Abstract
Members of the nitrilase 4 (NIT4) family of higher plants catalyze the conversion of β-cyanoalanine to aspartic acid and asparagine, a key step in cyanide detoxification. Grasses (Poaceae) possess two different NIT4 homologs (NIT4A and NIT4B), but none of the recombinant Poaceae enzymes analyzed showed activity with β-cyanoalanine, whereas protein extracts of the same plants clearly posses this activity. Sorghum bicolor contains three NIT4 isoforms SbNIT4A, SbNIT4B1, and SbNIT4B2. Individually, each isoform does not possess enzymatic activity whereas the heteromeric complexes SbNIT4A/B1 and SbNIT4A/B2 hydrolyze β-cyanoalanine with high activity. In addition, the SbNIT4A/B2 complex accepts additional substrates, the best being 4-hydroxyphenylacetonitrile. Corresponding NIT4A and NIT4B isoforms from other Poaceae species can functionally complement the sorghum isoforms in these complexes. Site-specific mutagenesis of the active site cysteine residue demonstrates that hydrolysis of β-cyanoalanine is catalyzed by the NIT4A isoform in both complexes whereas hydrolysis of 4-hydroxyphenylacetonitrile occurs at the NIT4B2 isoform. 4-Hydroxyphenylacetonitrile was shown to be an in vitro breakdown product of the cyanogenic glycoside dhurrin, a main constituent in S. bicolor. The results indicate that the SbNIT4A/B2 heterocomplex plays a key role in an endogenous turnover of dhurrin proceeding via 4-hydroxyphenylacetonitrile and thereby avoiding release of toxic hydrogen cyanide. The operation of this pathway would enable plants to use cyanogenic glycosides as transportable and remobilizable nitrogenous storage compounds. Through combinatorial biochemistry and neofunctionalizations, the small family of nitrilases has gained diverse biological functions in nitrile metabolism.
Keywords: 4-hydroxyphenylacetonitrile, β-cyanoalanine, cyanogenic glycosides, dhurrin
The toxic compound cyanide is produced in all higher plants during biosynthesis of the plant hormone ethylene (1, 2). In addition, many plants contain cyanogenic glycosides from which cyanide may be released in quite large amounts (for review, see refs. 3–5). Detoxification of cyanide in plants involves a reaction between cyanide and cysteine to form β-cyanoalanine and subsequent conversion of β-cyanoalanine into ammonia, asparagine and aspartic acid. These reactions are catalyzed by the enzymes β-cyanoalanine synthase and β-cyanoalanine hydratase (6–8) (Fig. 1). β-Cyanoalanine hydratase was recently demonstrated to be a nitrilase of the NIT4 family displaying a particular high nitrile-hydratase activity, thus converting β-cyanoalanine to asparagine and aspartic acid in ratios ranging from ≈1:1 up to ≈4:1 (9, 10). Homologs of NIT4 are known from Brassicaceae (11), Fabaceae (9), Solanaceae (12, 13), and Poaceae (14), and EST data provide evidence for a ubiquitous distribution of NIT4 genes in higher plants. In addition to NIT4 from Arabidopsis thaliana, the NIT4 homologs of Nicotiana tabacum and Lupinus angustifolius have been shown to be functional β-cyanoalanine-metabolizing enzymes (9, 10). In contrast, neither of the NIT4 homologs of Zea mays showed β-cyanoalanine-hydrolyzing activity in vitro (14) although maize possesses the ability to incorporate radioactively labeled hydrogen cyanide into asparagine (15). In this study, we report that NIT4 activity in Poaceae is entirely dependent on the formation of heterocomplexes consisting of two different NIT4 orthologs. Homolog composition determines whether the complex is able to metabolize β-cyanoalanine or additional substrates. The highly cyanogenic plant Sorghum bicolor has evolved a new NIT4 homolog, which, as part of a new NIT4 heterocomplex, may be involved in the endogenous turnover of the cyanogenic glucoside dhurrin without concomitant release of hydrogen cyanide.
Fig. 1.
Cyanide detoxification pathway in higher plants.
Results
NIT4 Homologs in Poaceae.
The rice genome contains two nitrilase genes and analyses of EST sequence data document that members of the Poaceae in general possess two or three nitrilase homologs with high sequence similarity to NIT4 of Arabidopsis thaliana (71–75% identity at amino acid level). Phylogenetic analyses using NIT4 sequences from Hordeum vulgare, Oryza sativa, Sorghum bicolor, Saccharum officinarum, Triticum aestivum, and Zea mays revealed that the two NIT4 homologs found in each of these plants (designated NIT4A and NIT4B) have arisen by an early NIT4 gene duplication, most likely as a result of a paleopolyploidization event during evolution of the Poaceae family (16) (Fig. 2). The median value of the synonymous substitutions per synonymous sites (Ks) between the six NIT4A/NIT4B sequence pairs was 0.90 and using an average synonymous substitution rate of 6.1–6.5 per 109 years (17, 18), the NIT4 gene-duplication event could be dated to 69–74 million years ago (MYA). This dating matches the 70 MYA calculated for the polyploidization event (16). A subsequent gene duplication of the NIT4B gene has occurred in the Z. mays/S. bicolor/S. officinarum lineage. The branching pattern of the phylogenetic tree suggests that this second gene duplication took place before the radiation of Z. mays and S. bicolor/S. officinarum [≈12–16.5 MYA (19, 20)], which was supported by a Ks calculation dating this second gene duplication to have taken place 34–37 MYA. Accordingly, S. bicolor and S. officinarum both possess two orthologous NIT4B genes designated NIT4B1 and NIT4B2. Analyses of EST and available genome data provided no evidence for the occurrence of a NIT4B2 gene in maize indicating that this gene might subsequently have been lost in this species.
