Abstract
Dactylaplasia, characterized by missing central digital rays, is an inherited mouse limb malformation that depends on two genetic loci. The first locus, Dac, is an insertional mutation around the dactylin gene that is inherited as a semidominant trait. The second locus is an unlinked modifier, mdac/Mdac, that is polymorphic among inbred strains. Mdac dominantly suppresses the dactylaplasia phenotype in mice carrying Dac. However, little is known about either locus or the nature of their interaction. Here we show that Dac is a LTR retrotransposon insertion caused by the type D mouse endogenous provirus element (MusD). This insertion exhibits different epigenetic states and spatiotemporally expresses depending on the mdac/Mdac modifier background. In dactylaplasia mutants (Dac/+ mdac/mdac), the LTRs of the insertion contained unmethylated CpGs and active chromatin. Furthermore, MusD elements expressed ectopically at the apical ectodermal ridge of limb buds, accompanying the dactylaplasia phenotype. On the other hand, in Dac mutants carrying Mdac (Dac/+ Mdac/mdac), the 5′ LTR of the insertion was heavily methylated and enriched with inactive chromatin, correlating with inhibition of the dactylaplasia phenotype. Ectopic expression was not observed in the presence of Mdac, which we refined to a 9.4-Mb region on mouse chromosome 13. We report a pathogenic mutation caused by MusD. Our findings indicate that ectopic expression from the MusD insertion correlates with the dactylaplasia phenotype and that Mdac acts as a defensive factor to protect the host genome from pathogenic MusD insertions.
Keywords: dactylin, ectrodactyly, LTR, split hand/split foot malformation, methylation
Dactylaplasia is an inherited mouse limb malformation that is characterized by missing central digital rays. The Dac mutation is inherited as a semidominant trait, evidenced by missing central digits in the fore- and hindlimbs of heterozygous mice and monodactyly in homozygous mice (1, 2). The Dac locus has been mapped to the distal end of chromosome 19. Two independent Dac mutant alleles, Dac1J and Dac2J, arose spontaneously in breeding colonies. Both are insertions residing within the same locus: Dac1J is located in the region upstream of the dactylin gene, and Dac2J is located in intron 5 of dactylin (3). Southern blot analysis indicated that both insertions are larger than 4.5 kb; however, “jumping PCR” identified only the LTR portion of the insertion (3). Partial PCR products terminating in the 5′ and 3′ LTRs cross-prime each other, resulting in amplified products that lack any of the internal sequence between LTRs. Therefore, these insertions were thought to be caused by an early transposon (ETn) element, which is a common mutagen in mice (4, 5).
Split hand/split foot malformation (SHFM) in humans is a congenital limb malformation that has an ectrodactyly phenotype analogous to that of the dactylaplasia mouse. SHFM is genetically heterogeneous; to date, five different loci, SHFM1 to SHFM5, have been mapped. Dactylaplasia is a mouse model of SHFM3 because the Dac locus is syntenic to the SHFM3 locus (10q24) (2, 6–9). Recently, 0.5-Mb tandem genomic duplications were identified at 10q24 in several SHFM3 families (10–13). The smallest duplicated region contained a disrupted extra copy of the dactylin gene and the LBX1, BTRC, POLL, and DPCD genes in their entirety. The dactylin gene encodes an F-box/WD40 repeat protein; members of this protein family commonly function in ubiquitin-dependent proteolytic pathways (14). Although the dactylin gene is considered to be the best candidate for SHFM3 and the mouse dactylaplasia phenotype, its specific function remains undetermined. In mice, the Dac2J insertion in intron 5 of dactylin results in the absence of the normal dactylin mRNA transcript; in contrast, the Dac1J insertion in the upstream region affects neither the size nor the amount of dactylin transcript (3). Furthermore, human SHFM3 patients demonstrate only partial dactylin duplications (10, 11). Therefore, the possibility exists that the dactylin gene itself is simply a bystander and that Dac insertions have long-range regulatory effects on neighboring genes.
The mouse dactylaplasia phenotype depends not only on the genotype at the mutated Dac locus but also on homozygosity for a recessive allele in another unlinked locus, mdac, which has been mapped to chromosome 13, between D13Mit10 and D13Mit99 (2). This locus is polymorphic among inbred strains, and two alleles have been identified. Inbred strains such as BALB/cJ, A/J, and 129/J carry mdac, which permits Dac expression; on the other hand, inbred strains such as CBA/J, C3H/J, and C57BL/6J carry the Mdac allele, which dominantly inactivates Dac (2). The Dac1J and Dac2J alleles are equally sensitive to Mdac (3). Therefore, the dactylaplasia phenotype is observed in only mice homozygous for mdac (Dac/+ mdac/mdac or Dac/Dac mdac/mdac) and is never observed in mice carrying Mdac, regardless of their Dac status (Dac/+ Mdac/mdac or Dac/Dac Mdac/mdac). Mutational insertions in the Dac locus have been identified and partially cloned; however, little is known about the insertional mutation or its modifier.
