Abstract
Recombination activating gene (RAG) 1 and RAG2 together catalyze V(D)J gene rearrangement in lymphocytes as the first step in the assembly and maturation of antigen receptors. RAG2 contains a plant homeodomain (PHD) near its C terminus (RAG2-PHD) that recognizes histone H3 methylated at lysine 4 (H3K4me) and influences V(D)J recombination. We report here crystal structures of RAG2-PHD alone and complexed with five modified H3 peptides. Two aspects of RAG2-PHD are unique. First, in the absence of the modified peptide, a peptide N-terminal to RAG2-PHD occupies the substrate-binding site, which may reflect an autoregulatory mechanism. Second, in contrast to other H3K4me3-binding PHD domains, RAG2-PHD substitutes a carboxylate that interacts with arginine 2 (R2) with a Tyr, resulting in binding to H3K4me3 that is enhanced rather than inhibited by dimethylation of R2. Five residues involved in histone H3 recognition were found mutated in severe combined immunodeficiency (SCID) patients. Disruption of the RAG2-PHD structure appears to lead to the absence of T and B lymphocytes, whereas failure to bind H3K4me3 is linked to Omenn Syndrome. This work provides a molecular basis for chromatin-dependent gene recombination and presents a single protein domain that simultaneously recognizes two distinct histone modifications, revealing added complexity in the read-out of combinatorial histone modifications.
Keywords: H3K4me, regulation, V(D)J recombination, H3R2me, SCID
V(D)J recombination is the site-specific DNA rearrangement that assembles antigen receptor genes from dispersed arrays of V, D, and J gene segments. Recombination is initiated by the lymphoid-specific recombination activating gene (RAG) 1 and RAG2 recombinase, which recognizes and cleaves the recombination signal sequences (1). V(D)J recombination is tightly regulated, occurring in a preferred temporal order and only in specific cell types and developmental stages. Ig heavy chain rearrangement precedes light chain rearrangement and Ig heavy-chain D to J joining precedes V to DJ recombination. In addition, Ig genes are fully rearranged only in B cells (not T cells), and T cell receptor genes are assembled in T but not B cells (1). Overexpression of RAG1 and RAG2 in nonlymphoid cells is sufficient to induce recombination of an artificial extrachromosomal substrate but does not support V(D)J recombination of endogenous loci (2). Therefore, the accessibility of these loci to the recombinase must be regulated (3). A large body of evidence suggests that the regulation of chromatin structure is involved in the regulation of V(D)J recombination (4–6).
Posttranslational modifications of histone tails, including methylation, acetylation, phosphorylation, ubiquitylation, sumoylation, and ADP ribosylation, have been found to play an important role in regulating chromatin structure and gene accessibility (7–10). Histone acetylation leads to transcription activation in general, and specific histone methylations have been implicated in either up- or down-regulation of associated genes. For example, trimethylation of lysine-4 on histone H3 (H3K4me3) is associated with transcription activation, whereas H3K9me3 is linked to transcription suppression (11). Histone methylation at antigen receptor loci has been reported at particular developmental stages, indicating that it may play a role in regulating recombination (12). Recognition of acetylated and methylated histone lysines thus becomes an essential part of chromatin-based gene regulation. Bromodomains have been shown to recognize acetylated lysine (13–15), and chromodomains of HP1 and Polycomb bind to methylated H3K9 and H3K27, respectively (16–19). More recently, a double chromodomain of CHD1 (20), double Tudor domains of JMJD2A (21), and the plant homeodomain (PHD) fingers in an array of proteins (22), including BPTF (23), ING2 (24), and Yng1 (25), have each been reported to recognize H3K4me3 with micromolar dissociation constants.
The observation that a single histone modification, H3K4me3, is recognized by multiple protein modules each with tens of homologues in one organism (7, 26, 27) raises the question of how specific targeting to distinctly modified chromatin is achieved. It has been proposed that additional modifications of adjacent or distal residues may influence histone recognition and that a combination of histone modifications may have synergistic or antagonistic effects (7, 26, 28). Asymmetric dimethylation of R2 or phosphorylation of T3 was reported to decrease binding of the double chromodomains of CHD1 to H3K4me3 (20). Similarly, binding of HP1 chromodomain to H3K9me is prevented by phosphorylation of the adjacent S10 (29). Although the homodimeric chromodomain of CMT3 preferably binds histone H3 methylated at both K9 and K27, the two modified methyl-lysines appear to bind to different subunits (28). A single protein domain that has increased affinity for a histone with more than one modification has yet to be identified.
RAG1 and RAG2 are large proteins with 500 to >1,000 residues, but only core regions of each are necessary and sufficient for DNA cleavage in vitro and recombination of extrachromosomal plasmid substrates in vivo (1). Outside of the core region, the C terminus of RAG2 contains a PHD finger, which is required for efficient V to DJ assembly of endogenous antigen receptor genes (30–32). When endogenous RAG2 in mice is replaced by the RAG2 core region only, V(D)J recombination occurs, but the numbers of mature B and T cells are substantially reduced (30, 32). Furthermore, five missense mutations in the RAG2-PHD finger have been identified in patients with severe combined immunodeficiency (SCID), including Omenn syndrome, which is characterized by restricted V(D)J recombination, oligoclonal T cells, and few or no B cells (33–35). Apparently, the full-length RAG2 with a mutation in the PHD finger can be more defective for in vivo recombination activity than the RAG2 core alone (36, 37). These observations suggest that RAG2-PHD may negatively regulate the RAG1 and RAG2 recombinase activity and restrict recombination to particular loci.