Fig. 2.
Phylogenetic analysis of Poaceae NIT4 homologs. Shown is the 50% consensus tree of a Bayesian analysis. Clade credibilities <1.00 are indicated. At, Arabidopsis thaliana; Hv, Hordeum vulgare; La, Lupinus angustifolius; Nt, Nicotiana tabacum; Os, Oryza sativa; Pt, Populus trichocarpa; Sb, Sorghum bicolor; So, Saccharum officinarum; Ta, Triticum aestivum; Zm, Zea mays.
Recombinant NIT4 Enzymes of Poaceae Do Not Hydrolyze β-Cyanoalanine.
Nitrilases of S. bicolor (SbNIT4A, B1, and B2), O. sativa (OsNIT4A), and H. vulgare (HvNIT4B) were expressed as C-terminal (His)6-tagged proteins in Escherichia coli and the purified enzymes were used for activity assays. All tested enzymes were homologs of the NIT4 enzymes from A. thaliana, N. tabacum, and L. angustifolius, which hydrolyze β-cyanoalanine (9, 10), but none of the enzymes from Poaceae were able to convert this substrate (Fig. 3 A and C). A similar observation had previously been made for two NIT4 homologs of Z. mays (14). These data prompted an investigation of the ability of the three NIT4 homologs of sorghum to metabolize other putative substrates (Table 1). SbNIT4A exhibited a weak enzymatic activity toward some nitriles, with the highest conversion rates being obtained with 4-hydroxyphenylacetonitrile and phenylacetonitrile but no activity [<0.1 nkat (mg protein)−1] toward β-cyanoalanine, benzonitrile, 4-hydroxybenzonitrile, butyronitrile, and 4-pentenenitrile. SbNIT4B1 and B2 showed no activity [<0.1 nkat (mg protein)−1] toward any of the tested compounds. The specific activities of SbNIT4A listed in Table 1 are in the same range as those reported for the Z. mays NIT2 (here designated ZmNIT4A), which is an ortholog of SbNIT4A (Fig. 2) (14). However, these activities are at least one to two orders of magnitude lower than the β-cyanoalanine-metabolizing activities reported for the NIT4 homologs of A. thaliana, N. tabacum (10) and L. angustifolius (9).
Fig. 3.
β-Cyanoalanine [Ala(CN)] hydrolyzing activity of different NIT4 homologs of grasses and combinations of these enzymes. The enzymes were incubated with β-cyanoalanine until full conversion of the substrate in the active samples was achieved. Aliquots of the assays were then separated by TLC. (A) NIT4 isoforms of S. bicolor. (B) Wild-type and mutated NIT4 isoforms of S. bicolor. (C) Combinations of NIT4 isoforms from S. bicolor, H. vulgare, and O. sativa. Asn, asparagine; Asp, aspartic acid; HvB, HvNIT4B; OsA, OsNIT4A. SbA, B1, and B2: SbNIT4A, B1, and B2, respectively. Sba, b1, and b2: SbNIT4A C195A, SbNIT4B1 C191A, and SbNIT4B2 C194A, respectively.
Table 1.
Substrate specificity of the recombinant SbNIT4A enzyme
| Substrate | Specific activity, nkat (mg protein)−1 |
|---|---|
| Pentanenitrile | 0.32 ± 0.12 |
| Indole-3-acetonitrile | 0.55 ± 0.06 |
| 4-Hydroxymandelonitrile* | 0.57 ± 0.23 |
| 5-Hexenenitrile | 0.70 ± 0.07 |
| 3-Butenenitrile | 0.76 ± 0.07 |
| 6-Heptenenitrile | 1.03 ± 0.06 |
| 3-Phenylpropionitrile | 1.32 ± 0.36 |
| 4-Phenylbutyronitrile | 2.50 ± 1.08 |
| 2-Phenylacetonitrile | 15.68 ± 1.60 |
| 4-Hydroxyphenylacetonitrile | 27.13 ± 5.68 |
Shown are the means ± SD from three independent experiments performed in triplicate.
*Measured at pH 6.0 because the compound is labile at pH 8.0.
Isolation of Nitrilases from Extracts of Sorghum bicolor.