The present study aimed to characterize Dac, Mdac, and interactions between the two loci to elucidate the pathological mechanism of dactylaplasia. Our study found that both Dac insertions are caused by a type D mouse endogenous provirus (MusD) element. We observed a correlation between the dactylaplasia phenotype and the epigenetic status of the MusD insertion, which is modulated by its modifier. Furthermore, we show ectopic expression of MusD and its related elements in Dac mutant limb buds. These observations demonstrate the impact of retrotransposon insertions in the genome, as well as a host defensive mechanism against retrotransposons.
Results
Characterization of Dac Insertions.
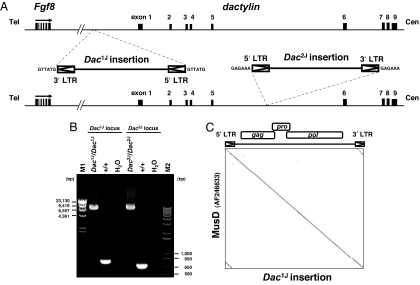
Two independent, spontaneously arising Dac mutant alleles, Dac1J and Dac2J, are caused by insertions around the dactylin gene. Each was previously partially cloned (3). To further characterize these insertions, we isolated each by PCR and sequenced them directly (Fig. 1A). Our analysis identified the insertions as LTR retrotransposons with lengths of 7,486 bp (Dac1J insertion; AB305072) and 7,473 bp (Dac2J insertion; AB305073), each containing 6-bp target-site duplications (Fig. 1B). Interestingly, the two sequences were 99.6% identical, and they shared sequence identity with an LTR retrotransposon (AF246633) that was originally reported as a MusD element (15) (Fig. 1C). The Dac1J insertion was integrated 10 kb upstream of the dactylin gene in antisense orientation, whereas the Dac2J insertion was integrated in intron 5 of dactylin in sense orientation (Fig. 1A). The Dac2J insertion site was identified at position 121,540 bp on AC003694 by inverse PCR before isolation of the Dac2J insertion site (data not shown). Each insertion had 100% identical 5′ and 3′ LTRs and contained intact ORFs for gag, pro, and pol genes.
Fig. 1.
Characterization of Dac insertions. (A) Insertions of LTR retrotransposons around the dactylin gene. The Dac1J insertion was integrated 10 kb upstream of the dactylin gene in antisense orientation. The Dac2J insertion was integrated into intron 5 of the dactylin gene in sense orientation. (B) PCR amplification of the insertions. The Dac1J and Dac2J insertions were 7,486 and 7,473 bp, respectively, and were 99.6% identical in sequence. M1, Lambda DNA-HindIII digest (New England Biolabs, Beverly, MA); M2, 1Kb Plus DNA Ladder (Invitrogen). (C) Dot plot DNA comparison of the Dac1J insertion and the MusD element (AF246633). The stringency of comparison was 19 of 23. Each Dac insertion shared 100% identity within the 5′ and 3′ LTRs and contained intact ORFs for the gag, pro, and pol genes.
DNA Methylation and Histone Modification of Dac Insertions.
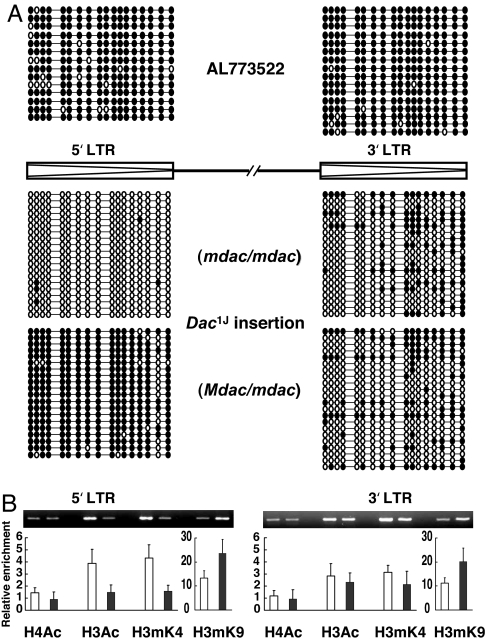
Dactylaplasia mice show a wide range of phenotypic variation despite their identical genotypes, suggesting an epigenetic effect on phenotype severity. To investigate the epigenetic status of these insertions, bisulfite sequencing and ChIP studies were performed for the Dac1J insertion by using freshly prepared embryonic tissues. An unrelated MusD element (AL773522) in the mouse genome was used as a control for each experiment. This element shows the greatest sequence similarity to Dac insertions in the mouse genome; its 5′ and 3′ LTRs are 100% identical, containing 18 CpGs, but it has an ORF-disrupting mutation in the pol gene due to a 1-bp deletion. It is known that most LTR retrotransposons in somatic cells are maintained in a heavily methylated and silent state (16). As expected, bisulfite sequencing showed that both the 5′ and 3′ LTRs of the AL773522 element were heavily methylated in Dac mutant mice (Dac1J/+ mdac/mdac) (Fig. 2A, Top). However, the Dac1J insertion carried an unmethylated 5′ LTR and relatively hypomethylated 3′ LTR even in somatic cells, exhibiting some interclone and intermouse variation (Fig. 2A, Middle). We saw no significant difference in DNA methylation status of the several tissue types analyzed for this study, including brain, liver, kidney, and tail (data not shown).
