Abstract
Terrestrial solar UVB radiation (≈295–320 nm) readily induces cyclobutane pyrimidine dimers (CPDs) in human skin DNA that result in characteristic mutations associated with nonmelanoma skin cancer. The proinflammatory cytokine TNFα is important in mouse skin chemical carcinogenesis and is thought to also play a role in UVR-induced skin cancer by its immunomodulatory properties. There is some in vitro evidence that CPDs initiate the production of TNFα, and we tested this hypothesis by comparing the wavelength dependence (action spectrum) for TNFα protein induction in human skin in vivo with our earlier in vivo action spectra for CPD induction in four different epidermal layers of human skin. Normal volunteers (n = 35) were irradiated with physiologically relevant doses of monochromatic UVB (290–320 nm), and TNFα concentration was assessed, by high-sensitivity ELISA, in exudates from skin suction blisters raised 8 h after irradiation. An action spectrum, constructed from the slopes of the dose–response curves at the different wavelengths, showed maximal efficacy at 300 nm. An excellent match was observed for TNFα and the CPD action spectrum for cells in the lower basal epidermis. These data strongly suggest that UVB-induced photodamage to DNA in the epidermal basal layer is a major trigger for TNFα production. The TNFα may originate directly from the keratinocytes in this layer or inflammatory cells that are rapidly recruited into the upper dermis (e.g., neutrophils) as a consequence of DNA photodamage to basal-layer keratinocytes.
Keywords: cytokine, DNA photodamage, photoimmunosuppression, cyclobutane pyrimidine dimer
Solar UV radiation (UVR) is known to have immunomodulatory effects in mice and humans (1, 2). The murine data unequivocally show that UVR-induced suppression of cell-mediated immunity is important in photocarcinogenesis and resistance to infectious agents. A single suberythemal exposure of solar simulated radiation (SSR) suppresses the induction phase of the contact hypersensitivity (CHS) response in skin cancer-prone human skin types I and II (2). In contrast, erythemal exposures were necessary for comparable levels of immunosuppression in skin types III and IV, which are less prone to skin cancer. The role of UVR-induced immunomodulation in humans remains unclear, but it is strongly suspected to play a role in human skin cancer (1) and photosensitivity disorders such as polymorphic light eruption (3). The immunomodulatory effects of UVR are mediated by the induction of molecular and cellular factors, including cytokines such as IL10 and TNFα (4, 5). We have shown that moderate single exposures of SSR readily induce high levels on TNFα mRNA (6) and protein (7) in human skin in vivo. Mouse studies also suggest that TNFα may play a role in chemical skin carcinogenesis (8) and photocarcinogenesis that is not related to its immunoregulatory properties (9).
An understanding of the mechanisms of UVR immunomodulation requires knowledge of the chromophores involved. There is a considerable body of evidence that stratum corneum urocanic acid (UCA) and epidermal DNA are major chromophores for the immunosuppressive effects of UVR in mouse studies in vivo, as measured, for example, by the suppression of the induction and elicitation phases of the CHS response. In vitro (10, 11) studies have provided evidence that DNA is an important chromophore for the induction of IL10 and TNFα protein and that this finding is mediated as a consequence of the formation of cyclobutane pyrimidine dimers (CPDs), which are readily induced in human skin in vivo by UVR (12, 13). To date, there has been no definitive evidence that DNA is a chromophore for TNFα in human skin in vivo.
Action spectra or wavelength-dependence studies are important because they may help to identify a chromophore and its location within the skin. Furthermore, action spectra also are important for hazard assessment, and therefore protection strategies, of specific parts of the solar UVR spectrum. This notion is important in sunscreen design because we have recently shown that the immune protection factor of a sunscreen on human skin may be considerably less than its sun protection factor, which is based on its ability to inhibit erythema (14). There are few action spectra data on the immunological effects of UVR. Mouse studies on the suppression of the CHS response implicate UVB in the solar UVR range (15). There are no comparable human data, and the role of UVA in the immunoregulatory effects of UVR remains uncertain (16). The aim of this study was to determine an action spectrum for the induction of TNFα protein in human skin in vivo. Our aim was to compare our TNFα data with our action spectra generated for CPD induction in different layers of human epidermis (13) to determine whether the CPD was the putative lesion for TNFα induction. Furthermore, the induction of TNFα is likely to reflect a human action spectrum for at least some of the immunological effects of UVR.
