Abstract
The family of interferon (IFN) regulatory factors (IRFs) encodes DNA-binding transcription factors, some of which function as modulators of virus-induced signaling. The IRF-3 gene is constitutively expressed in many tissues and cell types, and neither virus infection nor IFN treatment enhances its transcription. In infected cells, however, IRF-3 protein is phosphorylated at the carboxyl terminus, which facilitates its binding to the CBP/p300 coactivator. In the present study, we demonstrate that overexpression of IRF-3 significantly enhances virus-mediated transcription of the IFNA and IFNB genes in infected cells as well as IFN synthesis. IRF-3-mediated activation of IFN genes depends in part on carboxyl-terminal phosphorylation of a cluster of Ser/Thr residues, because a mutant with Ser/Thr to Ala substitutions activates the IFN promoter less efficiently. However, overexpression of IRF-3 in human 2FTGH cells alone results in the induction of an antiviral state, which depends on functional IFN signaling, because IRF-3 does not induce an antiviral state in mutant 2FTGH cells defective in either JAK-1 or p48 functions; also no antiviral effect of IRF-3 could be demonstrated in Vero cells that lack the IFNA and IFNB genes. This finding indicates that the observed antiviral activity of IRF-3 in 2FTGH cells results mainly from the induction of IFNs. Furthermore, E1A protein inhibited IRF-3-mediated stimulation of the IFNA4 promoter in transient expression assays; this inhibition could be reversed partially by overexpression of CBP/p300 and was not demonstrated with the mutant of E1A that does not bind p300. These results identify IRF-3 and CBP/p300 as integral components of the virus-induced complex that stimulates type 1 IFN gene transcription. The observation that adenovirus E1A antagonizes IRF-3 mediated activation suggests that E1A and IRF-3 may compete for binding to CBP/p300 and implicates a novel mechanism by which adenovirus may overcome the antiviral effects of the IFN pathway.
The interferon (IFN) regulatory factors (IRFs) play an important role in virus- and cytokine-induced signaling pathways (1). The first discovered IRF, IRF-1, was shown to activate the transcriptional activity of the IFNA and IFNB gene promoters, whereas the closely related IRF-2, which binds to the same sequences (IRF-E), generally functions as a repressor (2), although in the context of some promoters IRF-2 also can be an activator (3). In addition to IRF-1 and IRF-2, this growing family of transcription factors includes interferon consensus sequence-binding protein (ICSBP), ISGF3γ (p48), IRF-3, IRF-4 (PIP/LSIR), IRF-5, IRF-6, and IRF-7. All of these factors share homology in the amino terminus, which contains a DNA-binding domain (DBD) with the five characteristic tryptophan (W) repeats. The carboxyl-terminal regions of these proteins are much less conserved; IRF-1 and IRF-3 were shown to contain a transactivation domain in this region (4). ICSBP, p48, and IRF-4/PIP contain in this region an interaction domain, which mediates interaction with other transcription factors (5–7).
IFNA and IFNB promoters contain IRF-E within the virus-responsive element (VRE) (2) that play critical roles in virus-mediated inducibility. Transcription of IFNB gene is achieved by synergy among several transcription factors recruited to multiple, distinct DNA-binding sites in the VRE of the IFNB promoter. These factors form a multicomponent transcriptional enhancer complex, the enhanceosome, consisting of ATF2/c-Jun heterodimer, HMGY(I), NF-κB heterodimer, p50/p65, and IRF and interacting with the components of the general transcriptional machinery through association with the CBP/p300 coactivator (8, 9). The proteins regulating the virus-mediated induction of IFNA genes are less well defined; for virus-activated IFNA4 gene expression, two overlapping cis-acting elements (IRF-E and AF-1) are required (10) and point mutation in either site abolished virus-mediated activation. Although the presence of the IRF-E site was essential for transcriptional activity, the binding of IRF-1 to IRF-E in the inducible element (IE) of the IFNA4 gene promoter was not detected in the infected cells (11). In addition, homozygous deletion of IRF-1 in mice did not affect virus-mediated induction of IFNA and IFNB genes (12, 13), suggesting the possible involvement of other IRFs in these processes. The virus-induced factor that recognizes the IRF-E of the IFNA4 promoter but is distinct from IRF-1, IRF-2, and ISGF3γ has been described (14, 15).
We recently found a novel IRF, IRF-3, that binds to the IFN-stimulated response element of interferon-stimulated genes (ISG15) (16), as well as to IE of the IFNA4 gene promoter and the positive regulatory domain (PRD) III of the IFNB gene promoter (17). In the transient transfection assay, IRF-3 enhanced the Newcastle disease virus (NDV)-mediated transcriptional activation of the IFNA4 gene promoter (16) in L cells, but not in adenovirus E1A-transformed human kidney 293 cells, where overexpression of IRF-3 inhibited both IRF-1- and NDV-mediated transactivation of IFNA4 promoter. However, a fusion protein, consisting of an IRF-3 DNA-binding domain and relA(p65) activation domain efficiently stimulated transcription activity of the IFNA4 and IFNB promoters in 293 cells, further indicating the presence of IRF-3-binding sites in these promoters (17) and the possible involvement of the carboxyl-terminal part of IRF-3 in differential activation of IFNA4 promoters between infected L929 and 293 cells. Although transcription of the IRF-3 gene is not activated by viral infection, the carboxyl-terminal serine residues of IRF-3 protein are phosphorylated in infected cells, and this phosphorylation results in the translocation of IRF-3 to the nucleus and facilitates its binding to CBP/p300 (18–20).
The aim of present study was to further analyze the role of IRF-3 in transcriptional activation of the endogenous IFNA and IFNB genes. We demonstrate that (i) overexpression of IRF-3 in both rat and human fibroblasts enhances the levels of IFN synthesized in virus-induced cells and restricts virus replication; (ii) IRF-3-induced antiviral effects require activation of IFN genes and a functional IFN signaling pathway; and (iii) adenovirus E1A gene product down-modulates IRF-3-mediated transcriptional activation of the IFNA promoter.