Fig. 2.
Epigenetic modification of the Dac insertion. (A) DNA methylation profile for each CpG in the LTRs of the Dac1J insertion. Bisulfite-treated genomic DNA derived from Dac mutants in either the mdac/mdac or Mdac/mdac background was subjected to LTR amplification. PCR fragments were subcloned and sequenced. LTRs of the MusD element on AL773522 were analyzed as a control. Open circles, unmethylated CpGs; filled circles, methylated CpGs. Each line represents the sequence of one clone, and each block of clones represents the data from one mouse. (B) ChIP analysis of LTRs from the Dac1J insertion in either the mdac/mdac (white columns) or Mdac/mdac (black columns) background. Mononucleosomes were prepared from fresh embryos and subjected to ChIP by using antibodies against acetylated H4, acetylated H3, methylated H3-Lys-4 (H3mK4), and methylated H3-Lys-9 (H3mK9). Quantitative PCR analysis with SYBR Green was performed, and the data were summarized after normalizing either to the MusD element on AL773522 [acetylated histone H4 (H4Ac), acetylated histone H3 (H3Ac), and H3mK4] or Actb (H3mK9); levels for both were set to 1.0.
The ChIP assay revealed a correlation between DNA methylation and histone modification status within the LTRs of the Dac1J insertion. The unmethylated 5′ LTR of the Dac1J insertion (Dac1J/+ mdac/mdac) consisted of active chromatin enriched with acetylated histone and H3-Lys-4 methylation (Fig. 2B). We also performed bisulfite sequencing analysis on primary cultured embryonic fibroblasts but found that the DNA methylation status in fibroblasts differed from that of original fresh tissues, even after several passages (data not shown). Modified epigenetic status sometimes is observed during the establishment of a cell line, which does not represent the original status (17). Therefore, only embryonic tissue was used for both bisulfite sequencing and the ChIP assay.
Ectopic Expression of MusD Elements in Dactylaplasia Mutant Mice.
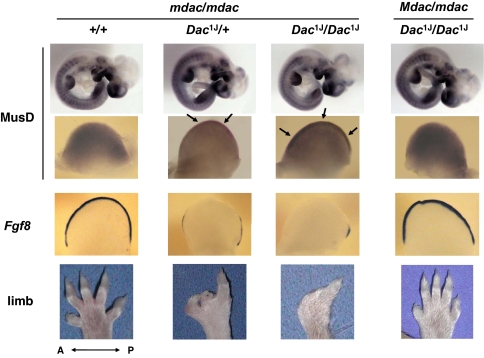
Recently, a tissue- and stage-specific expression pattern for MusD and ETn elements has been shown in organogenesis-stage mouse embryos (18). To examine the MusD expression pattern in dactylaplasia mice, we generated a MusD antisense strand probe from the 3′ UTR region of the MusD element for hybridization to embryos in various developmental stages [embryonic day (E)9.5–E11.5] (Fig. 3). Expression was observed in both fore- and hindlimb buds and confined to the mesenchyme in WT embryos, as reported previously (18). Interestingly, heterozygous dactylaplasia mutant (Dac1J/+ mdac/mdac) embryos exhibited aberrant MusD expression at the central portion of the apical ectodermal ridge (AER) in addition to its normal expression. In homozygous mutants (Dac1J/Dac1J mdac/mdac), this ectopic expression was more apparent and more widely dispersed across the AER. Expression was strongest in anterior limb buds on E10.5 and in posterior limb buds on E11 because there is a 0.5-day developmental delay of the hindlimbs with respect to the forelimbs (data not shown). These mutants showed no aberrant expression other than in limb buds.
Fig. 3.
Abnormal expression of MusD/ETn elements in dactylaplasia mutant limb buds. In situ hybridization to assess MusD/ETn and Fgf8 expression in whole embryos and their anterior limb buds collected at E10.5 and the representative phenotype of each genotype. Arrows indicate aberrant expression in the AER of the dactylaplasia mutant limb bud.
Effect of Mdac.
To investigate the effect of Mdac, dactylaplasia mice were crossed with C57BL/6J mice that carry Mdac. DNA from F1 hybrid embryos (Dac1J/+ Mdac/mdac) was subjected to bisulfite sequencing. Contrary to the unmethylated 5′ LTR seen in the dactylaplasia mutant (Dac1J/+ mdac/mdac), the 5′ LTR of the Dac1J insertion in the Mdac background was heavily methylated, although the 3′ LTR was unaffected (Fig. 2A, Bottom). We saw no significant difference in methylation status between the sense and antisense strands of either LTR (data not shown). The methylated 5′ LTR of the Dac1J insertion (Dac1J/+ Mdac/mdac) was depleted of histone acetylation but was enriched for methylated H3-Lys-9 (Fig. 2B). The 3′ LTR showed no change in histone modification status in the presence of Mdac, just as its DNA methylation status was unaffected.