Results
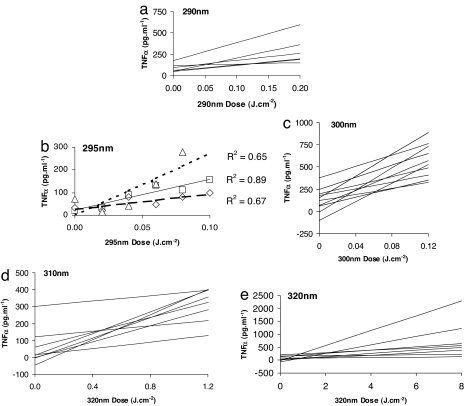
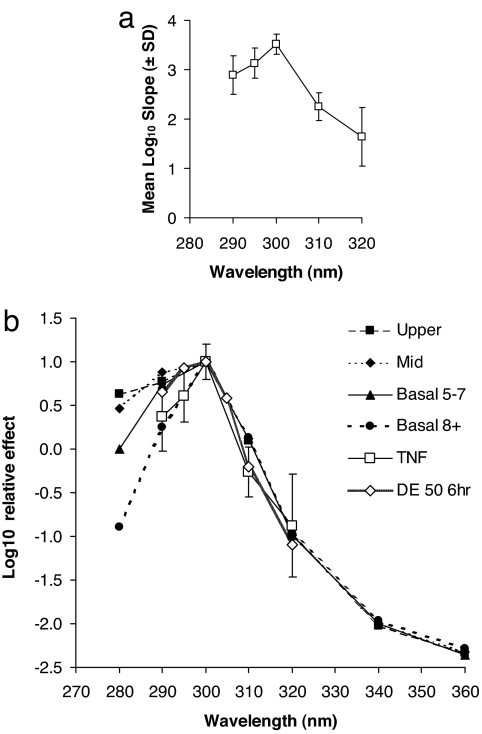
Individual linear regression analyses of dose–response data at 290 nm (n = 6), 295 nm (n = 3), 300 nm (n = 10), 310 nm (n = 8), and 320 nm (n = 8) are shown in Fig. 1. In the interests of clarity, individual data points and r2 values are given for 295 nm only because there were just three volunteers at this wavelength (Fig. 1b). The action spectrum for TNFα production is shown in Fig. 2a. This result also is plotted in Fig. 2b, after normalization at its peak of 300 nm, with previously published action spectra for CPD in four different layers of human epidermis of skin types I and II (13), which are also normalized at their 300-nm peak. The CPD action spectra also were based on slope analysis. The TNFα data were statistically compared with the CPD data in each epidermal layer at 290 nm because Fig. 2b shows a difference between the CPD basal 8+ data and the other epidermal layers. This analysis showed no difference between the values of TNFα and the basal 8+ data (P = 0.8), whereas there were differences between TNFα and the other epidermal layers (P = 0.01–0.05). Thus, the TNFα spectrum shows the best match with the lower basal layer (8+ cell layers) spectrum. Fig. 2b also shows the action spectrum for quantitatively assessed erythema at 6 h from the same volunteers as the TNFα study, normalized at it peak value of 300 nm. The UVR dose value has been set to represent a just-perceptible minimal erythema dose (MED), where delta-erythema (DE) is 50 (arbitrary units).
Fig. 1.
Linear regression analyses. (a) 290 nm. (b) 295 nm. (c) 300 nm. (d) 310 nm. (e) 320 nm. Inclusion criteria for all volunteers included a 300-nm MED of either 0.028 or 0.040 J·cm−2. Regressions are based on the mean TNFα value of two samples. (b) Individual data points and r2 values for the three volunteers in this group.
Fig. 2.
Action spectra for TNFα, DNA photodamage, and erythema. (a) Action spectrum for TNFα induction ± SD at 8 h. (b) TNFα spectrum (± SD) in comparison with action spectra for CPD in four different epidermal layers (upper, middle, and basal layers at two depths) at 0 h (13) and a just-perceptible erythema (DE = 50) measured by reflectance spectroscopy at 6 h in the current TNFα study volunteers. Note that (i) all data are normalized at 300 nm; (ii) TNFα was not measured at 305 nm because the manufacturer stopped production of the ELISA kits that we used; and (iii) basal 5–7 cells are found in layers 5–7, and basal 8+ is more than eight cell layers deep.
