Abstract
Rop/Rac small GTPases are central to diverse developmental and cellular activities in plants, playing an especially important role in polar growth of pollen tubes. Although it is established that a class of plant-specific RopGEFs promotes the activity of Rop/Rac through the catalytic PRONE (Plant-specific Rop nucleotide exchanger) domain, not much is known about how RopGEF function is controlled to allow a spatiotemporally regulated Rop activity. To understand such a process in pollen, we performed functional analysis with a pollen-specific RopGEF, AtRopGEF12. Overexpression of AtRopGEF12 had minimal phenotypic effects, whereas overexpression of a C-terminally truncated version disturbed tube growth, suggesting that the C terminus was inhibitory to GEF function. In contrast to non-pollen-expressed RopGEFs, pollen-expressed RopGEFs have conserved C termini. A phospho-mimicking mutation at an invariant serine within the C terminus of AtRopGEF12 resulted in loss of the C-terminal inhibition, suggesting that phosphorylation regulates GEF activity in vivo. The PRONE domain of AtRopGEF12 (PRONE12) was not sufficient to induce isotropic tube growth. We used mbSUS to show that AtRopGEF12 interacts with an Arabidopsis pollen receptor kinase AtPRK2a through its C terminus, and BiFC to show that they interact in pollen tubes. Coexpression of AtRopGEF12 and AtPRK2a caused isotropic growth reminiscent of that seen upon overexpression of a constitutively active (CA) Rop. Coexpression of AtPRK2a with an N-terminally truncated AtRopGEF12 did not induce isotropic growth, indicating a positive role for the N-terminal domain. Our results suggest a mechanism by which the noncatalytic domains of pollen-specific/enriched RopGEFs regulate PRONE function, leading to polarized pollen tube growth.
Keywords: phosphorylation, pollen tube growth, receptor kinase
Sexual reproduction in flowering plants is achieved by pollination and fertilization. Unlike mammals, whose sperm are motile, plant sperm are carried to the embryo sac by the pollen tube (1). Pollen tubes are tip-growing cells whose polarity is maintained by diverse cellular activities, most prominently by actin dynamics and a tip-focused Ca2+ gradient (2). Plant-specific Rop small GTPases, also named Rac GTPases, are orthologs of yeast and mammalian Rho GTPases. Rops have been reported to regulate many developmental processes, such as root hair growth (3, 4), leaf epidermal cell morphogenesis (5, 6), auxin and ABA signaling (7, 8), tolerance to oxygen deprivation (9), and pathogen resistance (10). The role of Rops in pollen tube growth has been extensively studied. Overexpression of a CA-Rop1 resulted in isotropic growth (11, 12), whereas reduced Rop levels inhibited pollen tube growth (13).
In yeast and mammals, three classes of regulators control the activity of Rho GTPases: GTPase activating proteins (GAPs), guanine nucleotide dissociation inhibitors (GDIs) and guanine nucleotide exchange factors (GEFs) (14). GAPs act as an “off” switch, promoting the innate GTPase activity to generate GDP-bound, inactivated Rho. GDIs anchor Rho in the cytosol, where GTP is abundant, and may have other functions such as transporting Rho GTPases to target sites. Both GAPs and GDIs of Rop have been identified in plant genomes based on conserved domain sequences with those of other eukaryotes (15, 16). RopGAPs and RopGDIs were shown to regulate the growth of pollen tubes (17, 18) and root hairs (15, 19), as well as being involved in tolerance during oxygen deprivation (9).
GEFs stimulate the exchange of GDP to GTP to switch “on” the Rho GTPases. No homologs of the typical Dbl type RhoGEFs were found in plant genomes. Recently, a family of plant-specific RopGEFs was identified (20), which contain a highly conserved PRONE domain for Rop binding and GEF activity, and variable N- and C-terminal regions. When several Arabidopsis RopGEFs were transiently overexpressed in tobacco pollen, AtRopGEF1 had the most dramatic phenotype, i.e., ballooning tips, whereas others had less severe effects (21). Based on the in vitro catalytic activity of full-length and truncated AtRopGEF1, an autoinhibition conferred by the C-terminal variable region was proposed (21), although both full-length and C-terminally truncated RopGEF1 versions expressed in tobacco pollen showed similar ballooning tips. PRONE1 localized at the plasma membrane and its overexpression also induced isotropic growth (21), suggesting that PRONE1 was sufficient for membrane association and Rop activation.
