ways, and the impact may have been extensive enough that the relationship between primates and ERVs could ultimately have to be viewed as symbiotic in a generalized sense.
Our findings provide support for Britten and Davidson's theory of regulatory network evolution through mobile elements. Several interesting corollaries stem from this hypothesis. First, the general mechanism, described here for primate-specific ERVs, should exist independently of any specific lineage or evolutionary time frame. ERVs or other mobile elements may have mediated the expansion of the p53 network several times in evolution. It is not unreasonable to speculate that such activity, much too distant in the past for us to recognize, may have contributed to the crowning of p53 as a master regulator. Second, p53 is unlikely to be the only transcription factor whose evolution has been impacted by this mechanism. In fact, the Britten–Davidson theory predicts that many master regulators may have used mobile elements to accelerate the establishment of their regimes. Third, some mobile elements may carry not just one binding site of a transcription factor, but an array of sites constituting an entire regulatory module to enhance their impact and tinker with the genome.
Technologies to interrogate the relationship between transcription factors and repetitive sequences are currently much limited. Most genomewide analyses of gene regulation, transcription factor binding sites, and other functional genomics, including the ENCODE project (27) designed to catalog all sequence elements in 1% of the human genome, tend to ignore the functional aspect of repetitive elements. The results reported here suggest that it may be valuable to take a deeper look at the role of these elements in the evolution of gene regulatory networks.
Materials and Methods
Cell Culture, ChIP, Quantitative RT-PCR, and Reporter Gene Assay.
HCT116 cells (p53+/+) and p53-null derivatives, provided by Bert Vogelstein (John Hopkins University, Baltimore), were grown in McCoy's 5A medium with 10% FBS at 37°C. ChIP assays with anti-p53 antibody DO-1 (Santa Cruz Biotechnology, Santa Cruz, CA) were performed per the Upstate Biotechnology (Lake Placid, NY) protocol after 5 × 106 HCT116 p53+/+ cells in 10-cm dishes were treated with 375 μM of 5-FU for 6 h. Gene expression studies were done with 3 × 106 HCT116 p53+/+ and p53−/− cells in 60-mm dishes, treated with 375 μM 5-FU or 0.6 μg/ml doxorubicin for 24 h or 60 J/m2 of UV irradiation with 24 h of recovery. RNA was isolated with TRIzol reagent (Invitrogen, Carlsbad, CA). cDNA was generated with SuperScriptIII reverse transcriptase (Invitrogen) and analyzed by real-time quantitative PCR. Individual LTRs were cloned into pp53-TA-Luc vector (Clontech, Mountain View, CA), transient transfections were performed by using FuGENE 6 (Roche, Indianapolis, IN), and reporter gene assays were performed with the Dual-Luciferase system (Promega, Madison, WI) (see SI Table 7 for details including primer information).
Sequence Data Sources.
The University of California, Santa Cruz assemblies and repeat libraries per species are: human (Mar2006/hg18/RM051101), chimp (Mar2006/panTro2/RM060120), macaque (Jan2006/rheMac2/RM20060120), and mouse (Feb2006/mm8/RM060120). See SI Text for details of assembling a database of nucleotide sequences of the tree of life. Coordinates of ERV LTRs were based on RepeatMasker annotated hg18. Coordinates of PET3+ loci were lifted to hg18 from annotation of human genome hg17 of (16).
p53 Binding Site and Matrix.
A p53 position-specific weight matrix for a 10-bp-long half site was trained on a total of 162 DNA binding sites collected from the TRANSFAC database and a literature search, the details of which as well as a LOGO of the matrix can be found in SI Text. The criteria we used to predict a p53 site require that a site contains at least two consecutive 10-bp half sites that both pass a lower cutoff (0.5) and at least one passes a higher cutoff (0.7). Predicted sites agree well with the canonical p53 consensus (two half sites of RRRCWWGYYY) (28) with at most two mismatches in either half site, none of which occurs in the middle four bases.
Estimating Age of an LTR.
For method 1, BLASTZ (38) was used to search LTR consensus sequences against the database of nucleotide from the tree of life. For method 2, 5′ and 3′LTR of the same ERV copy were aligned by using BLAST (39). A mutation rate of 2.3 × 10−9 substitutions per site per year was used to estimate the oldest possible insertion time (or upper bound), and 5 × 10−9 to estimate the youngest insertion time (or lower bound) (25). For method 3, genomic copies of an LTR family were aligned to a consensus sequence by using BLAST (39). Substitution rate was calculated with the Juke–Cantor formula (26). The model of neutral evolution was computed by PhyloP (40) from 4-fold degenerate sites in the ENCODE region (27).
Gene Ontology Enrichment.
University of California, Santa Cruz hg18 known genes (41) splice variants were combined into human gene loci. Each locus was defined by a representative transcription start site and a stop site. Gene Ontology annotation (42) terms for all splice variants were assigned to the gene locus, resulting in 14,626 annotated known gene loci. Enrichment was calculated by using the hypergeometric distribution, without correction for multiple hypotheses (43).
Supplementary Material
Acknowledgments
We thank Robert Baertsch, Fan Hsu, and Kate Rosenbloom for advice regarding data selection and analysis; Lan Truong, Hinrich Boeger, and Lauren Wenger for technical advice; Gary Stormo and Barak Cohen for critical reading of the manuscript; and Rick Wilson and the Genome Sequencing Center of Washington University (St. Louis, MO) for tarsier sequences. D.H. is a Howard Hughes Medical Institute Investigator. T.W. is a Helen Hay Whitney Fellow.
Abbreviations
- ERV
endogenous retrovirus
- 5-FU
5-fluorouracil.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703637104/DC1.