Discussion
Two main initiating photochemical events have been proposed for the immunoregulatory effects of UVR: (i) the formation of CPDs, in which DNA is the chromophore; and (ii) the formation of cis-UCA from stratum corneum-bound trans-UCA, in which the latter is the chromophore. It is probable that both mechanisms play a role depending on the specific immunological pathway (17). Furthermore, there also is evidence that UVR-induced oxidative stress plays a role (18), mediated by as yet unknown chromophores. The aim of our study was to determine the action spectrum for TNFα protein, a proinflammatory cytokine known to have immunoregulatory properties either directly, by its induction of other cytokines such as IL8 (19), or by the stimulation of the migration of Langerhans cells from the epidermis (20, 21). It would have been interesting to determine the action spectrum for IL10, but our previous studies showed no increase in protein at 8 h after irradiation and a relatively modest increase at 15 h (7), which is not a convenient sampling time point for volunteer studies.
Extensive pilot studies showed that it was necessary to select volunteers carefully to ensure obtaining data that could be analyzed with relatively small numbers of volunteers. This finding is because our unpublished studies showed that TNAα release was inversely related to SSR MED; the lower the MED range, the more TNFα was released per unit physical dose (J·cm−2). However, MED was a better predictor of TNFα response than skin type because, although MED is inversely correlated with skin type, there is considerable MED overlap between skin types (22). Thus, volunteers were selected from a narrow MED range (two adjacent increments in the test series) at 300 nm because this level is the peak of the erythema action spectrum (13). Fig. 1 shows that at some wavelengths (e.g., 300 and 310 nm), the regression lines seem to fall into two distinct families that were not related to 300-nm MED at the wavelength of interest or skin type. It is possible that response depends on TNFα polymorphisms.
Young et al. (13) showed that action spectra for CPD induction in human skin in vivo varied at wavelengths <300 nm with the epidermal layer in which they were assessed. This finding was not surprising because epidermal DNA and stratum corneum UCA (both with maximal absorbance in the UVC region) offer much greater attenuation at these shorter UVB wavelengths, and, in effect, these molecules act as natural sunscreens for target chromophores beneath them. Comparisons of our action spectrum for TNFα and CPD induction (Fig. 2b) showed that the best match was between TNFα and deep basal layer CPD. We also compared our TNFα action spectrum with that for the photoisomerization of UCA in human skin (23), and the results are markedly different especially between 300 and 320 nm. Thus, based on human action spectroscopy in vivo, our data strongly suggest that DNA, specifically in the basal layer, is an important chromophore for TNFα and supports the in vitro mouse keratinocyte studies of Kibitel et al. (11), in which T4 endonuclease V (T4N5)-enhanced CPD repair was shown to reduce TNFα protein production. A similar in vivo approach was made by Wolf et al. (24). However, these authors were not able to demonstrate any significant reduction of epidermal CPD by T4N5 or the presence of TNFα protein, although the T4N5 did reduce mRNA levels. Fig. 2b also shows that the 6-h erythema action spectrum is similar to the TNFα spectrum at wavelengths ≥300 nm, but has a better fit with the DNA damage in the regions above the upper basal layer at the shorter wavelengths, as we previously reported for 24-h erythema (13). This finding suggests that TNFα and erythema share DNA as a common chromophore, but that the location of this chromophore may be important for different endpoints.
The main source of the TNFα in our studies is likely to be basal layer keratinocytes. However, dermal DNA photodamage also may be important to mast cells that store preformed TNFα (25) and that can be released by UVB exposure (26). However, this notion seems unlikely given the low cellular content of the dermis at the time of irradiation and the substantial amounts of TNFα that are recovered in suction blister fluid. It also is possible that early recruitment of dermal inflammatory cells, such as neutrophils (27, 28), may contribute to the TNFα detected 8–9 h after irradiation.
Given that DNA is the likely major chromophore for erythema (13) and TNFα induction, and there is evidence that CPDs are important in the suppression of CHS in mice (29), one might expect the action spectra for erythema and suppression of the induction phase of CHS in humans to be similar. However, we recently concluded that these action spectra are different and that UVA is more immunosuppressive than it is erythemogenic in human skin (14). This finding suggests that other chromophores, such as UCA, also may be important, and this notion is supported by an action spectrum for the photoisomerization of trans- to cis-UCA in human skin in vivo, in which UVA had a more important role than it did for erythema (23). Some authors have suggested that cis-UCA can induce TNFα (30, 31), but others have not supported this theory (32–34). Our data support DNA, rather than UCA, as the main chromophore for TNFα. However, overall, the multiple individual events that contribute to functional measures of immunomodulation may well have different chromophores and, therefore, different action spectra (17).