Recently, the RAG2-PHD finger has been shown to interact with H3K4me3 and not with trimethylated K9, K23, K27, K36, or K79 of histone H3 (37). To gain additional insight into the molecular mechanisms that govern histone recognition and chromatin accessibility in V(D)J recombination, we have determined the crystal structures of RAG2-PHD alone and in complex with differently modified histone H3 N-terminal peptides. Our results provide the structural basis for recognition of the first six residues of H3K4me3 by RAG2-PHD. The structural results, together with pull-down assays and binding affinity measurements, demonstrate that contrary to what has been observed for other H3K4me3 binding domains, methylation of the adjacent arginine-2 does not inhibit but rather enhances the binding of RAG2-PHD to H3K4me3.
Results and Discussion
Structure of RAG2-PHD Complexed with an H3K4me3 Peptide.
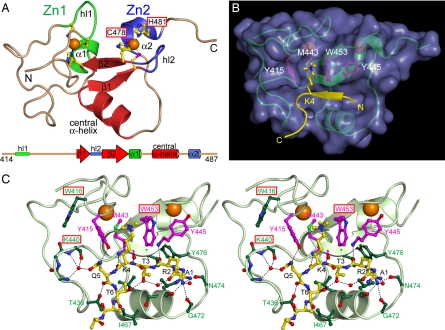
We determined the crystal structure of RAG2-PHD (residues 414–487 of 527) in the presence of H3K4me3 peptides at 2.4 Å (Table 1 and Fig. 1). In each asymmetric unit, there were three RAG2-PHD molecules, two bound to H3K4me3 and one free of the peptide. The structures of RAG2-PHD with or without the H3 peptide are superimposable with an rmsd of 0.6 Å over 66 pairs of Cα atoms (Fig. 2A). Despite the apparent differences from the NMR models (36) [supporting information (SI) Fig. 6], our crystal structures are fully compatible with the distance restraints measured by NMR spectroscopy (D. Ivanov, personal communication). As predicted by sequence analysis (38), RAG2-PHD is distinct from the zinc-binding RING finger and LIM domain. Like other PHD fingers (26, 39) (Fig. 3 A and B), RAG2-PHD consists of two nonconventional zinc fingers (36). Each Zn2+ ion is coordinated by a combination of four Cys and His residues, two from a distorted β-hairpin and two from the N terminus of a short α-helix (Fig. 1A). The two zinc fingers are interleaved, with residues 414–442 and 455–462 belonging to the first (Zn1) and residues 446–452 and 472–487 to the second (Zn2). A pair of antiparallel β-strands (β1: 443–446, β2: 452–455) and a central α-helix (463–471) linking the two zinc fingers form the core of RAG2-PHD and the binding site for the H3 peptide (Fig. 1).
Table 1.
Data collection and SAD phasing
| Wavelength, Å | 1.2818 |
| Space group | P21 |
| Unit cell | |
| a, b, c, Å | 54.8, 46.8, 57.0 |
| α, β, γ, ° | 90, 101.5, 90 |
| Resolution range, Å | 50–2.4 (2.49–2.4) |
| Rmerge,* | 0.126 (0.329) |
| Unique reflections | 10,187 |
| Redundancy | 3.2 (1.8) |
| Completeness, % | 88.9 (57.2) |
| I/σ(I) | 10.2 (2.2) |
| Figure of merit | 0.23 |
| Phasing power score† | 17.71 |
Data of the highest-resolution shell are shown in parentheses.
*Rmerge = ΣhΣi|Ihi − 〈Ih〉|/Σ〈Ih〉, where Ihi is the intensity of the ith observation of reflection h and 〈Ih〉 is the average intensity of redundant measurements of the h reflections.
†Determined by SOLVE.
Fig. 1.
Crystal structure of RAG2-PHD bound to H3K4me3. (A) Ribbon diagram of the RAG2-PHD structure. The Zn1 and Zn2 fingers are shown in green and blue. The Zn2+ ions (orange spheres) are coordinated by His (yellow–blue) and Cys (yellow–purple) shown as balls and sticks. The hairpin loops (hl1 and hl2), 310 α-helices (α1 and α 2), and all secondary structures are labeled. The interdigitating nature of the zinc fingers is illustrated in the linear diagram below it. (B) Binding of the H3K4me3 peptide to RAG2-PHD. The RAG2-PHD is shown as a green ribbon diagram encased in purple molecular surface, and the H3 peptide is highlighted in yellow. Four amino acids essential for the peptide binding are shown in magenta balls and sticks. (C) Stereoview of the detailed intermolecular contacts between RAG2-PHD and H3K4me3 peptide. The black dashed lines represent hydrogen bonds. The red boxes highlight those residues mutated in severe combined immunodeficiency (SCID) patients.
Fig. 2.
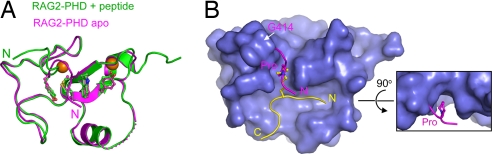
Structural comparison of RAG2-PHD with and without the H3 peptide. (A) Superposition of the RAG2-PHD crystal structures in the absence (magenta) and presence of H3K4me3 (green). Residues 471–473 are disordered in the apo form and represented by a dashed lane. The N terminus of the polypeptide chain is indicated. (B) The N-terminal peptide of RAG2-PHD partially occupies the H3-binding groove in the absence of H3 peptide. The molecular surface presentation of RAG2-PHD is shown with the bound H3K4me3 peptide (yellow wire), and superimposed on it is the N-terminal peptide of RAG2-PHD (pink wire) observed in the apo-structure. A proline residue, three residues N-terminal to the first residue of RAG2-PHD (G414), occupies the trimethyl ammonium-binding site.