MATERIALS AND METHODS
Plasmid Constructs and Oligonucleotides.
The IRF-3 expression plasmid was constructed by cloning the SalI-NotI IRF-3 cDNA fragment 3′ in-frame to hemagglutinin epitope in PHA-2 Hyb plasmid obtained from C. Dang (Johns Hopkins University, Baltimore). Mutant IRF-3 expression plasmids containing deleted proline cluster (amino acids 153–192), amino-terminal, or carboxyl-terminal fragments of IRF-3 were created by PCR and cloned into EcoRI-XhoI sites of pHA-2 Hyb. All plasmids were sequenced before use. The IRF-3(5A) plasmid containing the point mutations in carboxyl-terminal serines (S396A, S398A, S402A, and S504A) and threonine (T404A) was described recently (18). The IRF-1 and IRF-2 expression plasmids were a gift from T. Taniguchi (University of Tokyo). The indicator plasmids [IFNA4/chloramphenicol acetyltransferase (CAT) and IFNB/CAT] were described previously (21, 22). An E1A expression plasmid containing 13S E1A gene under the control of RSV promoter and the 12S E1A (Δp300) mutant that contains a 12- to 36-aa deletion were obtained from D. Kalvakolanu (University of Maryland, Baltimore); p300 expression plasmid was obtained from G. Nabel (23).
Cells and Transfections.
Rat embryo fibroblast (REF) cells, L cells, Vero cells, as well as 2FTGH cells and their mutants, obtained from G. Stark (Cleveland Clinic Foundation, Cleveland), were grown in DMEM supplemented with 10% fetal bovine serum (FBS). For the transient transfection assay, subconfluent cells (5 × 105/60-mm dish) were transfected with 1–2 μg of the reporter plasmids and indicated amounts of IRF expression plasmids using Superfect transfection reagent (Qiagen). The total amount of DNA used in each transfection was kept constant, and the β-galactosidase expressing plasmid (0.1 μg) was included as internal standard. IRF-3 levels encoded by the transfected IRF-3 expressing plasmid were determined by Western blot hybridization (24) with anti-IRF-3 antibodies or anti-HA antibodies. When indicated, infection with NDV was done 16 hr after transfection of REF cells; the cells were harvested for CAT assay 16 hr later.
CAT and Antiviral Assays.
The CAT assays were done as described previously (22), and CAT activity was normalized to constant transfection efficiency determined by β-galactosidase activity. For the antiviral assay, cells were transfected with 5 μg of IRF-3 expressing plasmid, and 24 hr after transfection cells were infected with vesicular stomatitis virus (VSV) [multiplicity of infection, 2]. Medium was collected 16 hr after infection, and the number of infectious particles was determined by a plaque assay with L cells as target cells. The levels of IFN in the medium were determined by a biological assay (25).
RNA Analysis.
Total RNA was isolated by the Trizol method (Life Technologies, Gaithersburg, MD). Ten micrograms of total RNA was analyzed by Northern blot hybridization with IFNA- and IFNB-specific riboprobes (26, 27). For the reverse transcription– PCR (RT-PCR) analysis, 2 μg of total RNA was reverse-transcribed to cDNA by using a sequence-specific 3′ primer. Five microliters of the reaction mixture was PCR-amplified by using Taq DNA polymerase (Life Technologies). The primers used for the cDNA synthesis and amplification of rat IFNA RNA were: 5′ primer ATG GCT CGG CTC TGT GCT and 3′ primer GCT CTC CAG ACT TCT GCT.
RESULTS
Overexpression of IRF-3 Stimulates Transcription of IFN Gene Promoters and Enhances Virus-Mediated Induction.
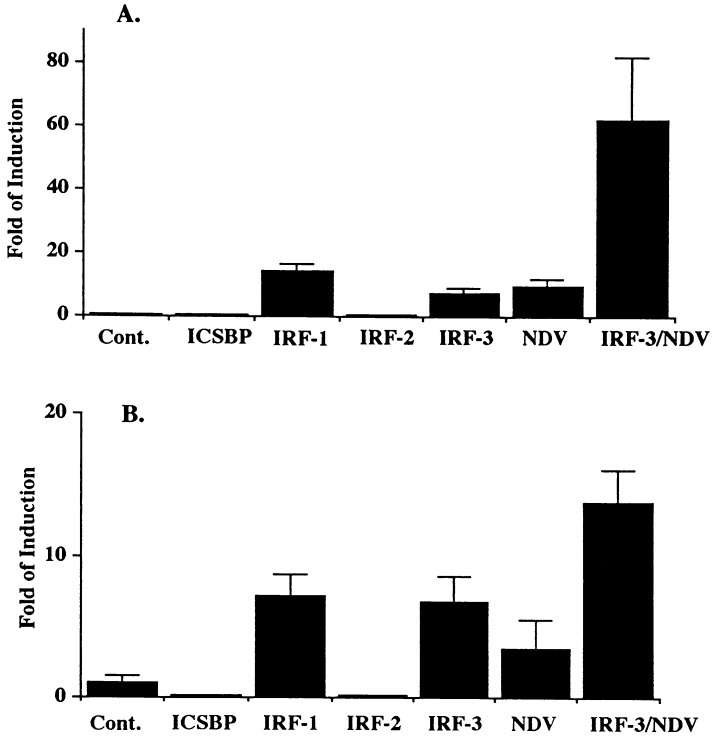
We recently showed that the recombinant amino-terminal IRF-3 peptide-(1–133) strongly binds to the IE of IFNA4 gene promoter and to the PRDIII region of the IFNB gene promoter (17). However, in 293 cells and mouse L cells, overexpression of IRF-3 alone did not enhance expression of IFNA4 or IFNB promoters in transient transfection assays (16, 17). To determine whether this lack of IRF-3-mediated activation is cell type-specific, IRF-3 expressing plasmid was cotransfected with the IFNA4 CAT reporter plasmid into REFs. The results in Fig. 1A show that overexpression of IRF-3 in these cells stimulated transcriptional activity of IFNA4 promoter; activation of this promoter by IRF-1 or IRF-2 served as positive and negative controls, respectively. Western blot analysis has shown that all the transfected IRFs were expressed at comparable levels (data not shown). We have shown previously that overexpression of IRF-1 enhances virus-mediated induction of IFNA promoter (11). Overexpression of IRF-3 also greatly enhanced the virus-mediated induction of this promoter, and the effect was more than additive. Both IRF-3 and IRF-1 also stimulated transcriptional activity of IFNB gene promoter, although to a lesser extent, and the synergism between IRF-3 and virus-activated transcription was lower when measured for IFNB promoter (3- to 4-fold enhancement when compared with virus alone) than for IFNA (6- to 7-fold enhancement). Both IRF-2 and ICSBP inhibited constitutive activity of the IFNB promoter.