In conclusion, our data provide human in vivo support for the pivotal role of DNA photodamage, especially CPD, in the release of TNFα by solar UVR. It is clear that DNA photodamage is important in the mutational effects (35) that give rise to nonmelanoma skin cancer. Such photodamage also may be important with regard to the immunological effects mediated by TNFα, which are thought to be related to skin cancer. Our data also add to the limited body of nonerythema human skin action spectra and provide a possible spectral weighting function for at least some of the immunological effects of UVR.
Materials and Methods
Monochromatic Radiation Spectra, Dosimetry, and Skin Irradiation.
Full details are described by Young et al. (13). We used monochromatic spectra at 290, 295, 300, 305, 310, and 320 nm from a monochromator with a grating blazed at 250 nm, which were delivered through a liquid light guide with a 5-mm exit diameter that made direct contact with the skin. A 5-nm full-width at half-maximum (FWHM) bandwidth (i.e., bandwidth at which 50% maximal irradiance occurs) was used for 290–310 nm and a 10-nm FWHM bandwidth was used for 320 nm; the latter also was filtered with 1-mm WG320 to attenuate any shorter wavelengths. The spectral maxima and their FWHM bandwidths were all verified spectroradiometrically before their use. These values are the same as those published by Young et al. (13), with the exception of the maxima of 295 and 305 nm, the shapes of which are similar to the other 5-nm FWHM spectra.
Volunteers and Study Protocol.
Healthy male and female (1:1) volunteers ages 18–32 years, with mainly skin type I or II determined by detailed questionnaire and interview, were fully informed of all procedures and gave written informed consent before taking in part in the studies, which were approved by the Ethics Committee of St. Thomas' Hospital and done in accordance with the declaration of Helsinki. The volunteers were screened on the basis of their just-perceptible MED at 300 nm by using a √2 dose series on previously unexposed buttock skin. They were recruited if their 300-nm MED was either 0.028 or 0.040 J·cm−2 (60% and 40% of volunteers, respectively, and evenly represented in the different wavelength groups). These inclusion criteria were based on our studies that showed that TNFα release per unit physical dose of UVR was an inverse function of MED, rather than skin type. Dose–response studies were done with each wavelength (n = 3–10) with a predetermined (by pilot studies) range of six physical UVR doses in duplicate, the highest being equivalent to approximately four MED depending on individual MEDs. The erythema response at each dose was quantified with a reflectance meter (Dia-Stron) at 6 h after irradiation, after which 5-mm suction blisters were raised (7) that took ≈2 h. The exudate (5–25 μl per blister) was collected at ≈8 h after irradiation and stored at −30°C. Comparisons of SSR-induced TNFα mRNA at 6 and 24 h showed higher levels at 6 h (6), whereas peak protein levels were seen at 15 h after three MED SSR, but a substantial rise was seen at 8 h (7). Suction blister fluid was analyzed in batches for TNFα by using high-sensitivity commercial ELISA kits (GE Healthcare).
Data Analysis.
The statistical approach that we used was based on Matthews et al. (36), who used one summary measure of serial measurements per subject. We used the slope of linear regression analysis of mean TNFα concentration (pg/ml) from duplicate samples, versus UVR dose (J·cm−2), which was determined for each volunteer within a given wavelength group; >70% of r2 values were >0.5, of which 40% were >0.75. Values <0.5 were evenly distributed across all wavelength subject groups. The mean r2 of all 35 regressions was 0.6 (SD ± 0.3), which was consistent with <25% of each individual measurement being random error. The mean log10 slope ± SD was plotted against wavelength to generate an action spectrum. This action spectrum was not quantum-corrected so that it could be directly compared with other nonquantum-corrected action spectra in the skin. The CPDs showed epidermal layer differences at wavelengths <300 nm (Fig. 2b). Therefore, comparisons of the TNFα and CPD data at the common wavelength of 290 nm were made by linear regression with dummy variables on the log of the slopes after normalization at 300 nm, correcting for unequal variance and repeated measurements per subject by Huber–White standard errors (37).