We previously showed (22) that a pollen-specific RopGEF homolog from tomato, LeKPP, specifically interacts with the cytoplasmic domain of LePRK2, implicating RopGEFs as missing links between perceiving extracellular cues and initiating small G protein-regulated intracellular signaling cascades in pollen tube polar growth. Here we present in vivo data revealing a distinct mechanism regulating the activity of AtRopGEF12, a LeKPP homolog. We show that the overexpression of a C-terminally truncated but not of a full-length AtRopGEF12 affected the polarity of pollen tube growth, suggesting a C-terminal inhibition of GEF activity in vivo. Unlike the highly variable C termini of RopGEFs whose expression was hardly detectable in pollen, the C termini of pollen-specific/enriched RopGEFs are conserved. A phosphor-mimicking mutation at an invariant serine residue in AtRopGEF12 resulted in release of the C-terminal inhibition. Although highly similar to PRONE1, PRONE12 was not sufficient to induce isotropic growth when overexpressed and had a cytoplasmic localization. This result, together with sequence alignment and phylogenetic analyses of PRONE domains, suggests that PRONEs can be classified into two groups that may have distinctive binding affinities to Rops. The subcellular localization and activity of pollen-specific/enriched RopGEFs may require the function of PRK2, in that coexpression of AtRopGEF12 with AtPRK2a induced isotropic growth, indicative of constitutive Rop activity. Our results reveal a signal relay in which AtPRK2a at the plasma membrane recruits AtRopGEF12 through its C terminus to maintain polar Rop activity in the pollen tube.
Results and Discussion
Identification of Pollen-Specific/Enriched RopGEFs in Arabidopsis.
There are 14 RopGEFs in Arabidopsis. Previous reports (21–23) of their expression patterns were not consistent. To determine their expression profiles, we did RT-PCR analysis. In three biological replicate RT-PCR experiments, we were able to detect the expression of each AtRopGEF in at least one tissue type [see supporting information (SI) Fig. 7], which is in accord with transcriptome results (23, 24). Each tissue tested expressed at least two AtRopGEFs, suggesting a potential redundancy to assure system flexibility. Among the 14 AtRopGEFs, AtRopGEF8, 9, 11, 12, and 13 displayed pollen-specific or enriched expression, whereas the others were not or hardly expressed in pollen, supporting the pollen transcriptome data (23). Of the five pollen-specific/enriched AtRopGEFs, AtRopGEF12 is the closest homolog of LeKPP. Therefore, we investigated the function of AtRopGEF12 in pollen tube growth.
Overexpression of a C-Terminally Truncated AtRopGEF12 Disturbed Pollen Tube Growth.
The C terminus of LeKPP was sufficient to bind LePRKs in a yeast two-hybrid screen (22), implying a potential regulatory role for the C terminus. To test the role of the C-terminal region on GEF activity in vivo, we generated constructs expressing either full-length or a C-terminally truncated AtRopGEF12 (AtRopGEF12ΔC), each fused with enhanced yellow fluorescent protein (eYFP) at the N terminus and under the control of the pollen-specific LAT52 promoter (ProLAT52) (25). The constructs were delivered into tobacco pollen by particle bombardment (26). Expression of ProLAT52:eYFP served as the control.
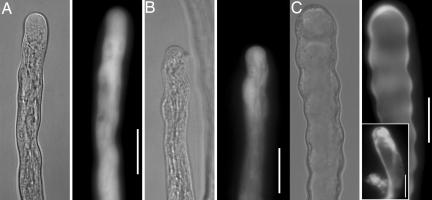
Overexpression of full-length AtRopGEF12 only increased pollen tube width slightly (Fig. 1B, and see SI Fig. 8) relative to the widths of pollen tubes transformed with eYFP (Fig. 1A, and see SI Fig. 8), as reported in ref. 21, whereas overexpression of AtRopGEF12ΔC (1–443) dramatically disturbed pollen tube morphology (Fig. 1C and see SI Fig. 8). Eighty percent of the tubes expressing AtRopGEF12ΔC had wavy margins, consistent with periodic stoppage and resumption of growth (Fig. 1C). Bifurcated tubes or additional membrane bulges were found at the shank of the remaining transformed pollen tubes (Fig. 1C Inset). Vacuoles occupied the very apical region in tubes expressing AtRopGEF12ΔC (Fig. 1C); in wild-type tubes, vacuoles are not apical. Multiple Arabidopsis transgenic lines expressing each construct were generated and these showed consistent and heritable pollen tube phenotypes similar to those seen in tobacco pollen (see SI Fig. 9). In our previous study (22), overexpression of LeKPP in tobacco disturbed pollen tube growth such that transformed tubes had wider, sometimes bulbous tips (21), whereas overexpression of its closest Arabidopsis homolog AtRopGEF12, had a milder although statistically significant effect on pollen tube width (Fig. 1 and SI Fig. 8). This difference may be due to different expression levels or to a species-specific difference. Overexpression of AtRopGEF12ΔC, but not the full length version, gave phenotypes reminiscent of overexpression of several Rops in pollen tubes (27), suggesting that the C-terminal truncation led to ectopic GEF activity.
Fig. 1.
C-terminally truncated AtRopGEF12 disturbed pollen tube growth. Bright-field images and fluorescent images are shown side-by-side. (A) A pollen tube overexpressing yGFP, as control. (B) A pollen tube overexpressing eYFP-AtRopGEF12. (C) A pollen tube overexpressing eYFP-AtRopGEF12ΔC. (Inset) Bifurcated phenotype. (Scale bars, 20 μm.)
Mutation at an Invariant Serine Residue within the C-Terminal Region of AtRopGEF12 Released the C-Terminal Inhibition in Vivo.