Figure 1.
IRF-3 activates IFNA and IFNB promoters and cooperates with viral infection. (A) REF (rat embryonic fibroblast) cells were cotransfected with 2 μg of various IRF expressing plasmids, 2 μg of IFN4/CAT reporter plasmid, and 100 ng of pCMV-β-galactosidase as an internal control. (B) Transfection was performed as in A except 1 μg of IFNB/CAT was used. When indicated, cells were infected with NDV for 16 hr as described in Materials and Methods. CAT activity was normalized to constant levels of β-galactosidase activity. SDs are shown by T bars.
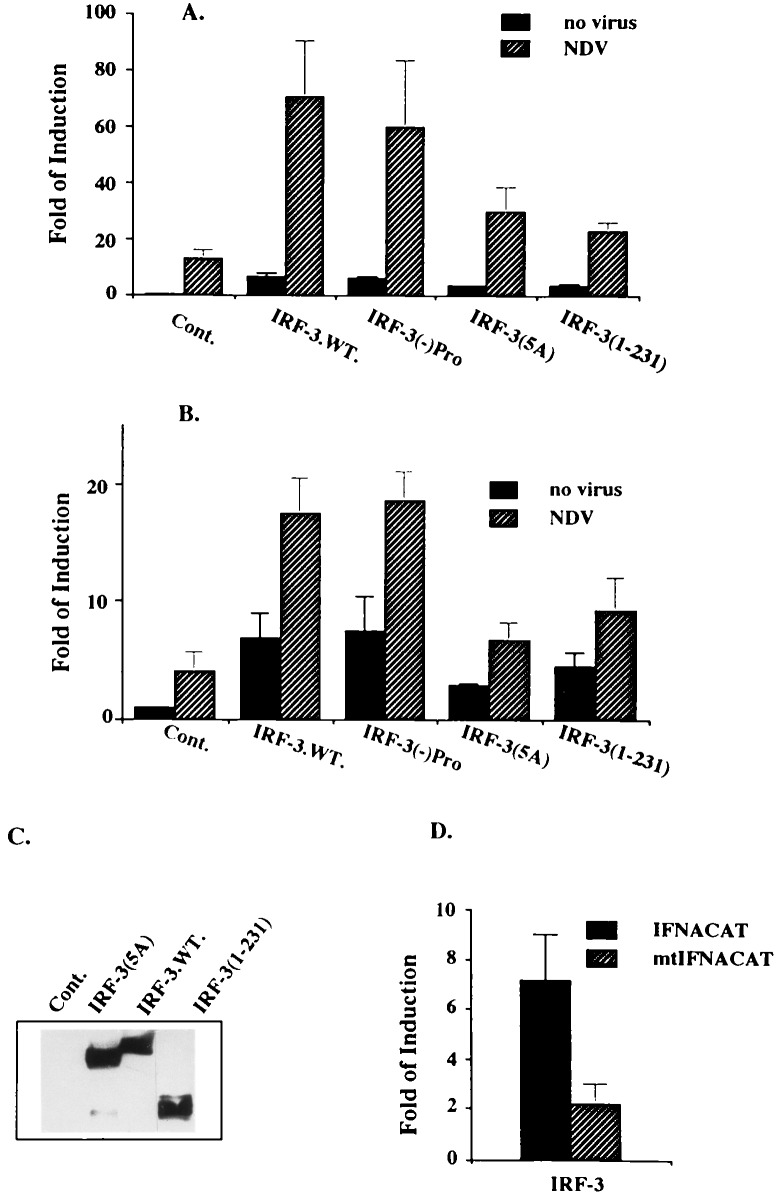
To define the region of IRF-3 essential for virus-mediated induction of these two promoters, the activation of IFNCAT reporter plasmids by various IRF-3 mutants was examined in the presence or absence of viral infection (Fig. 2). Deletion of the central proline-rich region of IRF-3 (−) Pro (153–192 aa), which we assumed to be important for the interaction of IRF-3 with other cellular proteins, did not affect the ability of IRF-3 to transactivate IFNA and IFNB promoters or to synergize with virus-mediated induction. We have shown recently (18) that virus infection induces phosphorylation of IRF-3 at the carboxyl-terminal end and that phosphorylation occurs at four serines and one threonine residue located between amino acids 395 and 407. Replacement of these residues by alanine [IRF-3(5A)] decreased but did not abolish the transactivating potential of IRF-3 both in infected and uninfected cells. Neither the amino-terminal DBD (amino acids 1–115) nor the carboxyl-terminal (amino acids 110–427) part of the IRF-3 alone is sufficient to activate the IFNA promoter (data not shown). However, the IRF-3 mutant containing the DBD and the central part of the protein (amino acids 1–231) retained low transactivating activity. Western blot analysis of the relative levels of IRF-3 encoded by IRF-3 and its mutants shows comparable levels of expression from the transfected plasmids (Fig. 2C). Taken together, these results indicate that (i) IRF-3 alone can function as a primary inducer of IFNA and IFNB promoters in REF cells; (ii) that the virus-mediated phosphorylation of IRF-3 up-regulates its transactivation activity; and (iii) the IRF-3 phosphorylation mutant IRF-3(5A) retains relatively low transcriptional activity.