Erythema.
Full details are given by Young et al. (13). Briefly, dose–response curves for erythema reflectance were generated as a logit function of log10 UVR dose. These values were used to calculate the dose needed to achieve a given level of erythema at 6 h that was expressed as DE, which represented the difference between the test site and an adjacent nonirradiated control. Mean log10 1/dose was plotted against wavelength to generate an action spectrum.
Acknowledgments
We thank Dr. Robert Barr for data analysis, John Sheehan for technical assistance, and Paul Seed for statistical advice. A.R.Y. raised funds for this project. This work was supported by European Commission contract no. ENV-4-CT97-0556.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Nishigori C, Yarosh DB, Donawho C, Kripke ML. J Invest Dermatol Symp Proc. 1996;1:143–146. [PubMed] [Google Scholar]
- 2.Kelly DA, Young AR, McGregor JM, Seed PT, Potten CS, Walker SL. J Exp Med. 2000;191:561–566. doi: 10.1084/jem.191.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van de Pas CB, Kelly DA, Seed PT, Young AR, Hawk JL, Walker SL. J Invest Dermatol. 2004;122:295–299. doi: 10.1046/j.0022-202X.2004.22201.x. [DOI] [PubMed] [Google Scholar]
- 4.Vincek V, Kurimoto I, Medema JP, Prieto E, Streilein JW. Cancer Res. 1993;53:728–732. [PubMed] [Google Scholar]
- 5.Rivas JM, Ullrich SE. J Leukocyte Biol. 1994;56:769–775. doi: 10.1002/jlb.56.6.769. [DOI] [PubMed] [Google Scholar]
- 6.Brink N, Szamel M, Young AR, Wittern KP, Bergemann J. Inflamm Res. 2000;49:290–296. doi: 10.1007/PL00000209. [DOI] [PubMed] [Google Scholar]
- 7.Barr RM, Walker SL, Tsang W, Harrison GI, Ettehadi P, Greaves MW, Young AR. J Invest Dermatol. 1999;112:692–698. doi: 10.1046/j.1523-1747.1999.00570.x. [DOI] [PubMed] [Google Scholar]
- 8.Moore RJ, Owens DM, Stamp G, Arnott C, Burke F, East N, Holdsworth H, Turner L, Rollins B, Pasparakis M, et al. Nat Med. 1999;5:828–831. doi: 10.1038/10552. [DOI] [PubMed] [Google Scholar]
- 9.Starcher B. Br J Dermatol. 2000;142:1140–1147. doi: 10.1046/j.1365-2133.2000.03539.x. [DOI] [PubMed] [Google Scholar]
- 10.Nishigori C, Yarosh DB, Ullrich SE, Vink AA, Bucana CD, Roza L, Kripke ML. Proc Natl Acad Sci USA. 1996;93:10354–10359. doi: 10.1073/pnas.93.19.10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kibitel J, Hejmadi V, Alas L, O'Connor A, Sutherland BM, Yarosh D. Photochem Photobiol. 1998;67:541–546. [PubMed] [Google Scholar]
- 12.Young AR, Chadwick CA, Harrison GI, Hawk JL, Nikaido O, Potten CS. J Invest Dermatol. 1996;106:1307–1313. doi: 10.1111/1523-1747.ep12349031. [DOI] [PubMed] [Google Scholar]
- 13.Young AR, Chadwick CA, Harrison GI, Nikaido O, Ramsden J, Potten CS. J Invest Dermatol. 1998;111:982–988. doi: 10.1046/j.1523-1747.1998.00436.x. [DOI] [PubMed] [Google Scholar]
- 14.Kelly DA, Seed PT, Young AR, Walker SL. J Invest Dermatol. 2003;120:65–71. doi: 10.1046/j.1523-1747.2003.12005.x. [DOI] [PubMed] [Google Scholar]
- 15.De Fabo EC, Noonan FP. J Exp Med. 1983;158:84–98. doi: 10.1084/jem.158.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phan TA, Halliday GM, Barnetson RS, Damian DL. Front Biosci. 2006;11:394–411. doi: 10.2741/1807. [DOI] [PubMed] [Google Scholar]