Ectopic expression seen in the dactylaplasia mutant (Dac1J/Dac1J mdac/mdac) was not observed in embryos carrying Mdac (Dac1J/Dac1J Mdac/mdac), correlating with its lack of a dactylaplasia phenotype (Fig. 3). Fgf8 was used as a control AER marker because dactylaplasia is known to demonstrate partial loss of Fgf8 expression in the AER of mutant limb buds (3, 19). Fgf8 expression was restored in embryos with Mdac, which exhibited no ectopic expression of MusD.
The Dac2J mutant shared features with the Dac1J mutant. The 5′ LTR of the Dac2J insertion also showed two contrasting epigenetic states dependent on the mdac/Mdac modifier background. In situ hybridization of Dac2J mutant embryos similarly showed aberrant MusD expression and loss of Fgf8 expression in the AER, which were restored by the presence of Mdac (data not shown).
The Dac2J mutation is characterized by the absence of the normal dactylin mRNA transcript (3). To see whether the dactylin transcript was restored in the presence of the Mdac, RNA derived from E10.5 embryos was subjected to quantitative real-time RT-PCR. To our surprise, Mdac did not affect the dactylin gene. The dactylin transcript in the Dac2J mutant was still absent despite a lack of dactylaplasia phenotype (Dac2J/Dac2J Mdac/mdac) [supporting information (SI) Fig. 5].
To investigate whether or not Mdac affects other loci, we looked for other unmethylated LTR retrotransposons in the mouse genome. A genome-wide in silico search for young elements, which have 100% identical 5′ and 3′ LTRs and display insertional polymorphisms in mouse strains, resulted in the identification of another unmethylated LTR retrotransposon (ETnII) on mouse chromosome 17. The ETnII was present in the Dac1J parental strain (NW_001030622) but was absent in the Dac2J parental strain and C57BL/6J (NT_039649). When compared with an unmethylated 5′ LTR of the ETnII in the Dac1J parental strain (mdac/mdac), the 5′ LTR was slightly methylated in F1 hybrids of the Dac1J strain and C57BL/6J (Mdac/mdac), exhibiting two distinct populations of PCR clones (SI Fig. 6).
Mapping of Mdac.
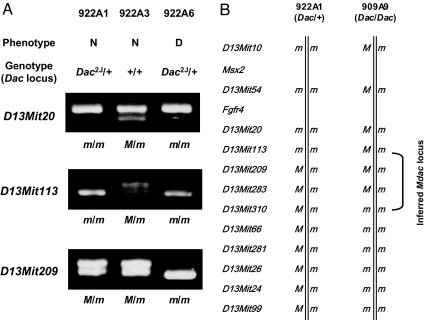
Mdac has been mapped to a 28-Mb region in the middle of chromosome 13, between D13Mit10 and D13Mit99 (2). To refine the Mdac locus, the dactylaplasia mouse (Dac/+ mdac/mdac) was crossed with C57BL/6J (+/+ Mdac/Mdac) to produce F1 hybrids (Dac/+ Mdac/mdac). The F1 hybrids were then backcrossed to the dactylaplasia parental strain (Dac/+ mdac/mdac). These backcrosses were typed for both Dac and microsatellite markers on chromosome 13, and phenotypes were compared. We analyzed 309 offspring obtained from the backcrossing test (207 of Dac1J line and 102 of Dac2J line). Eleven of 309 offspring exhibited recombination events that were informative for further mapping between D13Mit10 and D13Mit99. Among these, two independent recombination events placed Mdac within a 9.4-Mb interval region between D13Mit113 and D13Mit310 (Fig. 4). Two mice (922A1 and 909A9) exhibited a normal phenotype despite carrying the Dac mutant allele. These mice led us to exclude markers that were homozygous for the dactylaplasia parental strain, because both mice must have inherited Mdac from the C57BL/6J strain. Backcrossing of each Dac mutant line showed consistent results, indicating that Dac1J and Dac2J are both sensitive to Mdac.
Fig. 4.
Mapping of Mdac on mouse chromosome 13. F1 hybrids (Dac/+ Mdac/mdac) were produced by crossing the dactylaplasia mouse (Dac/+ mdac/mdac) and C57BL/6J (+/+ Mdac/Mdac) and then backcrossed to the dactylaplasia parental strain. These backcrosses were typed for both Dac and microsatellite markers on chromosome 13, and phenotypes were compared. (A) Recombination between D13Mit113 and D13Mit209 in a mouse 922A1. The mouse 922A1 exhibited normal phenotype despite carrying the Dac2J mutant allele. Microsatellite markers that were homozygous (D13Mit20 and D13Mit113) were excluded from the Mdac locus because this mouse must have the Mdac allele. N, normal; D, dactylaplasia; m, allele inherited from the dactylaplasia parental strain; and M, allele inherited from the C57BL/6J strain. (B) Recombination events in two offspring (922A1 and 909A9) placed Mdac within a 9.4-Mb interval region between D13Mit113 and D13Mit310. Both offspring exhibited a normal phenotype despite carrying the Dac mutant allele, indicating the presence of Mdac.