Figure 2.
Cooperation between IRF-3 or its mutants (described in Materials and Methods) and NDV in activation of the IFNA and the IFNB promoters. The experiments were carried out as described in Fig. 1 with IFNA4/CAT reporter (A) or IFNB/CAT reporter (B). (C) Expression of IRF-3 wild type (wt) and its mutants in transfected cells used for CAT assay determined by Western blotting. (D) Two micrograms of IRF-3 was cotransfected with 2 μg of either IFNA4/CAT plasmid or mutant IFNA4/CAT plasmid (mutations are at −94 and −103 nt).
As shown recently, recombinant IRF-3-(1–133) binds to the 3′ region of the IE of IFNA4 gene promoter (17), which contains the IRF-E and shows homology to PRDIII. As shown in Fig. 2D, two mutations in this region, at positions −103 and −94, that were shown previously to abolish the virus-activated IFNA promoter, substantially decreased IRF-3-mediated activation of −452-bp IFNA4 promoter. These results indicate that the GAAANN motif in the IRF-E plays a critical role in IRF-3-mediated transcriptional activation of the IFNA4 promoter.
Overexpression of IRF-3 Enhances Virus-Mediated Induction of Endogenous IFNA and IFNB Genes.
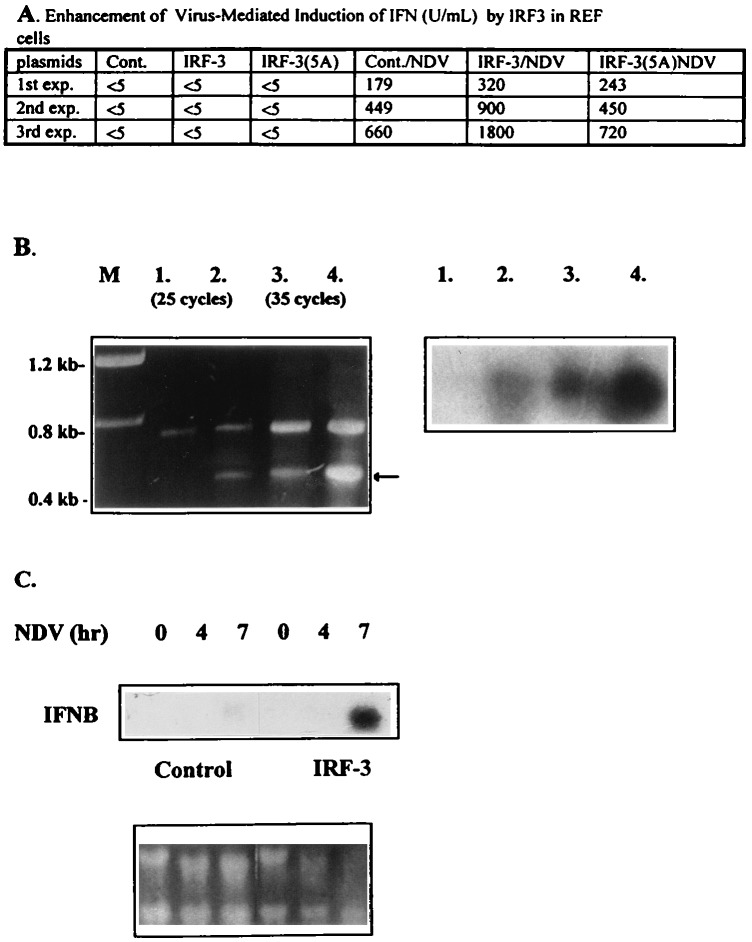
In a transient expression assay, IRF-3 enhances NDV-mediated induction of the IFNA and IFNB promoters. To determine whether IRF-3 also can stimulate induction of endogenous IFN genes, the levels of IFN synthesized in infected REF cells transfected with IRF-3 and in the untransfected controls were compared. As seen in Fig. 3A, overexpression of IRF-3 in REF cells enhanced virus-mediated stimulation of IFN synthesis (Fig. 3) by 2- to 3-fold. In contrast, there was no enhancement of IFN synthesis by the IRF-3(5A)-mutant. Western blot analysis of the relative levels of IRF-3 and IRF-3(5A) protein in transfected cells showed that these two proteins were expressed at comparable levels, indicating that the inability of IRF-3(5A) to stimulate IFN synthesis was not a result of the low levels of expression (data not shown). It should be noted that, under conditions of the transient transfection, only about 25–30% of cells were transfected. Thus, because not all cells are transfected, the observed synergism probably is underestimated.
Figure 3.
Overexpression of IRF-3 enhanced the NDV-mediated induction of endogenous IFNA4 and IFNB in REF cells. (A) Levels of biologically active IFN detected in media of infected cells. REF cells were transiently transfected with 2 μg of either pSp64 (cont.), IRF-3, or IRF-3(5A). Twenty-four hours after transfection, cells were infected with NDV for 16 hr. The media then was collected and IFN levels were determined (25). The data represent the results from three independent transfection experiments and assays. (B) RT-PCR. REF cells were transiently transfected with 4 μg of either pSp64 (lanes 1 and 3) or IRF-3 (lanes 2 and 4) by Superfect (Qiagen) transfection reagent. Two micrograms of purified RNA was amplified by RT-PCR method with specific primers for rat IFNA as described in Materials and Methods. Amplified fragments were separated on an agarose gel and visualized by ethidium bromide staining (Lt), or transferred onto nitrocellulose, and hybridized with the IFNA-specific riboprobe (Rt). (C) REF cells were transiently transfected with a control or IRF-3 plasmid. Twenty-four hours later, cells were infected for the time periods indicated. RNA was collected and analyzed by Northern hybridization to an IFNB-specific riboprobe.