Fig. 3.
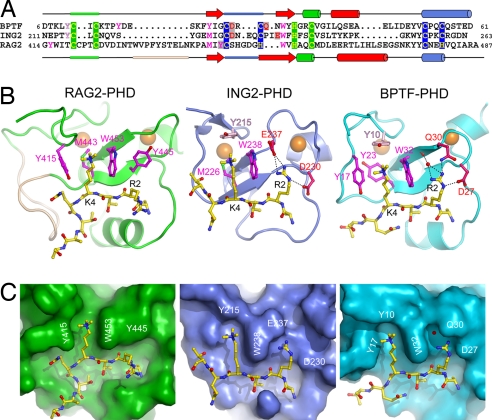
Comparison of RAG2-PHD with PHD fingers of ING2 and BPTF. (A) Sequence alignment of the three PHD fingers of RAG2, ING2, and BPTF. Zinc ligands (C or H) are highlighted in yellow on green (Zn1) or blue (Zn2) background; residues interacting with K4 are shown in magenta, and residues interacting with R2 are shown in red (native) or purple (methylated) on gray background. The tyrosine residue that closes the top of the aromatic cage in ING2 and BPTF and is absent in RAG2-PHD is shown in light pink. These residues are shown in the same color scheme in B. (B and C) Ribbon diagrams (B) and surface representation (C) of PHD fingers of RAG2, ING2 (PDB entry 2G6Q), and BPTF (PDB entry 2F6J) bound to H3K4me3 peptide. RAG2-PHD has an open aromatic channel and does not possess a negatively charged residue equivalent to E237 in ING2 or D27 in BPTF to interact with R2 of the H3 peptide.
The first six residues of the H3 peptide fit snugly in a sharply bent and branched binding groove on the surface of RAG2-PHD (Fig. 1B). The N-terminal amino group of the H3 peptide is hydrogen bonded with three carbonyl oxygens of RAG2-PHD, and the side chain of trimethylated K4 fits in a hydrophobic channel delimited by Y415 and W453 on the two sides and M443 in the back (Fig. 1 B and C). Residues R2-T3-K4me3 of the H3 peptide form a third β-strand adjacent to β1 and β2 of RAG2-PHD in a three-stranded antiparallel β-sheet. The H3 peptide is sharply kinked at Q5 like a saddle straddling the central helix of RAG2-PHD. The kinked H3 peptide is stabilized by the hydrogen bond between T3 and T6 within it. In addition, Q5 forms five direct or water-mediated hydrogen bonds with RAG2-PHD, including Y415 and T436 and the L438-N439-K440 loop (Fig. 1C).
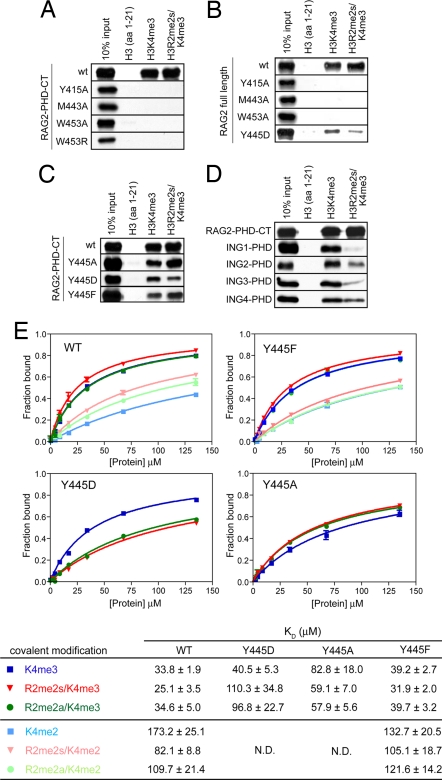
The trimethyl ammonium group of K4 is sandwiched between the aromatic side chains of Y415 and W453, which is reminiscent of the “aromatic cage” common among methyl-lysine-binding domains (8, 16, 17, 20, 21, 23–26, 40). Pull-down assays carried out with the RAG2-PHD-CT (residues 414–527) or with full-length RAG2 show that each of the three residues surrounding the K4me3 is essential for the H3 binding. Single substitution of Y415 or M443 with alanine, or W453 with alanine or arginine abolishes binding to H3K4me3 (Fig. 4 A and B).
Fig. 4.
RAG2-PHD prefers to bind H3 with the R2me2s and K4me3 double modification. (A–C) Pull-down assays of biotinylated native, singly (K4me3) and doubly modified (R2me2s/K4me3) H3 peptides by RAG2-PHD-CT fingers (residues 414–527) and mutations in the hydrophobic channel (A), full-length RAG2 (B), and RAG2-PHD-CT with the Y445 mutations (C). Mutations in the hydrophobic channel and of residue Y445 of RAG2 diminish the H3R2me2s/K4me3 binding. (D) Pull-down assay shows that methylation of R2 enhances binding of RAG2-PHD but decreases binding of various ING-PHD fingers. (E) Fluorescence anisotropy-based binding analysis of RAG2-PHD (414–487) and six differently modified H3 peptides (each with fluorescein label). The error bars represent deviations of triple measurements. The binding constants of wild-type, Y445D, Y445A, and Y445F mutant RAG2-PHD for the six H3 peptides are summarized in the table below the figure.