Overall sequence alignments of RopGEFs showed that the C-termini of RopGEFs were highly variable (20–22). However, alignment of the C-terminal domains of the five Arabidopsis pollen-specific/enriched RopGEFs and some RopGEFs from other higher plant species revealed previously unnoted conservation, highlighted by three potentially phosphorylatable residues (Fig. 2A). Our previous results (22) demonstrated that LeKPP extracted from pollen was phosphorylated and that its C-terminal domain bound to LePRKs. It was therefore tempting to postulate that the C-terminal inhibition would be released by phosphorylation. Phosphorylation release of GEF activity has been reported in mammalian RhoGEFs (14). Although the plant-specific RopGEFs have completely different catalytic domain sequences from mammalian RhoGEFs, similar regulatory mechanisms may exist.
Fig. 2.
An invariant serine in the C terminus of AtRopGEF12 is involved in the C-terminal inhibition of GEF activity in vivo. (A) Alignment of the C termini of the LeKPP clade RopGEFs. Three conserved phosphorylatable residues are highlighted with asterisks. Except for the Oryza sativa and Arabidopsis thaliana protein sequences, which were retrieved from TIGR (http://compbio.dfci.harvard.edu/tgi/tgipage.html), the C-terminal amino acid sequences of the five pollen-specific/enriched RopGEFs were used as queries for TBLASTN searches against the Expressed Sequence Tag (EST) database of National Center for Biotechnology Information (NCBI). Sequences can be found in GenBank under accession numbers AY730762 for LeKPP, CA819952 for Glycine max, BP031215 Lotus japonicus, EC937783 Vitis vinifera, DY535214 and DV533020 for Zea mays a and b, CO118031 for Gossypium raimondii, AU306277 for Zinnia elegans, CO052434 for Malus x domestica, CK757573 for Welwitschia mirabilis, BY858440 for Hordeum vulgare and CA294497 for Saccharum officinarum. Retrieved sequences were translated and aligned using Vector NTI (Invitrogen). (B) Pollen tube morphology and (C) average width upon overexpression of point-mutated AtRopGEF12. (Inset) S510D overexpression at 2 h. Asterisks indicate a significant difference from the yGFP control at the same data point (P < 0.05; t test). Double asterisks indicate a significant difference of S510D from other point-mutations (P < 0.05; t test). n ≈ 20–30 tubes. Error bars indicate standard deviation (SD). (Scale bars, 20 μm.)
To test whether the C-terminal conserved phosphorylatable residues were critical for C-terminal inhibition in vivo, we generated full-length AtRopGEF12 with point mutations at the three conserved phosphorylatable residues, T458, S500, and S510 (numbered as in AtRopGEF12), to either a phosphor-mimicking aspartic acid (D) or a nonphosphorylatable alanine (A) using site-directed mutagenesis. Fluorescent protein fusions of these proteins were transiently expressed in tobacco pollen.
Overexpression phenotypes of AtRopGEF12 with mutations at either T458 or S500 did not differ from those of wild-type AtRopGEF12 (Fig. 2 B and C), in that transformed tubes were slightly wavy and wider (Fig. 2B) than nontransformed tubes growing under the same conditions. The fluorescent signal was detected in the cytoplasm (Fig. 2B). Tubes transformed with AtRopGEF12-S510A were also similar to those overexpressing wild-type AtRopGEF12 except that vacuoles were often seen close to the subapical region (Fig. 2 B and C). In contrast, at early stages overexpression of AtRopGEF12-S510D resulted in wavy tubes, and the fluorescent signal was detected both in the cytoplasm and at the apical plasma membrane (Fig. 2B Inset). After a 4-h culture, all transformed tubes had vacuoles close to the tube apex and displayed significant tip swelling (Fig. 2 B and C). These results suggested that the invariant serine within the C terminus of the LeKPP clade RopGEFs was critical for the C-terminal inhibition, presumably through phosphorylation.
PRONEs Have at Least Two Functional Subgroups.
It has been shown (21) that PRONE1 was necessary and sufficient for membrane anchoring, and that its overexpression in pollen caused isotropic tube growth, indicative of constitutive Rop activity. Because the PRONE domains share high sequence identity and interact with the structurally conserved small GTPase Rop, we predicted that a similar isotropic growth would be induced by overexpression of PRONE12.
Surprisingly, overexpression of PRONE12 had only a mild phenotypic effect on pollen tube growth, similar to that of AtRopGEF12 overexpression (Fig. 3A, see SI Fig. 8), i.e., pollen tubes had vacuoles close to the subapical region and were slightly wavy (Fig. 3A). The fluorescent signal was mostly in the cytoplasm (Fig. 3A). Despite the fact that they share 50% identity and 74% similarity, the different overexpression phenotypes with PRONE1 and PRONE12 suggest that the PRONE domains have at least two distinctive functional subgroups. Phylogenetic analysis with PRONE domains obtained from diverse plant species also supports the existence of at least two PRONE types. As for the C termini, the PRONE domains of all five Arabidopsis pollen-specific/enriched PRONEs group within one clade (Fig. 3B). The difference between the two subgroups may lie in a few but distinctive amino acid changes (see SI Fig. 10), most prominently within the β-arm and subdomain 2 (28). A recent structural analysis of the PRONE8-Rop4-GDP complex revealed that Rop is captured by the β-arm of one PRONE8 and the main body of the other PRONE8 (28). It is likely that PRONE1 and PRONE12 have different binding affinities to Rop, with PRONE1 strong enough to anchor PRONE1 peripherally to the membrane, whereas PRONE12 is not.