To confirm these observations, the steady-state levels of IFN mRNA induced by NDV in the presence or absence of overexpressed IRF-3 were analyzed. Because the IFNA genes are expressed at very low levels in fibroblast cells upon NDV infection, the relative levels of IFNA mRNAs were evaluated by RT-PCR by using specific primers corresponding to rat IFNA gene (see Materials and Methods). The results in Fig. 3B show that the relative levels of correctly amplified fragment (marked by arrow) were substantially (about 5-fold) increased in infected IRF-3 expressing cells when compared with infected cells that did not overexpress IRF-3. Similarly, the relative levels of IFNB mRNA, compared by Northern blot analysis, at different times postinfection (Fig. 3C) were increased by 15-fold in IRF-3 transfected cells. These data suggest that IRF-3 has an important role in virus-mediated induction of both IFNA and IFNB genes. While this paper was being completed, Yoneyama et al. (19) showed that IRF-3 increases virus-mediated induction of IFNA and IFNB genes in L cells.
IFN Signaling Pathway Is Critical for IRF-3-Mediated Antiviral Effect.
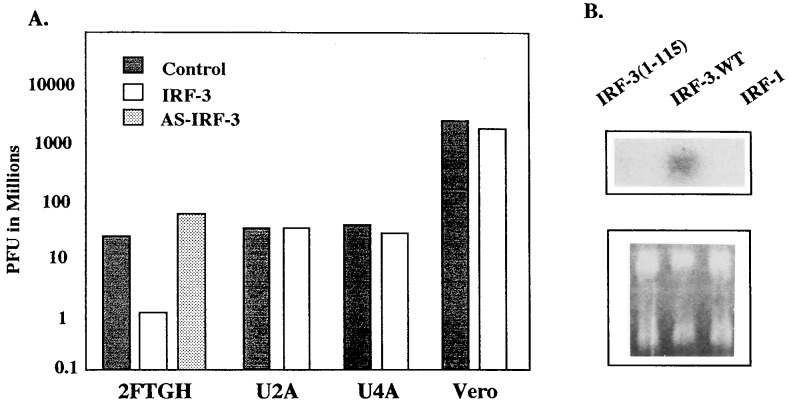
It was shown previously that overexpression of IRF-1 confers an antiviral state that is activated by an IFN-independent pathway (28). In a transient expression assay in L cells and 293 cells, IRF-3 enhances transcriptional activity of the ISG15 promoter (16, 18). To determine whether the antiviral effect can be induced by IRF-3 independent of IFN signaling, the human fibroblast line, 2FTGH, and mutant cell lines derived from these cells that lack one of the essential components of the IFN signaling pathway were used (29). Overexpression of IRF-3 in 2FTGH cells restricted VSV replication by more than 10-fold when compared with replication in 2FTGH cells transfected with the vector DNA (Fig. 4A). Analysis of the relative levels of IRF-3 in transfected 2FTGH cells shows the presence of IRF-3 in both the cytoplasm and nucleus (data not shown). Transfection with plasmid expressing IRF-3 antisense (IRF-3-AS) did not restrict VSV replication, and these cells were even more permissive to virus replication. These results further confirmed that the antiviral state is induced by IRF-3 and not by the transfected DNA. In contrast, overexpression of IRF-3 in U2A cells that have a nonfunctional p48 and thus are unable to form the ISGF3 transcriptional complex, or in U4A cells that are defective in JAK1 function, did not result in inhibition of VSV replication. Both of these cells are insensitive to the antiviral effect of IFN.
Figure 4.
Overexpression of IRF-3 activates the antiviral state in 2FTGH cells, but not in the mutants defective in IFN signaling. (A) The cells were transfected with 4 μg of IRF-3 expression plasmid and infected with VSV 16 hr posttransfection. VSV plaque assay was performed as described in the Materials and Methods. Representative experiment from three independent experiments is shown. The SD between individual experiments was less than 10%. (B) RNA was purified from 2FTGH cells transiently transfected with 4 μg of: IRF-3 (1–115), IRF-3, or IRF-1, 24 hr after transfection and analyzed by Northern blot hybridization with the IFNB-specific riboprobe.
By Northern hybridization, the presence of IFNB mRNA in IRF-3-transfected 2FTGH cells was detected, whereas no induction of IFNB mRNA was observed in cells transfected only with DBD of IRF-3 (amino acids 1–115) (Fig. 4B). Also, overexpression of IRF-1 did not induce expression of the IFNB gene in these cells. Under the same conditions, neither IRF-3 nor IRF-1 induced detectable levels of IFNA mRNA (data not shown). Taken together, these data indicate that the antiviral effect of IRF-3 requires a functional IFN signaling pathway and that, in 2FTGH cells even in the absence of viral infection, IRF-3 can be detected in the nucleus and can stimulate the transcription of IFNB, but not IFNA genes. However, because the expression of IFNA genes in these cells is not stimulated (or very little) even by viral infection, we cannot make a general conclusion that IRF-3 does not induce the IFNA genes.
To determine the contribution of direct activation of ISG with antiviral activity to the IRF-3-mediated induction of the antiviral state, Vero cells, which are sensitive to the antiviral effect of IFN but lack the endogenous IFNA and IFNB genes, were used. Because viral infection of Vero cells nonetheless can stimulate expression of the exogenously introduced IFNB promoter, this indicates that the viral-mediated signaling that leads to the activation of IFNB gene is not impaired in these cells (21). As shown in Fig. 4A, transient overexpression of IRF-3 in Vero cells did not induce the antiviral state in these cells, suggesting that the observed antiviral activity of IRF-3 in 2FTGH cells mainly results from direct induction of IFN gene expression.
Inhibition of IRF-3-Mediated Transactivation by E1A.