Comparison of PHD-H3K4me3 Structures.
Different proteins have evolved toward the same structural solution for binding di- or trimethylated lysine: the formation of a rigid cleft by a number of aromatic residues that stabilizes the methyl ammonium group by cation-π interactions. PHD fingers that bind methylated lysine all share a conserved tryptophan at the position equivalent to W453 (26) (Fig. 3A). This “aromatic cage” in BPTF-PHD is formed by four aromatic residues that interact with H3K4me3 from both sides, the back and the top (Fig. 3 B Right and C Right). ING2-PHD, on the other hand, lacks one aromatic residue on the side and has a methionine replacement in the back of the cage (Fig. 3 B Center and C Center). The aromatic cage of RAG2-PHD differs from all other structurally known methylated lysine-binding proteins in lacking a residue to close the cage beyond the tip of K4me3. Instead of a cage, the K4me3-binding site in RAG2-PHD is a channel that is open on the top and front (Fig. 3 B Left and C Left).
RAG2-PHD also differs from other H3K4me3-binding PHDs in its interactions with R2 of the H3 peptide. When complexed with the PHD finger of ING2 (24), BPTF (23), and Yng1 (25), the extended side chains of R2 and K4me3 of the H3 peptide occupy two adjacent parallel grooves separated by the conserved tryptophan equivalent to W453 of RAG2 (Fig. 3), and the guanidinium group of R2 forms salt bridges with a carboxylate, either Asp or Glu. Both the R2 binding groove and interacting carboxylate are absent in RAG2-PHD, and in their place is Y445. As predicted (26), the side chain of R2 extends toward solvent with no interaction with the RAG2-PHD (Fig. 3 B and C).
In total, 475 Å2 of the RAG2-PHD surface is buried upon the H3 peptide binding. This protein–peptide interface is ≈30% larger than those observed for ING2-PHD and BPTF-PHD (Fig. 3C). This difference is mainly due to an insertion of 10 residues in RAG2-PHD (depicted in wheat color in Fig. 3 A and B), which contains the L438-N439-K440 loop and interacts extensively with Q5 and T6 of the H3 peptide. As a result, the H3 peptide is sharply kinked at Q5 when bound to RAG2-PHD, whereas it is in an extended straight conformation in the complex with ING2 or BPTF (Fig. 3B). The H3-binding site of RAG2-PHD can thus be described as the crossroads of two grooves, one embracing the main chain of residues 1–4 and side chain of Q5, and the other holding the side chain of K4me3 and the main chain after Q5 (Fig. 3C).
RAG2-PHD Binds a “Cis-Peptide” in the Absence of H3 Peptide.
In the crystal structure of RAG2-PHD without the H3 peptide, the N-terminal eight residues carried over from the expression vector (SI Methods) occupy the K4Me3-binding site (Fig. 2). This “cis-peptide” enters the hydrophobic channel from the open end, and a proline residue mimics the trimethyl ammonium by interacting with Y415, W453, and M443 (Fig. 2). Binding of a proline between a tyrosine and tryptophan is commonly observed (41–43). The particular conformation of the cis-peptide may be stabilized by crystal lattice contacts because the same cis-peptide in the NMR model appeared to be disordered (36). However, several Pro residues in the RAG2 core are predicted to be on flexible loops, one of which may mimic the introduced proline and occupy the hydrophobic channel in solution. Mutations of R229 next to P230 on one such flexible loop have been widely observed in patients with Omenn syndrome (44).
The fact that a full-length RAG2 with mutations in the PHD finger is more deficient in V(D)J recombination than the RAG2 core without the zinc finger (30, 33–36), and the observations of an open K4me3 binding channel and its accommodation of a cis-peptide in the absence of H3K4me3 prompt us to postulate a potential mechanism for RAG2 auto-regulation. Binding of a self-peptide by RAG2-PHD may inhibit the RAG1/2 recombinase activity, and replacement of the self-peptide by the histone H3 methylated at lysine-4 may relieve such repression (see further discussion below).
RAG2-PHD Recognizes Symmetrically Dimethylated R2 of Histone H3K4me2/3.
The absence of a carboxylate in RAG2-PHD to form salt bridges with R2 leads us to hypothesize that the Y445, which replaces it, may interact with methylated R2. H3 peptides with monomethylation or dimethylation on the same (asymmetric, R2me2a) or different (symmetric, R2me2s) ω-N atoms of R2 and di- or trimethylation at K4 were synthesized (SI Table 2). Binding of H3 peptide with methylated lysine-4 and with doubly methylated R2 and K4 was first examined by pull-down assay. Both RAG2 and RAG2-PHD-CT appear to bind the H3K4me3/R2me2s slightly better than H3K4me3 (Fig. 4 A–D). The same assays showed that monomethylated arginine did not increase binding of RAG2-PHD-CT to H3K4me3 peptides (data not shown). The binding constants of these peptides to RAG2-PHD were then measured by two different fluorescence-based methods, the internal Trp fluorescence of W453 or fluorescence anisotropy of fluorescein attached to the H3 peptides (Fig. 4E and SI Fig. 7). Based on the fluorescence anisotropy, which gives stronger signals and higher precision, the lowest KD and hence the tightest binding was achieved with H3R2me2s/K4me3 (25.1 ± 3.5 μM), whereas H3K4me3 and H3R2me2a/K4me3 had slightly higher KD (both ≈34 μM). Such preference for H3 peptide methylated at both R2 and K4 is corroborated by the internal Trp fluorescence-based KD measurement, 31.0 ± 3.6 μM for H3R2me2s/K4me3 and 54.4 ± 7.3 μM for H3K4me3 (SI Fig. 7).