References
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M., FitzHugh W, et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 3.Bannert N, Kurth R. Annu Rev Genomics Hum Genet. 2006;7:149–173. doi: 10.1146/annurev.genom.7.080505.115700. [DOI] [PubMed] [Google Scholar]
- 4.Gifford R, Tristem M. Virus Genes. 2003;26:291–315. doi: 10.1023/a:1024455415443. [DOI] [PubMed] [Google Scholar]
- 5.Wessler SR. Proc Natl Acad Sci USA. 2006;103:17600–17601. doi: 10.1073/pnas.0607612103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bejerano G, Lowe CB, Ahituv N, King B, Siepel A, Salama SR, Rubin EM, Kent WJ, Haussler D. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- 7.Xie X, Kamal M, Lander ES. Proc Natl Acad Sci USA. 2006;103:11659–11664. doi: 10.1073/pnas.0604768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishihara H, Smit AFA, Okada N. Genome Res. 2006;16:864–874. doi: 10.1101/gr.5255506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lowe CB, Bejerano G, Haussler D. Proc Natl Acad Sci USA. 2007 doi: 10.1073/pnas.0611223104. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mikkelsen TS, Wakefield MJ, Aken B, Amemiya CT, Chang JL, Duke S, Garber M, Gentles AJ, Goodstadt L, Heger A, et al. Nature. 2007;447:167–177. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- 11.van de Lagemaat LN, Medstrand P, Mager DL. Genome Biol. 2006;7:R86. doi: 10.1186/gb-2006-7-9-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Medstrand P, van de Lagemaat LN, Mager DL. Genome Res. 2002;12:1483–1495. doi: 10.1101/gr.388902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bannert N, Kurth R. Proc Natl Acad Sci USA. 2004;101:14572–14579. doi: 10.1073/pnas.0404838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gould SJ, Vrba ES. Paleobiology. 1982;8:4–15. [Google Scholar]
- 15.Levine AJ. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 16.Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 17.Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, et al. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- 18.Qian H, Wang T, Naumovski L, Lopez CD, Brachmann RK. Oncogene. 2002;21:7901–7911. doi: 10.1038/sj.onc.1205974. [DOI] [PubMed] [Google Scholar]
- 19.Hoh J, Jin S, Parrado T, Edington J, Levine AJ, Ott J. Proc Natl Acad Sci USA. 2002;99:8467–8472. doi: 10.1073/pnas.132268899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clamp M, Cuff J, Searle SM, Barton GJ. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 21.Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 22.Kaneshiro K, Tsutsumi S, Tsuji S, Shirahige K, Aburatani H. Genomics. 2007;89:178–188. doi: 10.1016/j.ygeno.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 23.Villesen P, Aagaard L, Wiuf C, Pedersen FS. Retrovirology. 2004;1:32. doi: 10.1186/1742-4690-1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodman M, Porter CA, Czelusniak J, Page SL, Schneider H, Shoshani J, Gunnell G, Groves CP. Mol Phylogenet Evol. 1998;9:585–598. doi: 10.1006/mpev.1998.0495. [DOI] [PubMed] [Google Scholar]
- 25.Johnson WE, Coffin JM. Proc Natl Acad Sci USA. 1999;96:10254–10260. doi: 10.1073/pnas.96.18.10254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jukes TH, Cantor C. In: Mammalian Protein Metabolism. Munro HN, editor. New York: Academic; 1969. pp. 21–132. [Google Scholar]
- 27.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 29.Horvath MM, Wang X, Resnick MA, Bell DA. PLoS Genet. 2007;3:e127. doi: 10.1371/journal.pgen.0030127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prabhakar S, Poulin F, Shoukry M, Afzal V, Rubin EM, Couronne O, Pennacchio LA. Genome Res. 2006;16:855–863. doi: 10.1101/gr.4717506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takei Y, Ishikawa S, Tokino T, Muto T, Nakamura Y. Genes Chromosomes Cancer. 1998;23:1–9. [PubMed] [Google Scholar]
- 32.Zhao H, Granberg F, Elfineh L, Pettersson U, Svensson C. J Virol. 2003;77:11006–11015. doi: 10.1128/JVI.77.20.11006-11015.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis BP, Green RE, Brenner SE. Proc Natl Acad Sci USA. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McClintock B. Cold Spring Harb Symp Quant Biol. 1956;21:197–216. doi: 10.1101/sqb.1956.021.01.017. [DOI] [PubMed] [Google Scholar]
- 35.Britten RJ, Davidson EH. Q Rev Biol. 1971;46:111–138. doi: 10.1086/406830. [DOI] [PubMed] [Google Scholar]
- 36.Medstrand P, van de Lagemaat LN, Dunn CA, Landry JR, Svenback D, Mager DL. Cytogenet Genome Res. 2005;110:342–352. doi: 10.1159/000084966. [DOI] [PubMed] [Google Scholar]
- 37.de Parseval N, Heidmann T. Cytogenet Genome Res. 2005;110:318–332. doi: 10.1159/000084964. [DOI] [PubMed] [Google Scholar]
- 38.Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, Haussler D, Miller W. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 40.Siepel AC, Pollard KS, Haussler D. In: Apostolico A, Guerra C, Istrail S, Pevzner P, Waterman M, editors. Proceedings of the 10th International Conference on Research in Computational Molecular Biology; Heidelberg: Springer; 2006. pp. 190–205. [Google Scholar]
- 41.Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, Diekhans M, Furey TS, Harte RA, Hsu F, et al. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tavazoie S, Hughes JD, Campbell MJ, Cho RJ, Church GM. Nat Genet. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- 44.Smit AF. Curr Opin Genet Dev. 1996;6:743–748. doi: 10.1016/s0959-437x(96)80030-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.