As we have shown previously in 293 cells, IRF-3 was unable to transactivate IFNA and IFNB promoters, and IRF-3 overexpression did not enhance virus-mediated induction of the endogenous IFN genes in these cells (17). Because these cells express the adenovirus E1A oncogene product that inhibits IFNα responses (30), we examined whether E1A can also counteract IRF-3-mediated transactivation. As seen in Fig. 5A, expression of E1A resulted in 94% inhibition of IRF-3-mediated transactivation of the IFNA4 gene promoter, whereas E1A inhibited transactivation by IRF-3(5A) by only 15%. Finally, E1A also inhibited virus-mediated induction of the IFNA4 promoter, as well as the synergistic transactivation by virus and IRF-3 (Fig. 5B).
Figure 5.
Inhibition of IRF-3-mediated activation of the IFNA4 promoter by E1A. (A) REF cells were cotransfected with 2.0 μg of plasmids expressing IRF-1, IRF-3, or its mutants, 2 μg of IFNA4/CAT reporter plasmid, and 100 ng of pCMV-β-galactosidase in the presence and absence of E1A (2.0 μg) or E1A (Δp300) mutant. (B) The synergistic activation of IFNA promoter by IRF-3 overexpression and NDV infection did not circumvent E1A-mediated suppression. Transfection was carried out as described in A, and cells were infected 24 hr posttransfection with NDV for 16 hr as described in Materials and Methods. (C) Overexpression of p300 partially restored the IRF-3 activity from suppression by E1A. REF cells were transfected with 1.5 μg of IFNA4/CAT, 100 ng of pCMV-β-galactosidase, 1.2 μg of IRF-3 and E1A expressing plasmids, and increasing amounts (0, 0.5, 1.5 μg) of p300 expressing plasmid. (Inset) Western blotting. Expression of IRF-3 in REF cells cotransfected with IRF-3 and p300 (lanes 1–3) and E1A (lane 2) and infected with NDV (lane 3). (D) Induction of IFNA4/CAT by IRF-3 was suppressed by the cotransfection of carboxyl terminus of CBP (amino acids 1992–2441). Two micrograms of IFNA4/CAT, 100 ng of pCMV-β-galactosidase, and 1.5 μg of IRF-3 expressing plasmids were cotransfected with 0, 2, or 4 μg of Δ 5′ CBP plasmid. The CAT assay was done as described above.
The carboxyl-terminal phosphorylation of IRF-3 was shown recently to be important for interaction of IRF-3 with transcriptional adapters p300/CBP (18, 19) that are targeted by E1A (31). Transactivation of IFNA4 promoter by IRF-3 was inhibited by cotransfection with the carboxyl-terminal peptide (amino acids 1992–2441) of CBP that contains the IRF-3-binding site (18) (Fig. 5D). If inhibition by E1A is a result of competition for binding to limiting amounts of p300, then E1A mutant unable to bind p300 should not be inhibitory and overexpression of p300 may be expected to restore IRF-3-mediated transactivation in the presence of E1A. As seen in Fig. 5A, no inhibition of IRF-3 or virus-mediated activation of IFNA4 promoter was detected with E1A mutant [E1A(Δp300)] being unable to bind the transcriptional adapter p300/CBP (Fig. 5 A and B). Furthermore, overexpression of p300 partially reversed the E1A-mediated suppression. That the restoration of IRF-3 activity by p300 was incomplete indicates that an additional mechanism may be involved in E1A-mediated suppression. To exclude the possibility that the inhibitory effect of E1A on IRF-3 transactivation is a result of down-regulation of IRF-3 expression, we analyzed, by Western blot, the levels of IRF-3 in transfected cells in the presence and absence of E1A and found that the levels of IRF-3 were not affected by the presence of E1A (Fig. 5C).
DISCUSSION
The findings presented in this study show that IRF-3 can strongly enhance virus-stimulated expression of IFN genes in both mouse and human fibroblast cells. As shown previously (18), virus induces posttranslational phosphorylation of the carboxyl terminus of IRF-3 at residues Ser-396, -398, -402, and -405 as well as Thr-404. Mutations introduced into these phosphorylation sites [IRF-3(5A)] decreased the transactivating ability of IRF-3 by about 50% in a transient transfection assay in both infected and uninfected cells, and further abolished its ability to enhance the endogenous IFN synthesis in infected cells. These data indicate that the virus-induced phosphorylation of IRF-3 modulates transcriptional activity of IRF-3. However, IRF-3 overexpression alone can activate the transcriptional activity of IFNA and IFNB promoters, induce an antiviral state, and stimulate transcription of the endogenous IFNB gene in a cell type-specific manner (unpublished results). These data indicate that, in certain cell types when expressed at high levels, IRF-3 can overcome the requirement for the virus-mediated phosphorylation. Indeed, we have detected the presence of IRF-3 in the nucleus of 2FTGH cells overexpressing IRF-3 (data not shown), and a mouse cell line, in which expression of IRF-3 is regulated transcriptionally, produces high levels of IFN (500 units/ml) after induction of IRF-3 expression (unpublished results). These findings, together with the observation that mutants of IRF-3 that cannot be phosphorylated retain low transactivation activity, indicate that the basal transactivation activity of IRF-3 may be mediated by its transactivation domain. Nevertheless, the virus-mediated carboxyl-terminal phosphorylation of IRF-3 stimulates its transactivation potential, enhances its interaction with the transcriptional coactivator p300/CBP (18–20), and consequently stabilizes the activation complexes. The critical role of p300/CBP in IRF-3-mediated activation is further supported by three observations: (i) overexpression of the carboxyl-terminal region of CBP that interacts with IRF-3 (18) suppressed the IRF-3-mediated activation; (ii) IRF-3-stimulated activation of IFNA promoter was strongly (95%) inhibited by E1A, which interacts with the C/H3 domain of p300 as well as with the carboxyl-terminal region of p300 (32, 33); and (iii) E1A mutant [E1A(Δp300)], which does not bind p300, also does not modulate the IRF-3-mediated stimulation of the IFNA4 promoter. These results indicate that, when overexpressed, IRF-3 can bind p300/CBP. The Western blot analysis of IRF-3 in E1A-expressing cells and in the controls excluded the possibility that E1A targets IRF-3 for degradation. That overexpression of p300 was able to partially revert E1A repression of IRF-3 activity suggests that the inhibition may be a result of the competition for binding of E1A and IRF-3 to p300. The inhibition of IRF-3-mediated activation by E1A implies an alternative mechanism for viral mimicry, where adenovirus can overpass the antiviral effect of IFN by a direct interference with transcription of the IFNA and IFNB genes, in addition to the inhibition of the induction of ISG genes (30). It was observed previously that E1A inhibits induction of IFNB by double-stranded RNA (dsRNA); however, the mechanism of this inhibition was not clarified (34).