Because the difference in binding between singly and doubly modified peptides was <2-fold, the measurements were repeated multiple times to ascertain that the small differences are statistically significant relative to the experimental errors, deviations of measurements, and uncertainty of the curve fitting. The difference of KD due to symmetrically methylated R2 was increased to 2-fold when H3 contains K4me2, which binds RAG2-PHD less well than does K4me3 (Fig. 4E). It appears that the contribution of symmetrically methylated R2 to RAG2-PHD binding depends on the methylation state of K4. Both di- and trimethylated K4 of histone H3 have been detected at antigen receptor loci (12, 37). Recognition of R2 methylation by RAG2-PHD may enhance its weak association with H3K4me2 and allow it to compete against other H3K4me binding proteins, whose association with histone H3 is reduced by R2 methylation.
The KD of RAG2-PHD (25–50 μM) for the most preferred H3 peptide is ≈10-fold higher than that of the ING2-PHD and BPTF-PHD for the H3K4me3 peptide (2–5 μM) (23, 24). Because the number and chemical nature of interactions between the RAG2-PHD and H3 peptide are comparable if not greater than those made by ING2-PHD and BPTF-PHD (Fig. 3B), the relatively high KD is surprising. We suspect that the lack of a salt bridge between a carboxylate and H3R2 may account for the apparent high KD of RAG2-PHD. Disruption of the interaction with R2 in ING2-PHD or BPTF-PHD by replacing the carboxylate with Ala (D230A and D27A, respectively) reduced the affinity for H3K4m3 by ≈30-fold (23, 24). Similarly, when R2 is dimethylated, the affinity of ING2-PHD for H3K4me3 decreases ≈10-fold (SI Fig. 8). Competition by binding of the “cis-peptide” to RAG2-PHD may be another reason for the apparent low affinity. Furthermore, regions outside of the PHD finger may enhance binding of RAG2 to chromatin. As reported, a patch of acidic amino acids preceding the PHD finger is important for the interactions with histones, and point mutations in this region reduce endogenous V(D)J recombination (45).
Crystal Structures of RAG2-PHD Complexed with R2 and K4 Doubly Methylated H3.
To investigate the role of methylated R2, crystal structures of RAG2-PHD complexed with H3K4me3 peptides carrying monomethylated (H3R2me1/K4me3), asymmetric (H3R2me2a/K4me3), and symmetric dimethylated R2 (H3R2me2s/K4me3) and H3K4me2 with symmetric dimethylated R2 (H3R2me2s/K4me2) were determined at 2.0 Å or higher resolution (SI Table 3). By increasing the molar ratio of peptide to RAG2-PHD, the resulting crystals contain protein–peptide complexes only and no peptide-free proteins (see Materials and Methods).
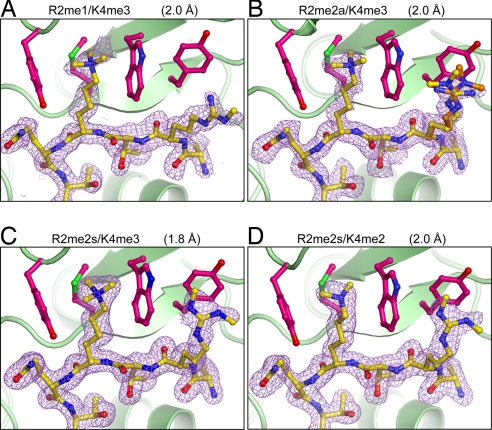
All of the structures are essentially identical except for the position of R2 on the H3 peptide. Monomethylated R2 behaves like unmethylated R2 and does not interact with RAG2 (Fig. 5A). Dimethylation of R2, whether symmetric or asymmetric, leads to interactions with Y445. In the H3R2me2a/K4me3 structure, the methylated guanidinium has to be modeled in two alternative conformations to best fit the diffuse electron density, which is within van der Waals contact distance of Y445 (Fig. 5B). The flexibility of R2me2a is interpreted as unstable association with RAG2-PHD. With R2me2s, the electron density defines a stable and discrete conformation for the methylated guanidinium (Fig. 5C). Identical interactions between Y445 and R2me2s are also observed in the complex of H3R2me2s/K4me2 (Fig. 5D). These structures together with the KD measurements suggest that RAG2-PHD prefers symmetrically dimethylated R2 and binds the R2me2s and K4me3 doubly modified H3 peptide the best. This is in clear contrast to the double chromodomains of CHD1 (20) and the ING2-PHD (Fig. 3A and SI Fig. 8), each of which binds the singly modified H3 peptide (K4me3) 4- to 5-fold better than H3 methylated at both R2 and K4. RAG2-PHD is an example of a single protein domain recognizing a doubly modified histone and preferred binding of H3 peptide with methylation at both R2 and K4. This is also the first example of a single protein domain recognizing a doubly modified histone.
Fig. 5.