Fig. 3.
PRONE domains have at least two distinct subgroups. (A) Overexpression of PRONE12 had a mild effect on pollen tube growth. (Scale bars, 20 μm.) (B) Phylogenetic analysis of PRONE domains from several plant species.
Coexpression of AtPRK2a with AtRopGEF12 Resulted in Isotropic Tube Growth.
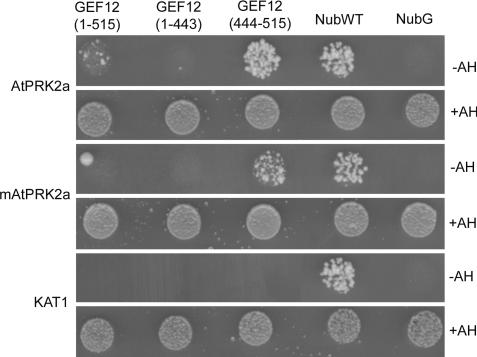
PRONE1 was membrane-localized, and its overexpression led to constitutive Rop activity (21), suggesting that its binding to Rop was sufficient for the membrane anchoring of PRONE1. In contrast, our results indicated that the noncatalytic domains of RopGEF12 may be required for GEF activity in vivo. We have shown that the C-terminal region of LeKPP binds specifically to the cytoplasmic domain of LePRK2 (22). Recently we characterized an Arabidopsis homolog of LePRK2, AtPRK2a (At2g07040). We first verified the physical interaction between AtPRK2a and AtRopGEF12 using the mating-based split-ubiquitin system (mbSUS) in yeast (29). We generated baits expressing either full-length AtPRK2a or a presumed kinase-inactive (K366R) mutant (mAtPRK2a); equivalent mutations abolished autophosphorylation of the LePRKs (30). Prey constructs expressing AtRopGEF12, AtRopGEF12ΔC, or the C terminus of AtRopGEF12 (AtRopGEF12-C) were generated. The C-terminal domain of AtRopGEF12 was necessary and sufficient for interaction with AtPRK2a (Fig. 4). The presumptive kinase-inactive mAtPRK2a still interacted with AtRopGEF12 through its C-terminal domain, albeit with a lower binding affinity than that of wild-type AtPRK2a (Fig. 4). A potassium channel protein (29) used as bait did not interact with AtRopGEF12 (Fig. 4).
Fig. 4.
The C-terminal domain of AtRopGEF12 is necessary and sufficient for interaction with AtPRK2a. Diploid yeast cells expressing a Cub-fused bait protein, AtPRK2a, mAtPRK2a or KAT1, and one of the three NubG-fused preys grew well on nonselective minimal plates supplemented with adenine and histidine (+AH), but only cells expressing AtPRK2a or mAtPRK2a with full-length AtRopGEF12 or AtRopGEF12 (444–515) grew on selection plates without adenine and histidine (−AH). Wild-type Nub (NubWT) and mutant Nub (NubG) served as positive and negative controls, respectively.
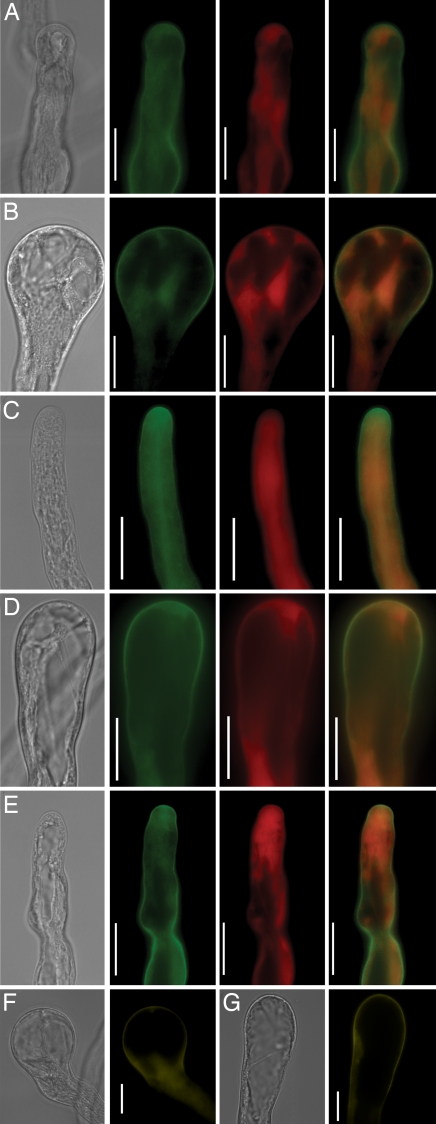
To test whether AtPRK2a binding could release the C-terminal inhibition, we co-expressed AtPRK2a-eCFP and eYFP-AtRopGEF12 in tobacco pollen. Co-expression of AtPRK2a-eCFP and eYFP, and of eCFP and eYFP-AtRopGEF12 were used as controls. Co-expression of AtPRK2a-eCFP and eYFP resulted in wider tubes (Fig. 5A, see SI Fig. 11). Coexpression of eCFP and eYFP-AtRopGEF12 resembled overexpression of AtRopGEF12 alone (data not shown). In contrast, coexpression of AtPRK2a-eCFP and eYFP-AtRopGEF12 resulted in isotropic growth (Fig. 5B, see SI Fig. 11), reminiscent of pollen tubes overexpressing a CA-Rop1 (12). AtRopGEF12 colocalized with AtPRK2a at the apical plasma membrane (Fig. 5A). We also tested whether the presumptive kinase inactive AtPRK2a (mAtPRK2a) could cause such an effect. Coexpression of mAtPRK2a-eCFP with eYFP did not affect pollen tube growth significantly (Fig. 5C, see SI Fig. 11), suggesting that kinase activity is necessary for the tip widening phenotype seen upon AtPRK2a overexpression (Fig. 5A). Coexpression of mAtPRK2a and AtRopGEF12 still induced tip swelling (Fig. 5D, see SI Fig. 11), although to a lesser extent (compare Fig. 5B).