The finding that many transcription factors activated in virus-infected cells, such as IRF-1, IRF-3, NF-κB, and STAT proteins, interact with p300 (23, 35–38) identifies p300 as an integral component of the virus-induced activation complex. Both IRF-3 and p300 have been found recently in the dsRNA-induced DRAF-1 (dsRNA-activated factor) and VA-IRF (virus-activated IRF) ternary complex that bind to IFN-stimulated response element and PRDI, respectively (19, 20).
Although IRF-1 was shown to effectively stimulate the IFNA4 promoter in transient transfection assays (10, 11), induction of an antiviral state in cells overexpressing IRF-1 was found to be independent of IFN induction (27). Also, induction of the ISG-561 gene by dsRNA was found to be a direct effect that requires STAT1, but not the other components of the IFN signaling pathway (44). In contrast, as shown in the present study, the induction of the antiviral state in cells overexpressing IRF-3 is mainly due to the induction of endogenous IFN genes. The significant enhancement of expression of IFNB genes and IFN synthesis in infected, IRF-3-overexpressing cells suggests that IRF-3 may be the limiting factor that determines the levels of IFN synthesized in infected cells. However, the low levels of IFNA gene expressed in infected fibroblasts could not be increased to the levels induced in infected B cells or macrophages (data not shown) by overexpression of IRF-3, suggesting a requirement for another, possibly lymphoid cell-specific, transcription factor(s). We have identified previously two proteins of 68 and 96 kDa that bind to the AF1 site of IFNA promoter (10), and ISGF3γ (p48) was implied to play a role in the induction of both IFNA and IFNB genes (39). Thus, the transcriptional activation of IFNA genes may require synergy among multiple transcription factors, such as that proposed for the IFNB gene promoter (8, 9, 40, 41). An important function of IRF-3 in the activation of endogenous IFN genes may relate to its ability to recruit CBP/p300 to the IFN enhanceosome. Several studies have now demonstrated the critical role of histone acetyltransferase activity of CBP/p300 and its associated factors (42) in the destabilizing and remodeling of nucleosomes and improving the accessibility of transcription factors to their DNA targets (43). While this paper was being reviewed, it was shown (45) that IRF-3 may indeed be a component of the enhanceosome that binds to the IFNB gene promoter.
In conclusion, the present studies have established that IRF-3 plays a direct role in virus-mediated signaling and have identified a mechanism by which IRFs modulate transcriptional activity of the endogenous genes. These studies also provide further evidence that interaction between transcriptional factors and the p300/CBP coactivator is essential for the integration of their functional activity in response to viral infection and that an alteration of these interactions by a virus-encoded protein may represent a novel mechanism by which some viruses down-regulate the transcription of the antiviral genes.
Acknowledgments
The authors thank Drs. G. Nabel, D. Kalvakolanu, and C. Dang for the p300, E1A, E1A(Δp300), and PHA-2 Hyb plasmids, respectively, Dr. G. Stark for the 2FTGH cells and their mutants, Dr. P. Talalay for editing the paper, and B. Schneider for her help with the manuscript. This work was supported by grants from the National Institutes of Health, National Institute of Allergy and Infectious Diseases (AI19737) (P.M.P.) and from the Medical Research Council of Canada (J.H.).
ABBREVIATIONS
- IFN
interferon
- ICSBP
IFN consensus sequence-binding protein
- IRF
IFN regulatory factor
- ISG
IFN-stimulated gene
- CAT
chloramphenicol acetyltransferase
- NDV
Newcastle disease virus
- PRD
positive regulatory domain
- DBD
DNA-binding domain
- REF
rat embryo fibroblast
- VSV
vesicular stomatitis virus
- dsRNA
double-stranded RNA
References
- 1.Nguyen H, Hiscott J, Pitha P M. Cyt Growth Fact Rev. 1997;8:293–312. doi: 10.1016/s1359-6101(97)00019-1. [DOI] [PubMed] [Google Scholar]
- 2.Harada H, Fujita T, Miyamoto M, Kimura Y, Maruyama M, Furia A, Miyata T, Taniguchi T. Cell. 1989;58:729–739. doi: 10.1016/0092-8674(89)90107-4. [DOI] [PubMed] [Google Scholar]
- 3.Vaughan P S, Aziz F, van Wijnen A J, Wu S, Harada H, Taniguchi T, Soprano K J, Stein J L, Stein G S. Nature (London) 1995;377:362–365. doi: 10.1038/377362a0. [DOI] [PubMed] [Google Scholar]
- 4.Lin R, Mustafa A, Nguyen H, Hiscott J. J Biol Chem. 1994;269:17542–17549. [PubMed] [Google Scholar]
- 5.Weisz A, Marx P, Sharf R, Appella E, Driggers P H, Ozato K, Levi B-Z. J Biol Chem. 1992;267:25589–25596. [PubMed] [Google Scholar]
- 6.Brass A L, Kehrli E, Eisenbeis C F, Storb U, Singh H. Genes Dev. 1996;10:2335–2347. doi: 10.1101/gad.10.18.2335. [DOI] [PubMed] [Google Scholar]
- 7.Veals S A, Schindler C, Leonard D, Fu X-Y, Aebersold R, Darnell J E, Jr, Levy D E. Mol Cell Biol. 1992;12:3315–3314. doi: 10.1128/mcb.12.8.3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim T K, Maniatis T. Mol Cell. 1997;1:119–129. doi: 10.1016/s1097-2765(00)80013-1. [DOI] [PubMed] [Google Scholar]