Interactions of RAG2-PHD with different R2 and K4 doubly modified H3 peptides. (A) H3R2me1/K4me3. (B) H3R2me2a/K4me3 (alternate conformations of R2 shown in yellow and orange). (C) H3R2me2s/K4me3. (D) H3R2me2s/K4me2. The RAG2-PHD is shown as green ribbons, and the four key binding residues are shown as magenta ball-and-sticks. The H3 peptide in yellow ball-and-stick is shown with the corresponding 2Fo − Fc omit electron density map contoured at 1 σ.
Mutational Analysis of H3K4me3/R2me2s Recognition.
The role of Y445 in R2me2s recognition by RAG2-PHD was further analyzed by mutagenesis. Y445 was replaced by either Asp (Y445D) or Ala (Y445A). Because Y445 is closely packed against W453 (Fig. 1C) and likely stabilizes W453 and the whole PHD finger, the Y445 mutant proteins may not fold properly. Nevertheless, we were able to express and purify well behaved mutant forms of RAG2-PHD and RAG2-PHD-CT. The Y445D and Y445A mutant proteins still bind R2 and K4 methylated H3 peptides but do so with reduced affinity across the board (Fig. 4 C and E), which may be attributed to the destabilized W453 and PHD structure. In contrast to the wild type, the Y445D protein prefers to bind the singly modified (K4me3) over doubly modified (K4me3/R2me2) H3 peptide by 3- to 4-fold (Fig. 4 C and E). This result was further confirmed by pull-down assays using Y445D substituted full-length RAG2 (Fig. 4B). The Asp substitution may favor the native R2 perhaps by forming salt bridges with it as observed in other PHD fingers. However, the Y445A mutant proteins still prefer doubly methylated H3 peptide (Fig. 4 C and E). This preference may be explained by the similar hydrophobic nature of Y445 and Y445A that complements doubly methylated R2.
Y445 is not absolutely conserved among RAG2 proteins and is substituted by phenylalanine in low vertebrate species. In our pull-down and fluorescence-based binding assays, the Y445F mutant protein binds H3K4me3 peptide slightly less well than wild type, but the Y445F RAG2-PHD binds doubly modified H3 peptide better than H3K4me3 like the wild-type protein (Fig. 4 C and E).
Disease-Associated Mutations in RAG2-PHD.
Of the five severe combined immunodeficiency (SCID) mutations found in RAG2-PHD (33–35), the C478Y and H481P mutations directly alter Zn2+ coordination (Fig. 1A) and most likely disrupt the PHD structure, which in turn may lead to degradation of RAG2 in vivo (46). SCID patients carrying these two mutations have no T or B lymphocytes. The remaining three mutations, W453R, which eliminates the interaction with K4me3, and W416L and K440N, which affect either the hydrogen bond network with the H3 peptide or the stability of Y415 in the hydrophobic channel for K4me3 binding (Fig. 1C), are likely not to change the PHD finger structure. Coincidentally, the latter three mutations lead to Omenn syndrome with oligoclonal T cells, indicating that these mutant proteins retain abnormal V(D)J recombination activity at restricted loci. The correlation of these mutations and their structural and functional involvement in methylated histone H3 binding supports our hypothesis that RAG2-PHD may play an important regulatory role in V(D)J recombination.
Concluding Remarks.
The structure of RAG2-PHD illustrates the versatility of a histone-binding module like the PHD fingers and explains how diverse modifications in histones might be recognized. For instance, a single substitution of an aspartate/glutamate with tyrosine (Y445) allows RAG2-PHD to recognize R2me2 in addition to K4me3; the absence of another tyrosine in RAG2-PHD transforms the “aromatic cage” into a channel that may enable RAG2 a self-regulatory mechanism.
Recent studies indicate that methylation of arginine in histones by protein arginine methyltransferases (PRMTs) plays an important role in gene regulation (10, 47). Identification of PRMTs, their histone targets, and the downstream consequences of arginine methylation are an emerging field. So far, in mammalians, CARM1 and PRMT1 have been shown to methylate arginine asymmetrically, and PRMT5 catalyzes the symmetric dimethylation of arginine. H3R2 is a substrate of CARM1, but it has not yet been observed that H3R2 can undergo symmetric dimethylation. The physiological relevance of doubly methylated H3 (R2me2s/K4me3) in V(D)J recombination and PRMTs that methylate H3R2 symmetrically in the absence or presence of K4me3 awaits identification.
Materials and Methods
Crystallization and Structure Determination.
Details of protein and peptide preparation, crystallization, and structure determination can be found in SI Methods. Briefly, the first crystal structure was solved from a single crystal of RAG2-PHD–H3K4me3 complex by the SAD method. The subsequent structures were solved by Molecular Replacement.
All of the structural figures were generated by using PyMOL (http://pymol.sourceforge.net).
Mutagenesis and Pull-Down Assays.
Point mutations were introduced by using QuikChange (Stratagene) and verified by sequencing. Biotinylated histone peptides (1 μg each) (SI Table 2) were incubated with 1 μg of GST-fused PHD fingers or Flag-tagged RAG2 in the binding buffer [50 mM Tris·HCl, (pH 7.5), 300 mM NaCl, 0.1% Nonidet P-40] overnight at 4°C. After a 1-h incubation with streptavidin beads (Amersham), beads were washed three times with the binding buffer, and bound proteins were analyzed by Western blotting using an anti-GST antibody (Santa Cruz Biotechnology) or anti-Flag antibody (Sigma). GST-RAG2-PHD (414–487) and GST-RAG2-PHD (414–527), and full-length RAG2 were expressed and purified as described in ref. 36. Expression and purification of the GST-PHD fingers of hING1 (200–279), hING2 (200–281), hING3 (347–418), and hING4 (184–249) was carried out as reported in refs. 48 and 49.