Discussion
We have characterized the Dac1J and Dac2J insertions from Dac mutant alleles on mouse chromosome 19. These insertions were identified as almost identical MusD elements, containing ORFs for gag, pro, and pol proteins of D-type virus. MusD is known to trigger the mobilization of ETn elements, which are the most active murine mobile elements and cause the majority of insertional mutations in mice (4, 5, 20, 21). Although the MusD itself has been shown previously to be autonomous for transposition in a tissue culture assay, no pathegnic MusD insertion has been identified to date (20). We have identified in vivo a MusD element as a de novo and pathogenic insertion.
It is noteworthy that two independent MusD insertions were identified at the same locus; however, it is still unclear how these MusD insertions lead to the dactylaplasia phenotype. In the Dac2J mutant allele, the MusD element was inserted at intron 5 of the dactylin gene. Owing to this mutation, dactylin transcripts are absent, suggesting that disruption of dactylin causes the dactylaplasia phenotype. On the other hand, the Dac1J insertion, which resides in the upstream region of dactylin, affects neither the amount nor the size of the dactylin transcript (3). Furthermore, the dactylin transcript was also absent in Dac2J mice carrying the Mdac allele (Dac2J/Dac2J Mdac/mdac), which show no dactylaplasia phenotype (SI Fig. 5). These data suggest that dactylin transcript levels are not essential for the dactylaplasia phenotype.
The dactylaplasia mouse is an ideal phenotypic and genotypic model for human ectrodactyly (SHFM3) because the responsible SHFM3 locus on human 10q24 is homologous to the Dac locus on mouse chromosome 19 (2, 6–9). Recently, genomic rearrangements have been identified in several unrelated SHFM3 families (10–13). Each was a tandem genomic duplication consisting of a disrupted extra copy of the dactylin gene and the entire LBX1, BTRC, POLL, and DPCD genes. No other mutations have been identified in SHFM3 patients to date. Overdosage of genes within the duplicated locus might be responsible for SHFM3. Similarly, the Dac insertion might cause overexpression of these genes. LTR retrotransposons are known to affect transcription of neighboring genes because the LTR has both sense and antisense promoter activity (22). We quantified LBX1, BTRC, POLL, and DPCD mRNA levels in dactylaplasia embryos but saw no significant changes in expression relative to WT (data not shown).
Aberrant expression of MusD elements in the AER was observed in dactylaplasia mutant embryos (Dac/+ mdac/mdac or Dac/Dac mdac/mdac) but not in WT embryos (+/+ mdac/mdac) or dactylaplasia embryos carrying Mdac (Dac/Dac Mdac/mdac). These observations suggest the possibility that the Dac insertion itself is expressed in the AER of mutant limb buds, although our probe detects not only transcripts from the Dac insertion but also other MusD and ETn transcripts because of sequence similarity. The Dac insertion is presumed to be poorly transcribed in the presence of Mdac because of its heavily methylated status. More interestingly, the ectopic expression seems to be correlated with spatiotemporal loss of Fgf8 expression in the AER (19). Decreased expression of Fgf8 in the dactylaplasia mouse is thought to be caused by degeneration of the AER. Fgf8-null mice result in early embryonic lethality, whereas heterozygous Fgf8 mutants show normal limb development. On the other hand, conditional knockout studies demonstrated that Fgf8 deficiency in the AER causes hypoplasia or aplasia of the limb (23, 24).
It is noteworthy that Fgf8 resides on mouse chromosome 19, only 70 kb away from the dactylin gene in head-to-tail orientation (Fig. 1A). An intracisternal A particle insertion in the nearby agouti gene, which controls coat color, is known to cause aberrant expression of the agouti gene. Different methylation states of the intracisternal A particle insertion in different cells lead to patchy coat color (25). Therefore, it cannot be ruled out that the active Dac insertion affects its neighboring gene, Fgf8. An external transcript from the Dac insertion toward Fgf8, which would be antisense for Fgf8, could cause Fgf8 repression in trans by the RNAi machinery. We investigated this possibility by conducting RT-PCR and whole mount in situ hybridization; however, we failed to detect antisense Fgf8 transcript or any transcription from the Dac insertion to Fgf8 (data not shown).
The 5′ LTR of the Dac insertion showed two contrasting epigenetic states that were dependent on the presence of Mdac. Although most retrotransposons are heavily methylated and inactivated in somatic cells, little is known about the mechanism by which the host genome suppresses transposable elements. Potential strategies include transcriptional silencing (DNA methylation and/or chromatin modification), posttranscriptional silencing (RNAi), and mutational inactivation (cytosine deaminases). Some apolipoprotein B mRNA editing complex (APOBEC) proteins are known to suppress LTR retrotransposons, including the intracisternal A particle and MusD, by either deaminase-dependent or unknown mechanisms (26–28). APOBEC proteins work primarily on transcribed LTR retrotransposons by inducing mutations in the transposed copies or by reducing cDNA levels. Thus, Mdac might differ from the APOBEC family because Mdac appears to work as a pretranscription barrier, causing DNA and/or histone methylation.