Fig. 5.
Isotropic growth is induced by coexpression of AtPRK2a or mAtPRK2a with AtRopGEF12. (A–E) From left to right, the first panel was taken under bright field, the second panel was taken under the CFP channel (false-colored green), the third panel was taken under the YFP channel (false-colored red), and the last panel is the merged image. Coexpression of AtPRK2a-eCFP and eYFP (A); AtPRK2a-eCFP and eYFP-AtRopGEF12 (B); mAtPRK2a-eCFP and eYFP (C); mAtPRK2a-eCFP and eYFP-AtRopGEF12 (D); and AtPRK2a-eCFP and eYFP-AtRopGEF12ΔN (E). (F and G). Bimolecular fluorescence complementation. Coexpression of YN-fused AtRopGEF12 and YC-fused AtPRK2a (F) or YC-fused mAtPRK2a (G) For F and G, bright-field images and YFP channel images are shown side by side. (Scale bars, 20 μm.)
Although the eCFP and eYFP signals colocalized at the plasma membrane, it was not proven that they directly interact in planta. Therefore, we used the bimolecular fluorescence complementation system (BiFC) (31) to demonstrate a direct interaction between AtPRK2a and AtRopGEF12 in transiently transformed tobacco pollen tubes. AtRopGEF12 interacted with both AtPRK2a (Fig. 5F) and mAtPRK2a (Fig. 5G) at the apical plasma membrane. Together, these results suggest that AtPRK2a may act as a scaffolding protein, recruiting AtRopGEF12 to the plasma membrane by binding to its C-terminal domain. The reduced binding affinity between mAtPRK2a and the C terminus of AtRopGEF12 may explain the less severe isotropic growth.
The N-Terminal Region of AtRopGEF12 Is Required for the Isotropic Growth Induced by Coexpression of AtPRK2a and AtRopGEF12.
That overexpression of the PRONE12 domain resulted in a milder phenotype than overexpression of AtRopGEF12ΔC suggested that the N-terminal region of AtRopGEF12 plays a positive role. We observed no detectable phenotypic effect by overexpressing AtRopGEF12-N (aa1–90), which was cytoplasmic (data not shown). This result indicates that the positive role the N-terminal region plays may not require trans factor binding because such a scenario would predict a dominant negative phenotype upon overexpressing the N-terminal domain. We also coexpressed an eYFP-fused N-terminally truncated AtRopGEF12 (eYFP-AtRopGEF12ΔN-91-516) with AtPRK2a-eCFP in transient assays. Unlike the isotropic growth induced by coexpression of eYFP-AtRopGEF12 and AtPRK2a-eCFP (Fig. 5B), pollen tube morphology upon coexpression of eYFP-AtRopGEF12ΔN and AtPRK2a-eCFP (Fig. 5E, see SI Fig. 11) did not significantly differ from that of eYFP and AtPRK2a-eCFP coexpression (Fig. 5A). This result supports a positive role for the N-terminal region of RopGEF12 in in vivo GEF activity.
Pollen-Specific/Enriched RopGEFs as the Missing Link Between Pollen Receptor Kinases and Rop-Mediated Intracellular Responses.
