Abstract
Homologs of Escherichia coli RecA recombination protein, which have been found throughout the living kingdom, promote homologous pairing and strand exchange. The nucleoprotein filament, within which strand exchange occurs, has been conserved through evolution, but conservation of the polarity of exchange and the significance of that directionality has not been settled. Using oligonucleotides as substrates, and assays based on fluorescence resonance energy transfer (FRET), we distinguished the biased formation of homologous joints at either end of duplex DNA from the subsequent directionality of strand exchange. As with E. coli RecA protein, the homologous Rad51 proteins from both Homo sapiens (HsRad51) and Saccharomyces cerevisiae (ScRad51) propagated DNA strand exchange preferentially in the 5′ to 3′ direction. The data suggest that 5′ to 3′ polarity is a conserved intrinsic property of recombination filaments.
Human Rad51 protein (HsRad51) is a member of the universally distributed class of RecA-like proteins that play important roles in homologous recombination and recombinational repair (1). In prokaryotes, the RecA proteins play roles in recombination, postreplication repair, and the repair of double-strand breaks (2). In eukaryotes, members of this class play roles in meiotic recombination, double-strand break repair (3, 4), and possibly in Ig switch recombination (5).
At the enzymological level, Rad51 from budding yeast and human cells, and human Dmc1, a RecA homolog specific to meiotic cells, have been shown to be DNA-dependent ATPases that carry out homologous pairing and strand-exchange reactions that resemble the reactions catalyzed by RecA protein (6–9).
RecA protein promotes a search for homology by a single strand and initiates an exchange between that strand and duplex DNA. The helical nucleoprotein filament that mediates those complicated interactions is assembled in a directional fashion on single-stranded DNA (ssDNA), has a distinct asymmetric shape, and promotes strand exchange in a polarized fashion, from 5′ to 3′ with respect to the single strand within the filament (10–14). Conservation of the recombination filament from eubacteria to mammals indicates the importance of the structure and the likelihood that its fundamental functions have been conserved (15, 16). Reports on RecA homologs from Ustilago maydis (17), Saccharomyces cerevisiae (18), and Homo sapiens (19) indicated that the polarity of strand exchange was 3′ to 5′, the inverse of that promoted by RecA protein. However, another study of yeast Rad51 protein revealed effects of overhanging complementary nucleotides at the ends of the duplex substrate that raised questions about the polarity assigned to the eukaryotic homologs of RecA (20).
Recently developed assays based on fluorescence resonance energy transfer (FRET) clearly distinguish homologous pairing from subsequent strand exchange and detect these reactions as they occur in solution without the deproteinization that occurs in many other assays in vitro (8, 21, 22). We combined those assays with a strategy to assess in oligonucleotide substrates the progression of strand exchange from one end of DNA to the other as a function of time, and hence to deduce the intrinsic directionality of exchange. We discuss the possible biological implications of these findings.
METHODS
Enzymes.
Human Rad51 protein (HsRad51) and human RPA were purified as described (8, 23). Rad51 protein from S. cerevisiae (ScRad51) was a generous gift from Patrick Sung (Univ. of Texas Health Science Center at San Antonio).
DNA Substrates.
The single-strand oligonucleotides used in this study were designated as either (−) or (+) strand, and the duplex was designated by a letter and digit code. For example, A16(−) was a single-stranded oligonucleotide, A16(+) was its complement, and A16 was the duplex form. To minimize secondary structure, we used GCG software and required that each 83-mer or 84-mer designed have fewer than 8 successive intrastrand hydrogen bonds separated by less than 4 unpaired bases. The following oligonucleotides were synthesized:
A16(−), W16(−), and their complements were 83-mer oligonucleotides whose sequences have been published (8, 21). A16(−) had 16% G and C content whereas W16(−) had 40% G and C.
RG1(−) was a chimeric 84-mer containing a 71% AT-rich segment (42-mer) at its 5′ end and a 71% GC-rich segment (42-mer) at its 3′ end: 5′-TTT ACT TGT ACT TCA TTC ATT CAC ATT CCT ATC ATG TTT CTA CGC CAC CTC CCG ACC CTC CCC ACC ACT CAC CCA ACC GCT ACC-3′.
RG2(−) was a chimeric 84-mer containing a 71% AT-rich segment (42-mer) at its 3′ end and a 71% GC-rich segment (42-mer) at its 5′ end: 5′-CCA TCG CCA ACC CAC TCA CCA CCC CTC CCA GCC CTC CAC CGC ATC TTT GTA CTA TCC TTA CAC TTA CTT ACT TCA TGT TCA TTT-3′.
A16(−), RG1(−), and RG2(−) contained a six-carbon linker terminated by a primary amine at the 3′ end whereas the respective complementary strands had the same linker at the 5′ end. Labeling of oligonucleotides with fluorescein (5-carboxy-fluorescein succinimidyl ester) and rhodamine (5,6 carboxy-tetramethyl rhodamine succinimidyl ester) at the linker was done as described (21). Fluorescein- and rhodamine-labeled oligonucleotides have been named by putting the letters F or R, respectively, at the end of the strand name. Duplexes were made by thermal annealing of complementary oligonucleotides.
Duplexes EG302/305 and EG303/304 were made by PCR of pEG47, a plasmid consisting of vector pIBI30 (IBI), into which the 83-mer A16 was inserted between XbaI and PstI sites.
Detection of Synaptic Complexes by Homology-Dependent Protection of a Restriction Site.