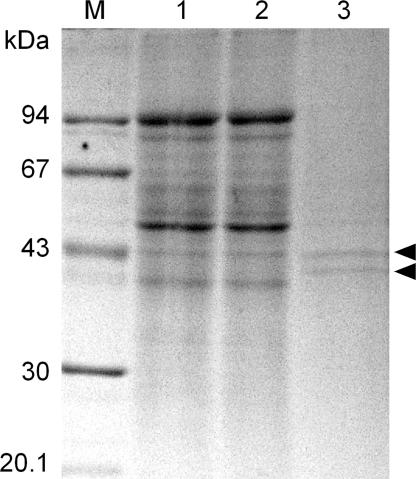
In contrast to the results obtained with the recombinant NIT4 enzymes, crude protein extracts from seedlings of S. bicolor were able to metabolize β-cyanoalanine (8). The reasons for this enzymatic activity could be (i) presence of other β-cyanoalanine metabolizing enzymes in the plant extracts, either additional NIT4 homologs or entirely different enzymes, or (ii) a specific need of Poaceae NIT4 homologs to be activated by posttranslational modification or by additional components. To discriminate between these possibilities, the enzymatically highly active sorghum protein extract was applied to a NIT4-immunoaffinity column. Passage over this column removed the β-cyanoalanine metabolizing activity from the protein extract and elution of the column provided two distinct proteins with approximate masses of 42 and 40 kDa (Fig. 4). Mass spectrometric sequencing identified the 42-kDa protein as SbNIT4B2 and the 40-kDa component as SbNIT4A (data not shown). These results demonstrated that the β-cyanoalanine-metabolizing activity is due to one or both of these two NIT4 homologs and that both isoforms are expressed at the same stage of seedling development.
Fig. 4.
Isolation of nitrilases from S. bicolor by immunoaffinity chromatography as monitored by SDS/PAGE and Coomassie staining. Lane 1, Nitrilase enriched preparation after ion-exchange chromatography (15 μl); lane 2, same extract after binding to the α-NIT4-affinity column (15 μl); lane 3, eluate from the α-NIT4-affinity column. The arrowheads point to the two isolated nitrilases with apparent molecular masses of 42 and 40 kDa. M, molecular mass standards.
NIT4A/B Heteromers of Poaceae Are Able to Metabolize β-Cyanoalanine, but Complex Composition Determines Substrate Specificity.
Nitrilases belong to the superfamily of C-N hydrolases (21, 22), members of which are known to form homomeric complexes consisting of 2–18 subunits (23–29). The copurification of SbNIT4A and B2 from sorghum seedlings raised the question whether these enzymes exist as heteromeric complexes in vivo and whether this interaction had an influence on their activity. Indeed, two component mixtures of the recombinant isoforms SbNIT4A and B1 as well as of SbNIT4A and B2 exhibited high β-cyanoalanine metabolizing activity, whereas a mixture of SbNIT4B1 and B2 remained inactive (Fig. 3A). The nitrile hydratase/nitrilase ratios for the SbNIT4A/B1 and SbNIT4A/B2 mixtures were 3.2 and 2.1, respectively, the latter being in agreement with a ratio of 2.3 obtained with the sorghum extract. The enzyme activities of the SbNIT4A/B1 and SbNIT4A/B2 heterocomplexes toward β-cyanoalanine were much higher (Table 2) than those observed with SbNIT4A or ZmNIT4A alone using other substrates (Table 1 and ref. 14). Both heterocomplexes had nearly identical Km values for β-cyanoalanine (Table 2), which were similar to those observed with the homomeric NIT4 enzymes from A. thaliana and L. angustifolius (9, 10). The direct interaction of SbNIT4A with SbNIT4B1 and B2 was demonstrated by pull-down assays (Fig. 5). These assays also revealed the presence of SbNIT4A and SbNIT4B1 homocomplexes and a weaker interaction of SbNIT4B1 with SbNIT4B2. Accordingly, the lack of activity of SbNIT4A and SbNIT4B1 alone as well as of SbNIT4B1/B2 mixtures was not due to a general failure of these proteins to interact.
Table 2.
Enzyme kinetic parameters for the recombinant SbNIT4A/B1 and SbNIT4A/B2 heterocomplexes
| Compound |
SbNIT4A/B1 |
SbNIT4A/B2 |
||
|---|---|---|---|---|
| Km, mM | Vmax, nkat (mg protein)−1 | Km, mM | Vmax, nkat (mg protein)−1 | |
| β-Cyanoalanine* | 0.86 ± 0.14 | 308 ± 51 | 0.89 ± 0.26 | 432 ± 32 |
| 2-Phenylacetonitrile | n.d. | n.d. | 0.19 ± 0.03 | 908 ± 108 |
| 4-Hydroxyphenylacetonitrile | n.d. | n.d. | 0.17 ± 0.01 | 940 ± 249 |
| Indole-3-acetonitrile | n.d. | n.d. | 0.16 ± 0.01 | 323 ± 31 |
The active enzyme component (SbNIT4A for β-cyanoalanine and SbNIT4B2 for the other substrates) was combined in a 1:10 ratio with the second enzyme component to favor that a high proportion of the active component was present in heterocomplexes. Shown are the means ± SD from two to three independent experiments performed in triplicate.
*Only nitrilase activity (release of NH4+) was measured. The total activity is 3- to 4-fold higher because of the high nitrile hydratase activity of the enzymes for this substrate (see text).
n.d., not determined.
Fig. 5.
Interaction of nitrilases from S. bicolor in vitro. Purified, (His)6-tagged SbNIT4A, B1, B2, and agmatine iminohydrolase from A. thaliana (AtAIH, as negative control) (55) were incubated with bacterial extracts containing c-myc-tagged SbNIT4A (Top), B1 (Middle), and B2 (Bottom). The (His)6-tagged proteins were purified and copurifying c-myc-tagged proteins were detected after SDS/PAGE by Western blotting.