The MusD insertion seems to act as a “controlling element” in the dactylin locus. Barbara McClintock first discovered transposable elements in the 1940s during studies of maize and called them “controlling elements” because they had the ability to alter normal patterns of gene expression in a variety of ways (29). These alterations are dependent on the activity state of the transposable element (30). Recent studies have provided details of the molecular events underlying these epigenetic phenomena not only in maize (31) but also in Arabidopsis (32, 33), Drosophila (34), and mice (25, 35, 36). Although many other controlling elements have been identified and characterized in different species, the mechanism by which the host genome regulates these controlling elements is not well understood.
Our backcross study has narrowed the Mdac/mdac locus to a 9.4-Mb interval between D13Mit113 and D13Mit310 on mouse chromosome 13. Bisulfite sequencing and ChIP studies suggest that Mdac is a DNA methylation, histone-modifying enzyme or a molecule involved in the RNAi pathway that recognizes retrotransposons; however, no known enzymes have been mapped in this region. The target of Mdac, or how Mdac recognizes it, is unclear. Most LTR retrotransposons are methylated and suppressed in mice regardless of Mdac status, suggesting that Mdac only recognizes active MusD and/or other LTR elements or that Mdac activity depends on genome position and is effective only around the dactylin gene.
During the study of another unmethylated ETnII element in the Dac1J parental strain, the 5′ LTR of the ETnII exhibited a slight increase in DNA methylation when crossed with C57BL/6J. Mdac may affect other active LTR retrotransposons regardless of their genomic locations; however, we cannot rule out the possibility that the C57BL/6J strain carries another modifier which recognizes this ETnII. Transgenic mouse study of Mdac in the mdac/mdac background would be necessary to investigate the effect of Mdac on other epigenetic sensitive loci.
Of great interest are the observations that mouse dactylaplasia and human SHFM3 are caused by a MusD insertion and genomic duplication, respectively. Further study is necessary to understand the link between dactylaplasia and SHFM3 and to elicit the mechanisms underlying these malformations. Moreover, identification of Mdac will shed light on the mechanisms by which host genomes silence transposable elements.
Materials and Methods
All primer sequences are listed in SI Table 1. All animal studies were performed in compliance with Osaka University guidelines.
Isolation and Sequencing of Dac Insertions.
We amplified each Dac insertion by using long primers that flanked the insertion. PCR was carried out in a final volume of 50 μl containing 1× LA PCR buffer II (Mg2+ free; Takara, Shiga, Japan), 200 nM each primer, 250 μM each dNTP, 500 ng of genomic DNA, 2.5 mM MgCl2, and 2.5 units of Ex TaqDNA polymerase (Takara) by using the GeneAmp PCR system 9700 (Applied Biosystems, Foster City, CA). Cycling conditions were as follows: initial denaturation at 95°C for 1 min, followed by the two-step profile: denaturation at 98°C for 10 s and annealing–extension at 68°C for 20 min for 10 cycles, then autoextension by 20-s per cycle for an additional 20 cycles. PCR products were purified by using the QIAquick PCR purification kit (Qiagen, Valencia, CA) and sequenced directly by using BigDye Terminator v3.1 (Applied Biosystems) and an internal primer.
Bisulfite Sequencing.
DNA (3 μg) extracted from embryonic tissue (E9–E11.5) was digested with EcoRI and treated with sodium bisulfite according to standard protocols. The bisulfite-treated DNA was resuspended in 30 μl of TE [10 mM Tris/1 mM EDTA (pH 8.0)], 2 μl of which was used in the PCR to amplify each LTR. PCR fragments were subcloned into the pCR4-TOPO vector (Invitrogen, Carlsbad, CA) and then sequenced by using M13Forward or M13Reverse standard primer.
Genotyping of Dac and Physical Mapping of Mdac.
Genotyping of the Dac allele was performed by using multiplex PCR analysis. DNA was isolated from the amniotic membrane or the tail. Noon of the day on which a vaginal plug was detected was considered E0.5. To refine the Mdac locus, the dactylaplasia mouse (Dac/+ mdac/mdac) was crossed with C57BL/6J (+/+ Mdac/Mdac) to produce F1 hybrids (Dac/+ Mdac/mdac). The F1 hybrids were then backcrossed to the dactylaplasia parental strain (Dac/+ mdac/mdac). These backcrosses were typed for both Dac and microsatellite markers on chromosome 13, and phenotypes were compared.
Microsatellite markers used for mapping of Mdac were D13Mit10, D13Mit54, D13Mit20, D13Mit113, D13Mit209, D13Mit283, D13Mit310, D13Mit66, D13Mit281, D13Mit26, D13Mit24, and D13Mit99, which were informative for the backcrossing test between dactylaplasia strains and C57BL/6J. One primer of each pair was labeled by 6-carboxyfluorescein at the 5′ end. Fluorescent PCR products were subjected to electrophoresis on the gel and/or analyzed by Automated Fluorescent DNA Sequencer by using GeneScan software (ABI 3100; Applied Biosystems). We excluded the following markers from the Mdac locus by the backcrossing test. (i) Homozygous markers of a mouse which exhibits normal phenotype despite carrying the Dac mutant allele. This mouse must have Mdac (Dac/+ Mdac/mdac or Dac/Dac Mdac/mdac). (ii) Heterozygous markers of a mouse which exhibits dactylaplasia phenotype. The mdac must be homozygous to exhibit dactylaplasia phenotype (Dac/+ mdac/mdac or Dac/Dac mdac/mdac). Primer sequences used to amplify microsatellite markers were derived from information available at Mouse Genome Informatics (www.informatics.jax.org).