Measurement of KD by Fluorescent Anisotropy.
The binding affinities between six fluorescein-labeled H3 peptides and RAG2-PHD (residues 414–487) without the GST tag were determined by fluorescence anisotropy as described in ref. 50. Briefly, serial 2-fold dilutions of RAG2-PHD in the binding buffer [20 mM Hepes (pH 7), 0.15 M NaCl, 1.4 mM 2-mercaptoethanol, 1 μM ZnSO4] were made to result in concentrations ranging from 150 μM to 1.2 μM. Each fluorescein-labeled peptide was added to the range of RAG2-PHD samples to a final concentration of 0.1–1 μM. After incubation at 22°C for 2 h, fluorescence anisotropy was measured by using the Photon Technology International Model C700 PTI-C700 at 22°C. The data were plotted as anisotropy versus the concentration of protein and fitted by using the program GraphPad Prism to the equation A = Af + (Ab − Af)[X/(KD + X)], where A is the measured anisotropy, Af the anisotropy of the peptide-free state, Ab is the anisotropy of the bound state, and X is the protein concentration. Measurements for each H3 peptide were repeated at least three times, and KD values were averaged over multiple independent experiments.
Measurement of KD by Tryptophan Fluorescence.
Association of RAG2-PHD and the H3 peptides used in crystallization was measured based on the internal fluorescence of W453 (RAG2-PHD) on a PTI-C700 at 22°C. Samples containing 10 μM RAG2-PHD in 20 mM Tris (pH 7.5), 0.15 M NaCl, 5% glycerol, 50 μM ZnSO4, and 1 mM DTT were mixed with increasing concentrations of each H3 peptide. After an incubation of 10–15 min, the fluorescence of W453 was measured with the incident wavelength of 295 nm, and the emission spectrum was collected at between 300 and 400 nm with a 0.5-nm step size and 0.3-s integration time, and averaged over three scans. The data were fitted by using GraphPad Prism, and KD values were averaged over three independent sets of data.
Supplementary Material
Acknowledgments
We thank Dr. S. Khorasanizadeh for generously providing the H3R2me2a/K4me3 peptide for initial structural studies, Drs. M. Gellert and D. Leahy for critical reading of the manuscript and insightful discussion, and Drs. K. Mizuuchi, T. Cellmer, and S. Vaiana for extensive help on fluorescence measurements. S.R.-M. thanks Drs. M. Nowotny and J. Y. Lee for help with diffraction data collection and structure determination. This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases/National Institutes of Health and grants from the National Institutes of Health (to M.A.O. and O.G.). O.G. is a recipient of a Burroughs Wellcome Career Award in Biomedical Sciences. A.G.W.M. is a Howard Hughes Medical Institute Predoctoral Fellow. S.R.-M. has been a recipient of a fellowship from the Human Frontier Science Program.
Abbreviations
- H3K4me3
histone H3 trimethylated at lysine-4
- H3R2me2s
histone H3 symmetrically dimethylated at arginine-2
- PHD
plant homeodomain.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors for the RAG2-PHD-H3 peptide complexes have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 2V83 (mixed apo and H3K4me3-bound crystal), 2V85 (H3R2me1/K4me3), 2V86 (H3R2me2a/K4me3), 2V88 (H3R2me2s/K4me2), and 2V87 (H3R2me2s/K4me3)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0709170104/DC1.
References
- 1.Gellert M. Annu Rev Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 2.Schatz DG, Oettinger MA, Baltimore D. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 3.Yancopoulos GD, Alt FW. Cell. 1985;40:271–281. [PubMed] [Google Scholar]
- 4.Golding A, Chandler S, Ballestar E, Wolffe AP, Schlissel MS. EMBO J. 1999;18:3712–3723. doi: 10.1093/emboj/18.13.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwon J, Imbalzano AN, Matthews A, Oettinger MA. Mol Cell. 1998;2:829–839. doi: 10.1016/s1097-2765(00)80297-x. [DOI] [PubMed] [Google Scholar]
- 6.Sleckman BP, Gorman JR, Alt FW. Annu Rev Immunol. 1996;14:459–481. doi: 10.1146/annurev.immunol.14.1.459. [DOI] [PubMed] [Google Scholar]
- 7.Jenuwein T, Allis CD. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 8.Khorasanizadeh S. Cell. 2004;116:259–272. doi: 10.1016/s0092-8674(04)00044-3. [DOI] [PubMed] [Google Scholar]
- 9.Shilatifard A. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 10.Wysocka J, Allis CD, Coonrod S. Front Biosci. 2006;11:344–355. doi: 10.2741/1802. [DOI] [PubMed] [Google Scholar]