Individual members within the two orthologous NIT4 groups in Poaceae (NIT4A and NIT4B) were functionally interchangeable as demonstrated by assays containing mixtures of NIT4 enzymes derived from barley (HvNIT4B), rice (OsNIT4A), and sorghum (SbNIT4A, SbNIT4B1 and SbNIT4B2) (Fig. 3C). All combinations of a NIT4A with a NIT4B isoform were active, irrespective of their origin, whereas combinations of either different NIT4A or NIT4B isoforms remained inactive.
Interestingly, SbNIT4A/B2 but not SbNIT4A/B1 was also able to use 2-phenylacetonitrile, 4-hydroxyphenylacetonitrile, indole-3-acetonitrile, and 6-heptenenitrile as substrates in order of decreasing activity (Fig. 6). The high activity of the SbNIT4A/B2 complex with arylacetonitriles was due to low Km and high Vmax values for these substrates (Table 2). Mixtures of SbNIT4B1 and SbNIT4B2 did not show activity toward any of the tested compounds.
Fig. 6.
Substrate specificities of SbNIT4A/B1, SbNIT4A/B2, and SbNIT4A/HvNIT4B complexes and identification of active subunits by site directed mutagenesis. Shown are the means ± SD from two to three independent experiments (n = 6–9). *, No activity measurable.
The Active Sites for the Different Substrates Are Different in SbNIT4A/B1 and SbNIT4A/B2 Heterocomplexes.
To further investigate the role of the individual isoforms for catalysis and substrate binding within the active heterocomplexes, the catalytically essential cysteine residue in the different isoforms (C195 in SbNIT4A, C191 in SbNIT4B1, and C194 in SbNITB2) was replaced by an alanine residue by site directed mutagenesis. The ability to metabolize β-cyanoalanine was restricted to heterocomplexes containing wild-type SbNIT4A, whereas mutations in SbNIT4B1 and B2 did not abolish this activity (Figs. 3B and 6). In contrast, hydrolysis of other substrates than β-cyanoalanine depended on a functional SbNIT4B2 isoform but was independent on a functional SbNIT4A isoform (Fig. 6). These experiments clearly demonstrate that the catalytic site for β-cyanoalanine hydrolysis resides on the NIT4A isoform and that the active site for the other substrates resides on the NIT4B2 isoform. These properties of the sorghum nitrilases offer a route to control and divert enzyme specificity by the use combinatorial biochemistry. The occurrence of two different active sites harbored at different subunits within the SbNIT4A/B2 heterocomplex was further substantiated by experiments in which the conversion rates obtained with separately or simultaneously administered β-cyanoalanine and 4-hydroxyphenylacetonitrile were shown to be additive (data not shown). We also analyzed the substrate specificity of the SbNIT4A/HvNIT4B heterocomplex. Most activities found for this complex could be attributed to the SbNIT4A isoform with the exception of 6-heptenenitrile, the conversion of which was due to the HvNIT4B isoform (Fig. 6). Thus, HvNIT4B adds another substrate specificity to the complex. The lower overall activity of the SbNIT4A/HvNIT4B mixture may be due to a lower amount of heterocomplex because of lower affinities of these enzymes for each other.
Formation of 4-Hydroxyphenylacetonitrile by Catabolism of Dhurrin.
Very high activity of the SbNIT4A/B2 complex was obtained by using 4-hydroxyphenylacetonitrile as substrate (Table 2 and Fig. 6). This compound has a high structural similarity to the cyanogenic glucoside dhurrin, which is present in high amounts in young seedlings of S. bicolor (30–33). However, dhurrin is not a direct substrate for the SbNIT4A/B2 complex (data not shown).
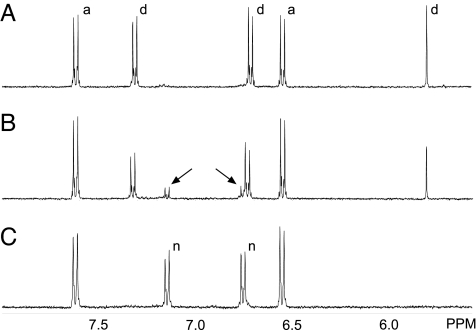
It is known that dhurrin rapidly decomposes under alkaline conditions giving rise to the formation of 4-hydroxymandelonitrile, 4-hydroxybenzaldehyde and cyanide (32, 34, 35). In our experiments, dhurrin decomposition occurred with similar rates in buffer alone (pH 7.5) and in buffer supplemented with DTT, the reducing agent typically used in nitrilase assays as enzyme stabilizer. LC-MS and NMR analyses confirmed that the degradation product of dhurrin in the absence of DTT was 4-hydroxymandelonitrile, which upon dissociation gave rise to the formation of 4-hydroxybenzaldehyde and hydrogen cyanide (Fig. 7A). However, in the presence of DTT, an additional product was formed, which by LC-MS and NMR analyses was identified as 4-hydroxyphenylacetonitrile (Fig. 7 B and C). As reported above, 4-hydroxyphenylacetonitrile is an excellent substrate for the SbNIT4A/B2 complex with the products being 4-hydroxyphenylacetic acid and ammonia (Fig. 8). Accordingly, incubation of the SbNIT4A/B2 complex with dhurrin in the presence of DTT resulted in the formation of 4-hydroxyphenylacetic acid (data not shown). β-Mercaptoethanol, glutathione, and l-cysteine did not support 4-hydroxyphenylacetonitrile production whereas dithioerythritol did (data not shown). This result indicated that a strong reductant containing vicinal thiol groups was necessary to facilitate the reaction.