Whole Mount in Situ Hybridization.
For whole mount in situ hybridization, embryos were collected and fixed in 4% paraformaldehyde in PBS. Digoxigenin-labeled probes were generated by transcription (Roche, Indianapolis, IN) from amplification products generated by RT-PCR from embryonic total RNA and containing T7 (sense strand) or SP6 (antisense strand) RNA polymerase promoters. In situ hybridizations were performed according to standard protocols. The sense strand probe was used as a negative control. A minimum of four embryos were analyzed for each genotype, and at least two separate experiments were conducted for each probe.
ChIP Assay and Quantitative Real-Time PCR.
The ChIP assay was performed according to the protocol provided by the manufacturer (Upstate Biotechnology, Lake Placid, NY), with slight modification. We prepared ≈3 × 107 cells from homogenized fresh embryos (E10.5) and fixed them in 1% formaldehyde. Antibodies used for ChIP were anti-acetylated histone H4, anti-acetylated histone H3, anti-dimethyl histone H3-Lys-4, and anti-dimethyl histone H3-Lys-9 (Upstate Biotechnology). DNA recovered from immunoprecipitated complexes was subjected to quantitative real-time PCR with SYBR Green PCR Master Mix by using an ABI PRISM 7900HT (Applied Biosystems) as described previously (37, 38). Primers were designed to cover each LTR and the Actb promoter region. The data were summarized after normalizing either to the MusD element on AL773522 (anti-acetylated histone H4, anti-acetylated histone H3, and anti-dimethyl histone H3-Lys-4) or to Actb (anti-dimethyl histone H3-Lys-9); levels for both were set to 1.0. ChIP was performed by using at least five embryos for each genotype, and PCRs were performed at least twice for each sample.
Supplementary Material
Acknowledgments
We thank Drs. Kazuhiro Kobayashi, Nobuhiro Fujikake, and Yoshitaka Nagai for helpful comments and Dr. Jennifer Logan for editing the manuscript. This work was supported by the 21st Century Centers of Excellence program of the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Abbreviations
- AER
apical ectodermal ridge
- E
embryonic day
- ETn
early transposon
- MusD
the type D mouse endogenous provirus element
- SHFM
split hand/split foot malformation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the DNA Data Bank of Japan (accession nos. AB305072 and AB305073).
See Commentary on page 18879.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705483104/DC1.
References
- 1.Chai CK. J Hered. 1981;72:234–237. doi: 10.1093/oxfordjournals.jhered.a109486. [DOI] [PubMed] [Google Scholar]
- 2.Johnson KR, Lane PW, Ward-Bailey P, Davisson MT. Genomics. 1995;29:457–464. doi: 10.1006/geno.1995.9981. [DOI] [PubMed] [Google Scholar]
- 3.Sidow A, Bulotsky MS, Kerrebrock AW, Birren BW, Altshuler D, Jaenisch R, Johnson KR, Lander ES. Nat Genet. 1999;23:104–107. doi: 10.1038/12709. [DOI] [PubMed] [Google Scholar]
- 4.Ostertag EM, Kazazian HH., Jr Annu Rev Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 5.Baust C, Baillie GJ, Mager DL. Mamm Genome. 2002;13:423–428. doi: 10.1007/s00335-002-2178-3. [DOI] [PubMed] [Google Scholar]
- 6.Nunes ME, Schutt G, Kapur RP, Luthardt F, Kukolich M, Byers P, Evans JP. Hum Mol Genet. 1995;4:2165–2170. doi: 10.1093/hmg/4.11.2165. [DOI] [PubMed] [Google Scholar]
- 7.Gurrieri F, Prinos P, Tackels D, Kilpatrick MW, Allanson J, Genuardi M, Vuckov A, Nanni L, Sangiorgi E, Garofalo G, et al. Am J Med Genet. 1996;62:427–436. doi: 10.1002/(SICI)1096-8628(19960424)62:4<427::AID-AJMG16>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 8.Raas-Rothschild A, Manouvrier S, Gonzales M, Farriaux JP, Lyonnet S, Munnich A. J Med Genet. 1996;33:996–1001. doi: 10.1136/jmg.33.12.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozen RS, Baysal BE, Devlin B, Farr JE, Gorry M, Ehrlich GD, Richard CW. Am J Hum Genet. 1999;64:1646–1654. doi: 10.1086/302403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Mollerat XJ, Gurrieri F, Morgan CT, Sangiorgi E, Everman DB, Gaspari P, Amiel J, Bamshad MJ, Lyle R, Blouin JL, et al. Hum Mol Genet. 2003;12:1959–1971. doi: 10.1093/hmg/ddg212. [DOI] [PubMed] [Google Scholar]