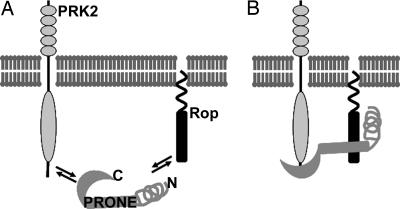
As depicted in Fig. 6, we propose a model to explain the regulatory mechanism of pollen-specific/enriched RopGEFs. The activity of pollen-specific/enriched RopGEFs is inhibited by their C-termini to prevent ectopic Rop activity outside of the apical region. Although AtPRK2a is plasma membrane-localized along the pollen tube (30, 32), Rop localization is restricted mostly to the apical plasma membrane of the pollen tube (11, 33) except in the cytoplasm, where Rop is probably sequestered by RopGDIs (17). Only at the apical plasma membrane, where both PRK2 and Rop are present, can the C termini of RopGEFs bind to the cytoplasmic domain of PRK2 to free the PRONE domain that can then be accessed by Rop. Phosphorylation at the invariant serine of RopGEFs is involved in this process. PRK2 may be the kinase that catalyzes phosphorylation at the invariant serine but we cannot exclude the involvement of other kinases. The N-terminal variable region may stabilize the complex by conformational effect. The binding between PRONE and Rop, together with the binding between PRK2 and the C terminus of RopGEF, contribute to the exchange of GDP to GTP, leading to Rop activation and downstream cellular responses.
Fig. 6.
Model depicting how localization and activity of pollen-selective RopGEFs require the presence of PRK2 and Rop. (A) Pollen-specific/enriched RopGEFs are inhibited by their C-terminal domain. In the absence of PRK2, the PRONE domain cannot access Rop. (B) When PRK2 and Rop are in close proximity, the C termini of pollen-specific/enriched RopGEFs interact with PRK2, exposing PRONE that is then accessible to Rop. The N-terminal variable region might stabilize the PRONE-Rop interaction through a conformational effect.
Several candidate ligands have been identified for LePRKs, such as LeLAT52, LeSHY, LeSTIG, etc. (34). Pollen tube growth was reduced by down-regulation of LeLAT52 (35) or the Petunia homolog of LeSHY (36), whereas pollen tube growth was promoted by addition of LeSTIG (37). A recent study by Wengier et al. (38) showed that an unknown component from tomato stylar extract dissociates a LePRK complex, leading to the hypothesis that the LePRK complex is inhibitory to pollen germination, whereas their dissociation releases such inhibition. In relation to our model, it is likely that a female-mediated PRK complex dissociation renders PRK2 available for RopGEF binding. Thus, Rop activation promoted by PRK-mediated GEF activity not only assures the maintenance of growth polarity over a relatively long distance, but can also allow rapid reorientation upon extracellular stimuli.
Materials and Methods
Plant Growth and Transformation.
Arabidopsis plants were grown in a 4:1:1 mix of Fafard 4P/perlite/pvermiculite under an 18-h-light/6-h-dark cycle at 21°C. To facilitate phenotypic analysis, the mutant quartet1-2 in the Col-0 ecotype (39) was used as wild type for stable transformation using the floral dipping method (40).
RNA Extraction and RT-PCR.
Total RNA from diverse tissues of ecotype Columbia (Col-0) was isolated using an RNeasy Plant miniprep kit (Qiagen) according to manufacturer's instructions. Oligo dT-primed cDNA was synthesized using SuperScript III Reverse Transcriptase with on-column DNase-treatment as recommended by the manufacturer (Invitrogen). Gene-specific RT-PCR primers were as described (22), but PCR was performed at an annealing temperature of 60°C, rather than at 54°C. Amplification of Arabidopsis ACTIN2 was used as the internal control.
DNA Manipulation.
All constructs except those used in mbSUS were generated using the Gateway system (Invitrogen). Primers are listed in SI Table 1. Entry vectors in pENTRY/SD/D-TOPO for the full-length and truncations of AtRopGEF12 were generated by RT-PCR from pollen cDNA using the combined primer pairs: Z177 and Z178 for AtRopGEF12, Z177 and Z180 for AtRopGEF12ΔC, Z179 and Z178 for AtRopGEF12-C, Z347 and Z178 for AtRopGEF12ΔN, Z177 and Z346 for AtRopGEF12-N, and Z352 and Z180 for PRONE12. To generate pollen-specific fluorescent protein fusion constructs for both transient and stable expression, LR reactions were conducted using LR Clonase II (Invitrogen) with the corresponding entry vectors and pollen-specific destination vectors (Y.Z. and S.M, unpublished data) adapted from pB7WG or pB7GW series (41).
Mutations at the C-terminal region of AtRopGEF12 were generated using the Quick change site-directed mutagenesis kit (Stratagene, La Jolla, CA) with the following primer pairs: Z284A and Z284B for T458D, Z285A and Z285B for T458A, Z260A and Z260B for S500D, Z261A and Z261B for S500A, Z241A and Z241B for S510D, and Z242A and Z242B for S510A (SI Table 1). A K336R mutation at AtPRK2a was generated similarly using the primer pair Z281A and Z281B. The wild-type entry vectors were used as the template for mutagenesis. Fluorescent protein fusion expression vectors were generated similarly using LR reactions.
Constructs for the mbSUS were generated by in vivo recombination (29) using the following primer pairs: Z392 and Z393 for AtPRK2a and mAtPRK2a, Z394 and Z395 for AtRopGEF12, Z394 and Z397 for AtRopGEF12ΔC and Z395 and Z396 for AtRopGEF12-C. The pollen-specific BiFC constructs were generated using Gateway-compatible destination vectors (Y.Z. and S.M., unpublished data) based on a previously reported 35S promoter-driven BiFC system (31).