Fig. 7.
In vitro-breakdown of dhurrin as monitored by 1H-NMR. (A) Spectrum recorded 40 min after dissolving dhurrin (4 mM) in 100 mM KDCO3 (pH 8.3). Approximately half the amount of dhurrin is converted into 4-hydroxybenzaldehyde. (B) Spectrum recorded 20 min after addition of DTT (2 mM). 4-Hydroxyphenylacetonitrile formation is apparent (arrows). (C) As in B, except that the spectrum was recorded after 5 h. All dhurrin has now been converted into 4-hydroxybenzaldehyde and 4-hydroxyphenylacetonitrile. Signals in the 5.5- to 8.0-ppm region are shown. Signals assigned to dhurrin (d), 4-hydroxybenzaldehyde (a), and 4-hydroxyphenylacetonitrile (n) are labeled.
Fig. 8.
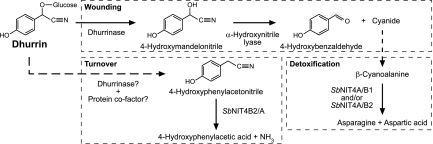
Degradation of dhurrin in S. bicolor as a response to wounding and the proposed pathway for in vivo turnover of dhurrin avoiding concomitant release of hydrogen cyanide with the NIT4B2/A heterocomplex as a key enzyme.
Dhurrin content reaches maximum in 2-day-old dark grown sorghum seedlings after which time degradation proceeds faster than de novo synthesis (30). Analyses of 2- and 4-day-old seedlings by LC-MS showed the presence of a component of m/z 359, the intensity of which was three times higher in 4-day-old compared with 2-day-old seedlings. In MS2 spectra, this component provided a fragment ion m/z 315 [M − 44] corresponding to loss of CO2. These properties matches those of 4-glucosyloxy phenylacetic acid (m/z 359 = [M + 2Na − H]), the glucoside of 4-hydroxyphenylacetic acid. This assignment was further substantiated by an exact mass measurement of the compound, giving a mass of 337.0888 Da, only 2 ppm off the expected mass of 337.0894 Da. Free 4-hydroxyphenylacetic acid was not detected in the seedlings.
Discussion
In this study we have shown that the NIT4 enzymes of Poaceae, in contrast to the nitrilases of other plant families (9, 10, 36), must form heteromeric complexes to gain high catalytic activity. All Poaceae species analyzed contain orthologous genes for two NIT4 isoforms (NIT4A and NIT4B), which most likely have arisen by an ancient Poaceae whole genome duplication ≈70 MYA (16). Accordingly, the promotors of the NIT4A and NIT4B genes were initially identical and would have ensured coexpression of both isoforms, a prerequisite for heterocomplex formation. Under this condition mutations may then have accumulated, which prevented activation of homomeric complexes whereas heteromeric complexes remained active. Thereafter, loss of one of the two NIT4 genes was not longer possible without losing all NIT4 activity. The NIT4A isoform retained the ability to hydrolyze β-cyanoalanine whereas NIT4B lost this activity but retained the capability to interact with and activate NIT4A. In parallel, neofunctionalization of the NIT4B isoform could take place. This scheme of NIT4 evolution in the Poaceae is supported by analyses of ratios of nonsynonymous to synonymous mutation rates, which shows a strong purifying (= negative) selection on the NIT4A isoforms, whereas this pressure is relaxed on the NIT4B isoforms (data not shown). One of the NIT4B isoforms of S. bicolor was neofunctionalized and gained the potential to use structurally very different nitrile substrates. These nitriles include indole-3-acetonitrile, a possible precursor of the auxin indole-3-acetic acid. Indeed, the SbNIT4A/B2 complex showed the highest indole-3-acetonitrile-hydrolyzing activity ever reported for a plant nitrilase. However, even higher activity was obtained with 4-hydroxyphenylacetonitrile and we propose that this compound is the physiological substrate for the SbNIT4A/B2 heterocomplex in vivo. 4-Hydroxyphenylacetonitrile has so far not been reported as a genuine constituent in S. bicolor. In this study we have demonstrated that in the presence of DTT, in vitro decomposition of the cyanogenic glucoside dhurrin results in the formation of 4-hydroxyphenylacetonitrile. In young S. bicolor seedlings, dhurrin constitutes 6% of the dry weight, shows in vivo turnover (37, 38) and is catabolized in older seedlings (30, 31), but the catabolic route is not known. In principle, dhurrin can be metabolized to cyanide and 4-hydroxybenzaldehyde by the sequential action of the β-glucosidase dhurrinase and α-hydroxynitrile lyase. This process, known as cyanogenesis, takes place when the compartmentalized organization at the subcellular level is disrupted e.g., by pathogens or herbivores (Fig. 8) (4, 39, 40). Based on the observations that dhurrin can decompose to 4-hydroxyphenylacetonitrile in vitro, we propose that this reaction also proceeds in vivo and may be carried out by a new type of β-glucosidase or by dhurrinase in combination with a protein cofactor (Fig. 8), analogous to the glucosinolate/myrosinase/epithiospecifier system in the Brassicaceae (41). The 4-hydroxyphenylacetonitrile formed from dhurrin will subsequently be converted into ammonia and 4-hydroxyphenylacetic acid by the SbNIT4A/B2 complex. In accordance with this model, an increased amount of 4-hydroxyphenylacetic acid (as glucose conjugate) was found to be present in older seedlings of S. bicolor. This route of dhurrin catabolism avoids the release of cyanide and subsequent demands for its detoxification. Instead, the nitrogen atom of the nitrile function is directly recovered as ammonia. Although many grass species are known to contain cyanogenic glycosides (42), S. bicolor is unique in having a cyanogenic glycoside content that is two to four orders of magnitude higher compared with other grass species (43–45). The possibility to catabolize these large amounts of dhurrin in an efficient way without the intermediate release of toxic compounds may have been one driving force for the neofunctionalization of the NIT4B2 isoform in S. bicolor. Dhurrin is also present in high amounts in sugarcane (4–5 mg per gram of leaf fresh weight)¶, and the presence of a NIT4B2 ortholog in this plant suggests that an endogenous dhurrin catabolism was already present in the progenitor of sorghum and sugarcane.