- 11.Kano H, Kurosawa K, Horii E, Ikegawa S, Yoshikawa H, Kurahashi H, Toda T. Hum Genet. 2005;118:477–483. doi: 10.1007/s00439-005-0074-0. [DOI] [PubMed] [Google Scholar]
- 12.Everman DB, Morgan CT, Lyle R, Laughridge ME, Bamshad MJ, Clarkson KB, Colby R, Gurrieri F, Innes AM, Roberson J, et al. Am J Med Genet A. 2006;140:1375–1383. doi: 10.1002/ajmg.a.31246. [DOI] [PubMed] [Google Scholar]
- 13.Lyle R, Radhakrishna U, Blouin JL, Gagos S, Everman DB, Gehrig C, Delozier-Blanchet C, Solanki JV, Patel UC, Nath SK, et al. Am J Med Genet A. 2006;140:1384–1395. doi: 10.1002/ajmg.a.31247. [DOI] [PubMed] [Google Scholar]
- 14.Ianakiev P, Kilpatrick MW, Dealy C, Kosher R, Korenberg JR, Chen XN, Tsipouras P. Biochem Biophys Res Commun. 1999;261:64–70. doi: 10.1006/bbrc.1999.0963. [DOI] [PubMed] [Google Scholar]
- 15.Mager DL, Freeman JD. J Virol. 2000;74:7221–7229. doi: 10.1128/jvi.74.16.7221-7229.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoder JA, Walsh CP, Bestor TH. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 17.Jaenisch R, Bird A. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 18.Loebel DA, Tsoi B, Wong N, O'Rourke MP, Tam PP. Gene Expr Patterns. 2004;4:467–471. doi: 10.1016/j.modgep.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Crackower MA, Motoyama J, Tsui LC. Dev Biol. 1998;201:78–89. doi: 10.1006/dbio.1998.8938. [DOI] [PubMed] [Google Scholar]
- 20.Ribet D, Dewannieux M, Heidmann T. Genome Res. 2004;14:2261–2267. doi: 10.1101/gr.2924904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maksakova IA, Mager DL. J Virol. 2005;79:13865–13874. doi: 10.1128/JVI.79.22.13865-13874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Druker R, Whitelaw E. J Inherit Metab Dis. 2004;27:319–330. doi: 10.1023/B:BOLI.0000031096.81518.66. [DOI] [PubMed] [Google Scholar]
- 23.Moon AM, Capecchi MR. Nat Genet. 2000;26:455–459. doi: 10.1038/82601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewandoski M, Sun X, Martin GR. Nat Genet. 2000;26:460–463. doi: 10.1038/82609. [DOI] [PubMed] [Google Scholar]
- 25.Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Nat Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- 26.Esnault C, Heidmann O, Delebecque F, Dewannieux M, Ribet D, Hance AJ, Heidmann T, Schwartz O. Nature. 2005;433:430–433. doi: 10.1038/nature03238. [DOI] [PubMed] [Google Scholar]
- 27.Chen H, Lilley CE, Yu Q, Lee DV, Chou J, Narvaiza I, Landau NR, Weitzman MD. Curr Biol. 2006;16:480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 28.Esnault C, Millet J, Schwartz O, Heidmann T. Nucleic Acids Res. 2006;34:1522–1531. doi: 10.1093/nar/gkl054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fincham JR, Sastry GR. Annu Rev Genet. 1974;8:15–50. doi: 10.1146/annurev.ge.08.120174.000311. [DOI] [PubMed] [Google Scholar]
- 30.Whitelaw E, Martin DI. Nat Genet. 2001;27:361–365. doi: 10.1038/86850. [DOI] [PubMed] [Google Scholar]
- 31.Martienssen R, Baron A. Genetics. 1994;136:1157–1170. doi: 10.1093/genetics/136.3.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rangwala SH, Elumalai R, Vanier C, Ozkan H, Galbraith DW, Richards EJ. PLoS Genet. 2006;2:270–281. doi: 10.1371/journal.pgen.0020036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rangwala SH, Richards EJ. Genetics. 2007;176:151–160. doi: 10.1534/genetics.107.071092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gdula DA, Gerasimova TI, Corces VG. Proc Natl Acad Sci USA. 1996;93:9378–9383. doi: 10.1073/pnas.93.18.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duhl DM, Vrieling H, Miller KA, Wolff GL, Barsh GS. Nat Genet. 1994;8:59–65. doi: 10.1038/ng0994-59. [DOI] [PubMed] [Google Scholar]
- 36.Rakyan VK, Chong S, Champ ME, Cuthbert PC, Morgan HD, Luu KV, Whitelaw E. Proc Natl Acad Sci USA. 2003;100:2538–2543. doi: 10.1073/pnas.0436776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su RC, Brown KE, Saaber S, Fisher AG, Merkenschlager M, Smale ST. Nat Genet. 2004;36:502–506. doi: 10.1038/ng1351. [DOI] [PubMed] [Google Scholar]
- 38.Hashimshony T, Zhang J, Keshet I, Bustin M, Cedar H. Nat Genet. 2003;34:187–192. doi: 10.1038/ng1158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.