All PCR amplifications used Phusion hot start high-fidelity DNA polymerase with the annealing temperature and extension time recommended by the manufacturer (Finnzyme, Ipswich, MA) and were sequenced using an ABI 3300 sequencer. Sequences were analyzed using Vector NTI (Invitrogen). QIAquick PCR purification kit, QIAprep Spin miniprep kit, and Qiagen TIP-100 kit (Qiagen) were used for PCR product recovery, DNA minipreps, and DNA midipreps, respectively.
Sequence Analysis and Phylogeny.
Sequence alignment was performed with ClustalW (www.ebi.ac.uk/clustalw), and the phylogeny was built by neighbor joining using Molecular Evolutionary Genetics Analysis (MEGA3.1) (42), calculated by using bootstrap analysis (replicates = 500).
Pollen Germination and Transient Expression.
Arabidopsis in vitro pollen tube growth was conducted at 22.5°C, as described (43). All Arabidopsis pollen tube growth experiments were repeated at least three times. Transient expression assays in tobacco pollen were conducted as described (22, 26). Images were captured from 2–8 h after germination. Each construct was tested in two independent bombardments, and except where noted, 100–120 tubes were scored. Pollen tube width was measured as the diameter of the widest region in the apical or subapical region, using the measuring function of Axiovision software. Twenty to 30 transient pollen tubes were measured to determine the average tube width, and a t test was performed to determine whether the difference was significant (P < 0.05, t test).
Microscopy.
Microscopic imaging was performed using an inverted Axiophot microscope (Zeiss) with either bright-field or epifluorescence optics. Images were captured using a Spot digital camera (Diagnostic Instruments, Sterling Heights, MI), exported using AxioVision (Zeiss), and processed using Photoshop 7.0 (Adobe).
Supplementary Material
Acknowledgments
We appreciate the help of Leonor Boavida with pollen germination, and thank Waichoi Shek and Colleen Lau for technical support. Vectors and yeast strains used in mbSUS and vectors used in BiFC were obtained from the ABRC (www.biosci.ohio-state.edu/∼plantbio/Facilities/abrc/abrchome.htm). We thank Shaul Yalovsky, Jorge Muschietti, and Weihua Tang for comments on the manuscript. This work was supported by U.S. Department of Agriculture Current Research Information System 5335-21000-030-00D.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705874104/DC1.
References
- 1.Taylor LP, Hepler PK. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:461–491. doi: 10.1146/annurev.arplant.48.1.461. [DOI] [PubMed] [Google Scholar]
- 2.Hepler PK, Vidali L, Cheung AY. Annu Rev Cell Dev Biol. 2001;17:159–187. doi: 10.1146/annurev.cellbio.17.1.159. [DOI] [PubMed] [Google Scholar]
- 3.Jones MA, Shen JJ, Fu Y, Li H, Yang Z, Grierson CS. Plant Cell. 2002;14:763–776. doi: 10.1105/tpc.010359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Molendijk AJ, Bischoff F, Rajendrakumar CS, Friml J, Braun M, Gilroy S, Palme K. EMBO J. 2001;20:2779–2788. doi: 10.1093/emboj/20.11.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloch D, Lavy M, Efrat Y, Efroni I, Bracha-Drori K, Abu-Abied M, Sadot E, Yalovsky S. Mol Biol Cell. 2005;16:1913–1927. doi: 10.1091/mbc.E04-07-0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gifford ML, Robertson FC, Soares DC, Ingram GC. Plant Cell. 2005;17:1154–1166. doi: 10.1105/tpc.104.029975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tao LZ, Cheung AY, Wu HM. Plant Cell. 2002;14:2745–2760. doi: 10.1105/tpc.006320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng ZL, Nafisi M, Tam A, Li H, Crowell DN, Chary SN, Schroeder JI, Shen J, Yang Z. Plant Cell. 2002;14:2787–2797. doi: 10.1105/tpc.005611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baxter-Burrell A, Yang Z, Springer PS, Bailey-Serres J. Science. 2002;296:2026–2028. doi: 10.1126/science.1071505. [DOI] [PubMed] [Google Scholar]
- 10.Fujiwara M, Umemura K, Kawasaki T, Shimamoto K. Plant Physiol. 2006;140:734–745. doi: 10.1104/pp.105.068395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kost B, Lemichez E, Spielhofer P, Hong Y, Tolias K, Carpenter C, Chua NH. J Cell Biol. 1999;145:317–330. doi: 10.1083/jcb.145.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Lin Y, Heath RM, Zhu MX, Yang Z. Plant Cell. 1999;11:1731–1742. doi: 10.1105/tpc.11.9.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin Y, Yang Z. Plant Cell. 1997;9:1647–1659. doi: 10.1105/tpc.9.9.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt A, Hall A. Genes Dev. 2002;16:1587–1609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- 15.Bischoff F, Vahlkamp L, Molendijk A, Palme K. Plant Mol Biol. 2000;42:515–530. doi: 10.1023/a:1006341210147. [DOI] [PubMed] [Google Scholar]