This study demonstrates that the substrate specificity of nitrilases is defined and extended by combinatorial biochemistry and constitutes the second case where recruitment of nitrilases from primary metabolism (cyanide detoxification) into secondary metabolism is observed. The other examples are the nitrilase-1 homologs in Brassicaceae, which also have evolved from a NIT4 progenitor and the function of which most likely lies within in the catabolism of glucosinolates (36), secondary compounds found in this family (for review, see ref. 46).
Cyanogenic glycosides have previously been proposed to serve an important role as mobile reservoirs of reduced nitrogen (47). It remains to be investigated whether non-Poaceae species like lotus, flax, and cassava, which also contain high amounts of cyanogenic glycosides, also possess additional nitrilases that could be involved in a noncyanogenic catabolism of cyanogenic glycosides. Likewise, nitrilases may also be involved in endogenous turnover of other nitrogenous natural products like the nitrile glucosides in lotus (48) or ricinine of castor bean (49).
Materials and Methods
Detailed descriptions of primer sequences, cloning steps,and purification of nitrilases from S. bicolor are given in supporting information (SI) Materials and Methods.
Cloning, Mutagenesis, and Expression of Nitrilases from Poaceae.
ORFs of Poaceae nitrilases were amplified by PCR from EST clones (SbNIT4A, GenBank accession no. BE595164; SbNIT4B2, GenBank accession no. CB929027; OsNIT4A, GenBank accession no. D15299; HvNIT4B, GenBank accession no. BF620977) or from a cDNA library (SbNIT4B1) (50). Cloning of the ORFs in pET-21b(+) (Novagen, Darmstadt, Germany) for expression of C-terminally (His)6-tagged proteins was performed as described (9, 10). Site-directed mutagenesis was based on the QuikChange protocol with the modifications published by Zheng et al. (51). Expression and purification of recombinant nitrilases from E. coli BL21 CodonPlus (DE3) RIL (Stratagene) using Ni2+-NTA agarose (Qiagen) is described in ref. 10. For expression of c-myc-tagged nitrilases, a c-myc-tag adapter including a terminal stop codon was introduced into the NotI/XhoI restricted pET-21b(+), resulting in pET-21b(+)myc. This vector was then used for cloning.
In Vitro Interaction of Nitrilases.
Crude protein extracts of E. coli expressing c-myc-tagged nitrilases were enriched by ammonium-sulfate precipitation (40% saturation). Purified (His)6-tagged nitrilases (5 μg) were added to 10 μl of these enriched extracts in a total volume of 600 μl of binding buffer (50 mM sodium phosphate, pH 8.0, 300 mM NaCl) including 10 mM imidazole. After 30-min incubation, the (His)6-tagged nitrilases were purified by Ni-NTA spin columns (Qiagen). Washing was done three times with binding buffer, including 40 mM imidazole and elution with 200 μl of binding buffer including 250 mM imidazole. The eluted proteins were precipitated by trichloroacetic acid, dissolved in SDS sample buffer and separated by SDS/PAGE. Copurifying myc-tagged nitrilases were detected by Western blotting with a monoclonal α-c-myc-tag antibody.
Purification of Nitrilases from Seedlings of S. bicolor and Mass Spectrometric Sequencing.
Nitrilases of S. bicolor (L.) Moench var. Redland were purified from leaves of 9-day-old seedlings by a combination of ammonium-sulfate precipitation, ion-exchange chromatography, and immunoaffinity chromatography on an α-NIT4-antibody column. Sequencing of the eluted proteins by mass spectrometry was performed as described (9).
Activity Assays.
Standard assays (1 ml, 37°C) were carried out in 50 mM KPi (pH 8.0), 1 mM DTT, 2.5–5 mM substrate, and 2–10 μg of purified enzyme. Activity was measured as ammonia release (10). Assays were conducted with at least two different enzyme preparations and performed in triplicate with an additional background control containing heat denatured enzyme. For analyses of enzyme complexes, the combined enzymes were incubated (30 min, room temperature) to facilitate heterocomplex formation. Calculation of specific activity of enzyme complexes was based on the amount of the active enzyme component only (SbNIT4A for β-cyanoalanine and SbNIT4B2 for the other substrates). Km and Vmax values were determined at 30°C by weighted nonlinear regression of the data to the Michaelis-Menten equation (52). Separation of β-cyanoalanine, asparagine, and aspartic acid by TLC has been described (9).
Phylogenetic Analyses.