- 9.Merika M, Williams A J, Chen G, Collins T, Thanos D. Mol Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- 10.Au W-C, Su Y, Raj N B K, Pitha P M. J Biol Chem. 1993;268:24032–24040. [PubMed] [Google Scholar]
- 11.Au W-C, Raj N B K, Pitha P M. Nucleic Acids Res. 1992;20:2877–2884. doi: 10.1093/nar/20.11.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuyama T, Kimura T, Kitagawa M, Pfeffer K, Kawakami T, Watanabe N, Kundig T M, Amakawa R, Kishihara K, Wakeham A, et al. Cell. 1993;75:83–97. [PubMed] [Google Scholar]
- 13.Reis L F, Ruffner H, Stark G, Aguetand M, Weissmann C. EMBO J. 1994;13:4798–4806. doi: 10.1002/j.1460-2075.1994.tb06805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Genin P, Braganca J, Darracq N, Doly J, Civas J. Nucleic Acids Res. 1995;23:5055–5063. doi: 10.1093/nar/23.24.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braganca J, Genin P, Bandu M T, Darracq N, Vignal M, Casse C, Doly J, Civas A. J Biol Chem. 1997;272:22154–22162. doi: 10.1074/jbc.272.35.22154. [DOI] [PubMed] [Google Scholar]
- 16.Au W-C, Moore P A, Lowther W, Juang Y-T, Pitha P M. Proc Natl Acad Sci USA. 1995;92:11657–11661. doi: 10.1073/pnas.92.25.11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schafer S L, Lin R, Moore P A, Hiscott J, Pitha P M. J Biol Chem. 1998;273:2714–2720. doi: 10.1074/jbc.273.5.2714. [DOI] [PubMed] [Google Scholar]
- 18.Lin R, Heylbroech C, Pitha P, Hiscott J. Mol Cell Biol. 1998;18:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. EMBO J. 1998;17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weaver B K, Kumar K P, Reich N C. Mol Cell Biol. 1998;18:1359–1368. doi: 10.1128/mcb.18.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mosca J D, Pitha P M, Hayward G S. J Virol. 1992;66:3811–3822. doi: 10.1128/jvi.66.6.3811-3822.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raj N B K, Au W-C, Pitha P M. J Biol Chem. 1991;266:11360–11365. [PubMed] [Google Scholar]
- 23.Perkins N K, Felzien L K, Betts J C, Leung K, Beach D H, Nabel G J. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- 24.Popik W, Pitha P M. Mol Cell Biol. 1996;16:6532–6541. doi: 10.1128/mcb.16.11.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raj N B K, Cheung S C, Rosztoczy I, Pitha P M. J Immunol. 1992;148:1934–1940. [PubMed] [Google Scholar]
- 26.Bisat F, Raj N B K, Pitha P M. Nucleic Acids Res. 1988;16:6067–6083. doi: 10.1093/nar/16.13.6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pitha P M, Biegel D, Yetter R A, Morse H C., III J Immunol. 1988;141:3611–3616. [PubMed] [Google Scholar]
- 28.Pine R. J Virol. 1992;66:4470–4478. doi: 10.1128/jvi.66.7.4470-4478.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darnell J E J, Kerr I M, Stark G R. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 30.Kalvakolanu D V R, Bandyopadhyay S K, Harter M L, Sen G S. Proc Natl Acad Sci USA. 1991;88:7459–7463. doi: 10.1073/pnas.88.17.7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. Nature (London) 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 32.Nakajima T, Fukamizu A, Takahashi J, Gage F H, Fisher T, Blenis J, Montminy M R. Cell. 1996;86:465–474. doi: 10.1016/s0092-8674(00)80119-1. [DOI] [PubMed] [Google Scholar]
- 33.Kurokawa R, Kalafus D, Ogliastro M-H, Kioussi C, Xu L, Torchia J, Rosenfeld M G, Glass C K. Science. 1998;279:700–703. doi: 10.1126/science.279.5351.700. [DOI] [PubMed] [Google Scholar]
- 34.Ackill A M, Foster G R, Laxton C D, Flavell D M, Stark R G, Kerr I M. Nucleic Acids Res. 1991;19:4387–4394. doi: 10.1093/nar/19.16.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H G, Moran E, Yaciuk P. J Virol. 1995;69:7917–7924. doi: 10.1128/jvi.69.12.7917-7924.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J J, Vinkemeier U, Gu W, Chakravarti D, Horvath C M, Darnell J E., Jr Proc Natl Acad Sci USA. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhattacharay S, Eckner R, Grossman S, Oldread E, Arany Z, D’Andrea A, Livingston D M. Nature (London) 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- 38.Horvai A E, Xu L, Korzus E, Brard G, Kalafus D, Mullen T M, Rose D W, Rosenfeld M G, Glass C K. Proc Natl Acad Sci USA. 1997;94:1074–1079. doi: 10.1073/pnas.94.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harada H, Matsumoto M, Sato M, Kashiwazaki Y, Kimura T, Kitagawa M, Yokochi T, Sok-PinTin R, Takasugi T, Kadokawa Y, et al. Genes Cells. 1996;1:995–1005. doi: 10.1046/j.1365-2443.1996.870287.x. [DOI] [PubMed] [Google Scholar]
- 40.Garoufalis E, Kwan I, Lin R, Mustafa A, Pepin N, Roulston A, Lascote J, Hiscott J. J Virol. 1994;68:4707–4715. doi: 10.1128/jvi.68.8.4707-4715.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thanos D, Maniatis T. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 42.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 43.Kadonaga J T. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 44.Bandyopadhyay S K, Leonard G T, Jr, Bandyopadhyay T, Stark G R, Sen G C. J Biol Chem. 1995;270:19624–19629. doi: 10.1074/jbc.270.33.19624. [DOI] [PubMed] [Google Scholar]
- 45.Wathelet M G, Lin C H, Parekh B S, Ronco L V, Howley P M, Maniatis T. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]