- 16.Wu G, Li H, Yang Z. Plant Physiol. 2000;124:1625–1636. doi: 10.1104/pp.124.4.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klahre U, Becker C, Schmitt AC, Kost B. Plant J. 2006;46:1018–1031. doi: 10.1111/j.1365-313X.2006.02757.x. [DOI] [PubMed] [Google Scholar]
- 18.Klahre U, Kost B. Plant Cell. 2006;18:3033–3046. doi: 10.1105/tpc.106.045336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carol RJ, Takeda S, Linstead P, Durrant MC, Kakesova H, Derbyshire P, Drea S, Zarsky V, Dolan L. Nature. 2005;438:1013–1016. doi: 10.1038/nature04198. [DOI] [PubMed] [Google Scholar]
- 20.Berken A, Thomas C, Wittinghofer A. Nature. 2005;436:1176–1180. doi: 10.1038/nature03883. [DOI] [PubMed] [Google Scholar]
- 21.Gu Y, Li S, Lord EM, Yang Z. Plant Cell. 2006;18:366–381. doi: 10.1105/tpc.105.036434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaothien P, Ok SH, Shuai B, Wengier D, Cotter R, Kelley D, Kiriakopolos S, Muschietti J, McCormick S. Plant J. 2005;42:492–503. doi: 10.1111/j.1365-313X.2005.02388.x. [DOI] [PubMed] [Google Scholar]
- 23.Pina C, Pinto F, Feijo JA, Becker JD. Plant Physiol. 2005;138:744–756. doi: 10.1104/pp.104.057935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Twell D, Yamaguchi J, McCormick S. Development (Cambridge, UK) 1990;109:705–713. doi: 10.1242/dev.109.3.705. [DOI] [PubMed] [Google Scholar]
- 26.Twell D, Klein TM, Fromm ME, McCormick S. Plant Physiol. 1989;91:1270–1274. doi: 10.1104/pp.91.4.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheung AY, Chen CY, Tao LZ, Andreyeva T, Twell D, Wu HM. J Exp Bot. 2003;54:73–81. doi: 10.1093/jxb/erg044. [DOI] [PubMed] [Google Scholar]
- 28.Thomas C, Fricke I, Scrima A, Berken A, Wittinghofer A. Mol Cell. 2007;25:141–149. doi: 10.1016/j.molcel.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 29.Obrdlik P, El-Bakkoury M, Hamacher T, Cappellaro C, Vilarino C, Fleischer C, Ellerbrok H, Kamuzinzi R, Ledent V, Blaudez D, et al. Proc Natl Acad Sci USA. 2004;101:12242–12247. doi: 10.1073/pnas.0404467101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muschietti J, Eyal Y, McCormick S. Plant Cell. 1998;10:319–330. doi: 10.1105/tpc.10.3.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bracha-Drori K, Shichrur K, Katz A, Oliva M, Angelovici R, Yalovsky S, Ohad N. Plant J. 2004;40:419–427. doi: 10.1111/j.1365-313X.2004.02206.x. [DOI] [PubMed] [Google Scholar]
- 32.Kim HU, Cotter R, Johnson S, Senda M, Dodds P, Kulikauska R, Tang W, Ezcura I, Herzmark P, McCormick S. Plant Mol Biol. 2002;50:1–16. doi: 10.1023/a:1016077014583. [DOI] [PubMed] [Google Scholar]
- 33.Lin Y, Wang Y, Zhu JK, Yang Z. Plant Cell. 1996;8:293–303. doi: 10.1105/tpc.8.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang W, Ezcurra I, Muschietti J, McCormick S. Plant Cell. 2002;14:2277–2287. doi: 10.1105/tpc.003103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muschietti J, Dircks L, Vancanneyt G, McCormick S. Plant J. 1994;6:321–338. doi: 10.1046/j.1365-313x.1994.06030321.x. [DOI] [PubMed] [Google Scholar]
- 36.Guyon V, Tang WH, Monti MM, Raiola A, Lorenzo GD, McCormick S, Taylor LP. Plant J. 2004;39:643–654. doi: 10.1111/j.1365-313X.2004.02162.x. [DOI] [PubMed] [Google Scholar]
- 37.Tang W, Kelley D, Ezcurra I, Cotter R, McCormick S. Plant J. 2004;39:343–353. doi: 10.1111/j.1365-313X.2004.02139.x. [DOI] [PubMed] [Google Scholar]
- 38.Wengier D, Valsecchi I, Cabanas ML, Tang WH, McCormick S, Muschietti J. Proc Natl Acad Sci USA. 2003;100:6860–6865. doi: 10.1073/pnas.0631728100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Francis KE, Lam SY, Copenhaver GP. Plant Physiol. 2006;142:1004–1013. doi: 10.1104/pp.106.085274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clough SJ, Bent AF. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 41.Karimi M, Inze D, Depicker A. Trends Plants Sci. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- 42.Kumar S, Tamura K, Nei M. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 43.Boavida LC, McCormick S. Plant J. 2007 doi: 10.1111/j.1365–313x.2007.03248.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.