Full-length coding cDNA sequences of the nitrilase 4 homologs (Fig. 2) were obtained directly or assembled from EST clones from GenBank using the DNAMAN program (Lynnon Biosoft). The encoded amino acid sequences were used to align the cDNA sequences with ClustalW implemented in the MEGA 3.1 program (53) using standard parameters (SI Fig. 9). Extreme diverse 5′ and 3′ sequences were deleted from the alignment. Phylogenetic inference was performed by Bayesian analysis using the MrBayes software (54) (version 3.1.2, general time-reversible model, variable rates for each codon position, 2 million generations, burn-in: 25% of saved trees). Ks values were calculated with MEGA 3.1 (pairwise distance, complete deletion of gaps, Nei-Gojobori method with Jukes-Cantor correction).
Mass Spectrometry and NMR Analyses of Dhurrin Metabolites.
Dhurrin metabolites and decomposition products were verified by LC-MS and NMR using authentic standards.
Supplementary Material
Acknowledgments
We thank Marie-Michele Cordonnier-Pratt (University of Georgia, Athens, GA) and the National Institute of Agrobiological Sciences (Ibaraki, Japan) for providing EST clones from S. bicolor and O. sativa, respectively. This project was funded by Deutsche Forschungsgemeinschaft Grant PI 424/1-2 and by the Danish National Research Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709315104/DC1.
De Rosa VE, Jr, Nogueira FTS, Mazzafera P, Landell MGA, Arruda P, XXVIth Congress of the International Society of Sugarcane Technologists, July 29–August 2, 2007, Durban, South Africa, poster abstract.
References
- 1.Peiser GD, Wang T-T, Hoffman NE, Yang SF, Liu H-W, Walsh CT. Proc Natl Acad Sci USA. 1984;81:3059–3063. doi: 10.1073/pnas.81.10.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pirrung MC. Bioorg Chem. 1985;13:219–226. [Google Scholar]
- 3.Vetter J. Toxicon. 2000;38:11–36. doi: 10.1016/s0041-0101(99)00128-2. [DOI] [PubMed] [Google Scholar]
- 4.Zagrobelny M, Bak S, Rasmussen AV, Jørgensen B, Naumann CM, Møller BL. Phytochemistry. 2004;65:293–306. doi: 10.1016/j.phytochem.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 5.Bak S, Paquette SM, Morant M, Morant AV, Saito S, Bjarnholt N, Zagrobelny M, Jørgensen K, Osmani S, Hamann T, et al. Phytochem Rev. 2006;5:309–329. [Google Scholar]
- 6.Hendrickson HR, Conn EE. J Biol Chem. 1969;244:2632–2640. [PubMed] [Google Scholar]
- 7.Blumenthal SG, Hendrickson HR, Abrol YP, Conn EE. J Biol Chem. 1968;243:5302–5307. [PubMed] [Google Scholar]
- 8.Castric PA, Farnden KJ, Conn EE. Arch Biochem Biophys. 1972;152:62–69. doi: 10.1016/0003-9861(72)90193-2. [DOI] [PubMed] [Google Scholar]
- 9.Piotrowski M, Volmer JJ. Plant Mol Biol. 2006;61:111–122. doi: 10.1007/s11103-005-6217-9. [DOI] [PubMed] [Google Scholar]
- 10.Piotrowski M, Schönfelder S, Weiler EW. J Biol Chem. 2001;276:2616–2621. doi: 10.1074/jbc.M007890200. [DOI] [PubMed] [Google Scholar]
- 11.Bartel B, Fink GR. Proc Natl Acad Sci USA. 1994;91:6649–6653. doi: 10.1073/pnas.91.14.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dohmoto M, Tsunoda H, Isaji G, Chiba R, Yamaguchi K. DNA Res. 2000;7:283–289. doi: 10.1093/dnares/7.5.283. [DOI] [PubMed] [Google Scholar]
- 13.Dohmoto M, Sano J, Tsunoda H, Yamaguchi K. DNA Res. 1999;6:313–317. doi: 10.1093/dnares/6.5.313. [DOI] [PubMed] [Google Scholar]
- 14.Park WJ, Kriechbaumer V, Müller A, Piotrowski M, Meeley RB, Gierl A, Glawischnig E. Plant Physiol. 2003;133:794–802. doi: 10.1104/pp.103.026609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oaks A, Johnson FJ. Phytochemistry. 1972;2:3465–3471. [Google Scholar]
- 16.Paterson AH, Bowers JE, Chapman BA. Proc Natl Acad Sci USA. 2004;101:9903–9908. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lynch M, Conery JS. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 18.Gaut BS, Morton BR, McCaig BC, Clegg MT. Proc Natl Acad Sci USA. 1996;93:10274–10279. doi: 10.1073/pnas.93.19.10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaut BS, Doebley JF. Proc Natl Acad Sci USA. 1997;94:6809–6814. doi: 10.1073/pnas.94.13.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swigonova Z, Lai J, Ma J, Ramakrishna W, Llaca V, Bennetzen JL, Messing J. Genome Res. 2004;14:1916–1923. doi: 10.1101/gr.2332504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bork P, Koonin EV. Protein Sci. 1994;3:1344–1346. doi: 10.1002/pro.5560030821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pace HC, Brenner C. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-1-reviews0001. reviews0001.1–0001.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pace HC, Hodawadekar SC, Draganescu A, Huang J, Bieganowski P, Pekarsky Y, Croce CM, Brenner C. Curr Biol. 2000;10:907–917. doi: 10.1016/s0960-9822(00)00621-7. [DOI] [PubMed] [Google Scholar]