- 17.Kim TH, Moodycliffe AM, Yarosh DB, Norval M, Kripke ML, Ullrich SE. Photochem Photobiol. 2003;78:228–234. doi: 10.1562/0031-8655(2003)078<0228:votadw>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 18.Halliday GM. Mutat Res. 2005;571:107–120. doi: 10.1016/j.mrfmmm.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 19.Raingeaud J, Pierre J. FEBS Lett. 2005;579:3953–3959. doi: 10.1016/j.febslet.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 20.Cumberbatch M, Griffiths CE, Tucker SC, Dearman RJ, Kimber I. Br J Dermatol. 1999;141:192–200. doi: 10.1046/j.1365-2133.1999.02964.x. [DOI] [PubMed] [Google Scholar]
- 21.Cumberbatch M, Kimber I. Immunology. 1995;84:31–35. [PMC free article] [PubMed] [Google Scholar]
- 22.Harrison GI, Young AR. Methods. 2002;28:14–19. doi: 10.1016/s1046-2023(02)00205-0. [DOI] [PubMed] [Google Scholar]
- 23.McLoone P, Simics E, Barton A, Norval M, Gibbs NK. J Invest Dermatol. 2005;124:1071–1074. doi: 10.1111/j.0022-202X.2005.23731.x. [DOI] [PubMed] [Google Scholar]
- 24.Wolf P, Maier H, Mullegger RR, Chadwick CA, Hofmann-Wellenhof R, Soyer HP, Hofer A, Smolle J, Horn M, Cerroni L, et al. J Invest Dermatol. 2000;114:149–156. doi: 10.1046/j.1523-1747.2000.00839.x. [DOI] [PubMed] [Google Scholar]
- 25.Gordon JR, Galli SJ. Nature. 1990;346:274–276. doi: 10.1038/346274a0. [DOI] [PubMed] [Google Scholar]
- 26.Walsh LJ. Immunol Cell Biol. 1995;73:226–233. doi: 10.1038/icb.1995.37. [DOI] [PubMed] [Google Scholar]
- 27.Hawk JL, Murphy GM, Holden CA. Br J Dermatol. 1988;118:27–30. doi: 10.1111/j.1365-2133.1988.tb01746.x. [DOI] [PubMed] [Google Scholar]
- 28.Teunissen MB, Piskin G, di Nuzzo S, Sylva-Steenland RM, de Rie MA, Bos JD. J Immunol. 2002;168:3732–3739. doi: 10.4049/jimmunol.168.8.3732. [DOI] [PubMed] [Google Scholar]
- 29.Kripke ML, Cox PA, Alas LG, Yarosh DB. Proc Natl Acad Sci USA. 1992;89:7516–7520. doi: 10.1073/pnas.89.16.7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurimoto I, Streilein JW. J Immunol. 1992;148:3072–3078. [PubMed] [Google Scholar]
- 31.Prater MR, Blaylock BL, Holladay SD. Photodermatol Photoimmunol Photomed. 2003;19:287–294. doi: 10.1046/j.1600-0781.2003.00058.x. [DOI] [PubMed] [Google Scholar]
- 32.Zak-Prelich M, Norval M, Venner TJ, Bisset Y, Walker C, Rafferty TS, Sauder DN, McKenzie RC. Photochem Photobiol. 2001;73:238–244. doi: 10.1562/0031-8655(2001)073<0238:cuadni>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 33.Amerio P, Toto P, Feliciani C, Suzuki H, Shivji G, Wang B, Sauder DN. Br J Dermatol. 2001;144:952–957. doi: 10.1046/j.1365-2133.2001.04181.x. [DOI] [PubMed] [Google Scholar]
- 34.Redondo P, Garcia-Foncillas J, Cuevillas F, Espana A, Quintanilla E. Photodermatol Photoimmunol Photomed. 1996;12:237–243. doi: 10.1111/j.1600-0781.1996.tb00211.x. [DOI] [PubMed] [Google Scholar]
- 35.Wikonkal NM, Brash DE. J Investig Dermatol Symp Proc. 1999;4:6–10. doi: 10.1038/sj.jidsp.5640173. [DOI] [PubMed] [Google Scholar]
- 36.Matthews JNS, Altman DG, Campbell MJ, Royston P. Br Med J. 1990;300:230–235. doi: 10.1136/bmj.300.6719.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huber PJ. Proceedings of the Fifth Berkeley Symposium in Mathematical Statistics and Probability; Berkeley: Univ California Press; 1967. pp. 221–233. [Google Scholar]