- 11.Kouzarides T. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 12.Morshead KB, Ciccone DN, Taverna SD, Allis CD, Oettinger MA. Proc Natl Acad Sci USA. 2003;100:11577–11582. doi: 10.1073/pnas.1932643100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 14.Jacobson RH, Ladurner AG, King DS, Tjian R. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 15.Owen DJ, Ornaghi P, Yang JC, Lowe N, Evans PR, Ballario P, Neuhaus D, Filetici P, Travers AA. EMBO J. 2000;19:6141–6149. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacobs SA, Khorasanizadeh S. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 18.Min J, Zhang Y, Xu RM. Genes Dev. 2003;17:1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nielsen PR, Nietlispach D, Mott HR, Callaghan J, Bannister A, Kouzarides T, Murzin AG, Murzina NV, Laue ED. Nature. 2002;416:103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- 20.Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 21.Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Science. 2006;312:748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- 22.Shi X, Kachirskaia I, Walter KL, Kuo JH, Lake A, Davrazou F, Chan SM, Martin DG, Fingerman IM, Briggs SD, et al. J Biol Chem. 2007;282:2450–2455. doi: 10.1074/jbc.C600286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, Li H, Baker L, Boyle J, Blair LP, Chait BT, et al. Mol Cell. 2006;24:785–796. doi: 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruthenburg AJ, Allis CD, Wysocka J. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y. Nat Struct Mol Biol. 2006;13:572–574. doi: 10.1038/nsmb0706-572. [DOI] [PubMed] [Google Scholar]
- 28.Lindroth AM, Shultis D, Jasencakova Z, Fuchs J, Johnson L, Schubert D, Patnaik D, Pradhan S, Goodrich J, Schubert I, et al. EMBO J. 2004;23:4286–4296. doi: 10.1038/sj.emboj.7600430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 30.Akamatsu Y, Monroe R, Dudley DD, Elkin SK, Gartner F, Talukder SR, Takahama Y, Alt FW, Bassing CH, Oettinger MA. Proc Natl Acad Sci USA. 2003;100:1209–1214. doi: 10.1073/pnas.0237043100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirch SA, Rathbun GA, Oettinger MA. EMBO J. 1998;17:4881–4886. doi: 10.1093/emboj/17.16.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang HE, Hsu LY, Cado D, Cowell LG, Kelsoe G, Schlissel MS. Immunity. 2002;17:639–651. doi: 10.1016/s1074-7613(02)00448-x. [DOI] [PubMed] [Google Scholar]
- 33.Noordzij JG, de Bruin-Versteeg S, Verkaik NS, Vossen JM, de Groot R, Bernatowska E, Langerak AW, van Gent DC, van Dongen JJ. Blood. 2002;100:2145–2152. [PubMed] [Google Scholar]
- 34.Sobacchi C, Marrella V, Rucci F, Vezzoni P, Villa A. Hum Mutat. 2006;27:1174–1184. doi: 10.1002/humu.20408. [DOI] [PubMed] [Google Scholar]
- 35.Villa A, Sobacchi C, Notarangelo LD, Bozzi F, Abinun M, Abrahamsen TG, Arkwright PD, Baniyash M, Brooks EG, Conley ME, et al. Blood. 2001;97:81–88. doi: 10.1182/blood.v97.1.81. [DOI] [PubMed] [Google Scholar]
- 36.Elkin SK, Ivanov D, Ewalt M, Ferguson CG, Hyberts SG, Sun ZY, Prestwich GD, Yuan J, Wagner G, Oettinger MA, Gozani OP. J Biol Chem. 2005;280:28701–28710. doi: 10.1074/jbc.M504731200. [DOI] [PubMed] [Google Scholar]
- 37.Matthews AGW, Kuo AJ, Ramón-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, et al. Nature. 2007 doi: 10.1038/nature0643/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Callebaut I, Mornon JP. Cell Mol Life Sci. 1998;54:880–891. doi: 10.1007/s000180050216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bienz M. Trends Biochem Sci. 2006;31:35–40. doi: 10.1016/j.tibs.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 40.Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang X, Poy F, Zhang R, Joachimiak A, Sudol M, Eck MJ. Nat Struct Biol. 2000;7:634–638. doi: 10.1038/77923. [DOI] [PubMed] [Google Scholar]
- 42.Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP. Nat Struct Biol. 2000;7:639–643. doi: 10.1038/77929. [DOI] [PubMed] [Google Scholar]
- 43.Zarrinpar A, Bhattacharyya RP, Lim WA. Sci STKE. 2003;2003:re8. doi: 10.1126/stke.2003.179.re8. [DOI] [PubMed] [Google Scholar]
- 44.Marrella V, Poliani PL, Casati A, Rucci F, Frascoli L, Gougeon ML, Lemercier B, Bosticardo M, Ravanini M, Battaglia M, et al. J Clin Invest. 2007;117:1260–1269. doi: 10.1172/JCI30928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.West KL, Singha NC, De Ioannes P, Lacomis L, Erdjument-Bromage H, Tempst P, Cortes P. Immunity. 2005;23:203–212. doi: 10.1016/j.immuni.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 46.Li Z, Dordai DI, Lee J, Desiderio S. Immunity. 1996;5:575–589. doi: 10.1016/s1074-7613(00)80272-1. [DOI] [PubMed] [Google Scholar]
- 47.Bedford MT, Richard S. Mol Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 48.Gozani O, Karuman P, Jones DR, Ivanov D, Cha J, Lugovskoy AA, Baird CL, Zhu H, Field SJ, Lessnick SL, et al. Cell. 2003;114:99–111. doi: 10.1016/s0092-8674(03)00480-x. [DOI] [PubMed] [Google Scholar]
- 49.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, et al. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jacobs SA, Fischle W, Khorasanizadeh S. Methods Enzymol. 2004;376:131–148. doi: 10.1016/S0076-6879(03)76009-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.