- 24.Kumaran D, Eswaramoorthy S, Gerchman SE, Kycia H, Studier FW, Swaminathan S. Proteins. 2003;52:283–291. doi: 10.1002/prot.10417. [DOI] [PubMed] [Google Scholar]
- 25.Sakai N, Tajika Y, Yao M, Watanabe N, Tanaka I. Proteins. 2004;57:869–873. doi: 10.1002/prot.20259. [DOI] [PubMed] [Google Scholar]
- 26.Nagasawa T, Wieser M, Nakamura T, Iwahara H, Yoshida T, Gekko K. Eur J Biochem. 2000;267:138–144. doi: 10.1046/j.1432-1327.2000.00983.x. [DOI] [PubMed] [Google Scholar]
- 27.Jandhyala D, Berman M, Meyers PR, Sewell BT, Willson RC, Benedik MJ. Appl Environ Microbiol. 2003;69:4794–4805. doi: 10.1128/AEM.69.8.4794-4805.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piotrowski M, Janowitz T, Kneifel H. J Biol Chem. 2003;278:1708–1712. doi: 10.1074/jbc.M205699200. [DOI] [PubMed] [Google Scholar]
- 29.Sewell BT, Berman MN, Meyers PR, Jandhyala D, Benedik MJ. Structure (London) 2003;11:1413–1422. doi: 10.1016/j.str.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 30.Halkier BA, Møller BL. Plant Physiol. 1989;90:1552–1559. doi: 10.1104/pp.90.4.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Busk PK, Møller BL. Plant Physiol. 2002;129:1222–1231. doi: 10.1104/pp.000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akazawa T, Miljanich P, Conn EE. Plant Physiol. 1960;35:535–538. doi: 10.1104/pp.35.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunstan WR, Henry TA. Philos Trans R Soc London. 1902;199:399–410. [Google Scholar]
- 34.Mao CH, Anderson L. J Org Chem. 1965;30:603–607. [Google Scholar]
- 35.Mao CH, Blocher JP, Anderson L, Smith DC. Phytochemistry. 1965;4:297–303. [Google Scholar]
- 36.Vorwerk S, Biernacki S, Hillebrand H, Janzik I, Müller A, Weiler EW, Piotrowski M. Planta. 2001;212:508–516. doi: 10.1007/s004250000420. [DOI] [PubMed] [Google Scholar]
- 37.Bough WA, Gander JE. Phytochemistry. 1971;10:67–77. [Google Scholar]
- 38.Adewusi SR. Plant Physiol. 1990;94:1219–1224. doi: 10.1104/pp.94.3.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poulton JE. Plant Physiol. 1990;94:401–405. doi: 10.1104/pp.94.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nahrstedt A. Plant Syst Evol. 1985;150:35–47. [Google Scholar]
- 41.Lambrix V, Reichelt M, Mitchell-Olds T, Kliebenstein DJ, Gershenzon J. Plant Cell. 2001;13:2793–2807. doi: 10.1105/tpc.010261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones DA. Phytochemistry. 1998;47:155–162. doi: 10.1016/s0031-9422(97)00425-1. [DOI] [PubMed] [Google Scholar]
- 43.Lehmann G, Zinsmeister HD, Erb N, Neunhoeffer O. Z Ernährungswiss. 1979;18:16–22. doi: 10.1007/BF02026532. [DOI] [PubMed] [Google Scholar]
- 44.Zinsmeister HD, Erb N, Lehmann G. Z Lebensm Unters Forsch. 1980;171:170–173. [Google Scholar]
- 45.Nielsen KA, Olsen CE, Pontoppidan K, Møller BL. Plant Physiol. 2002;129:1066–1075. doi: 10.1104/pp.001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Halkier BA, Gershenzon J. Annu Rev Plant Biol. 2006;57:303–333. doi: 10.1146/annurev.arplant.57.032905.105228. [DOI] [PubMed] [Google Scholar]
- 47.Selmar D, Lieberei R, Biehl B. Plant Physiol. 1988;86:711–716. doi: 10.1104/pp.86.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Forslund K, Morant M, Jørgensen B, Olsen CE, Asamizu E, Sato S, Tabata S, Bak S. Plant Physiol. 2004;135:71–84. doi: 10.1104/pp.103.038059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tuson RV. J Chem Soc. 1864:195–197. [Google Scholar]
- 50.Koch BM, Sibbesen O, Halkier BA, Svendsen I, Møller BL. Arch Biochem Biophys. 1995;323:177–186. doi: 10.1006/abbi.1995.0024. [DOI] [PubMed] [Google Scholar]
- 51.Zheng L, Baumann U, Reymond JL. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hernandez A, Ruiz MT. Bioinformatics. 1998;14:227–228. doi: 10.1093/bioinformatics/14.2.227. [DOI] [PubMed] [Google Scholar]
- 53.Kumar S, Tamura K, Nei M. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 54.Ronquist F, Huelsenbeck JP. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 55.Janowitz T, Kneifel H, Piotrowski M. FEBS Lett. 2003;544:258–261. doi: 10.1016/s0014-5793(03)00515-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.