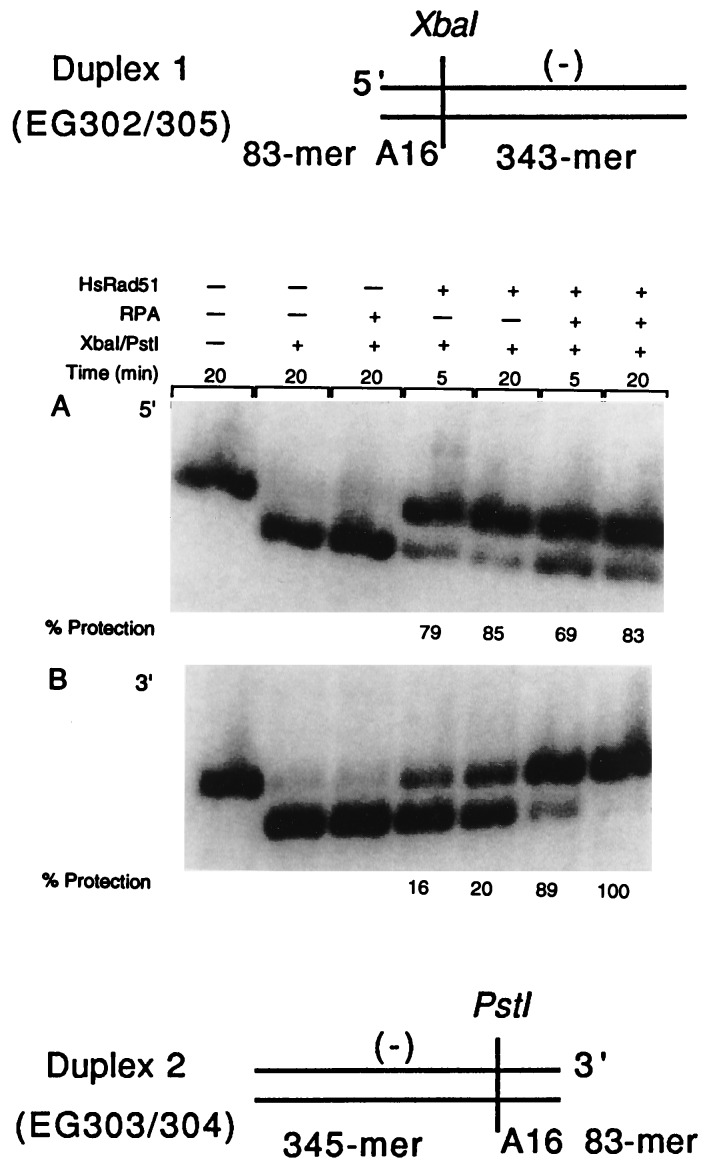
Duplex DNA substrates used for the restriction site protection assay were EG302/305 (426-mer) and EG303/304 (428-mer). EG302/305 contained the AT-rich A16 sequence at its 5′ end and EG303/304 had the A16 sequence at its 3′ end, the ends being named in relation to the sequence of the single strand in the nucleoprotein filament (Fig. 1, duplexes 1 and 2). HsRad51 (1.2 μM) was preincubated with 3 μM A16 (83-mer oligonucleotides) at 37°C for 5 min in 10 μl reaction buffer R (final concentration of 25 mM Hepes⋅KOH, pH 7.4/100 μg BSA/ml/1 mM DTT/2 mM ATP/1 mM MgCl2). RPA then was added to a final concentration of 0.075 μM, and incubation was continued for 4 min. The concentration of MgCl2 was increased to 30 mM followed by the addition of 3 μM 32P-labeled EG302/305 or EG303/304 duplex DNA. At 5-min and 20-min time intervals, 3 μl of reaction mixture (from total of 10 μl) was transferred to 7 μl of buffer A (10 mM Tris, pH 7.9/10 mM MgCl2/50 mM NaCl/1 mM DTT/100 μg BSA/ml) and 20 units of restriction enzyme was added. XbaI and PstI were the restriction enzymes used to cleave EG302/305 and EG303/304, respectively. Reactions were continued for 3 min and were stopped by the addition of 2 μl of stop solution (final concentration of 0.5% SDS/50 mM EDTA/0.04% bromophenol blue/0.04% xylene cyanol/5% glycerol). After an incubation on ice for 5 min, the reactions were subjected to 1.0% agarose gel electrophoresis in TAE buffer at 5 V/cm for 2 hr. After the gel was dried, the bands were analyzed and quantitated on a PhosphorImager by using imagequant software (Molecular Dynamics).
Figure 1.
Formation of homologous joints at either end of duplex DNA in the presence of RPA. For this assay, the presynaptic filament was formed on oligonucleotide A16(−) and the duplex DNAs were duplex 1 (EG302/305) and duplex 2 (EG303/304). Duplexes 1 and 2 possessed AT-rich A16 sequences at the 3′ end or 5′ end of the displaced strand, respectively. Duplex 1 (426-mer), used for the experiment presented in A, tests for pairing at the 5′ end whereas duplex 2, a 428-mer duplex (B) detects pairing at the 3′ end. We monitored protection of the XbaI site resulting from the reaction between A16 and EG302/305 and protection of the PstI site resulting from the reaction between A16 and EG303/304. Restriction enzymes were used in an amount that was sufficient to cleave the substrate DNA in 30 sec in absence of pairing. 32P-labeled 426-mer duplex 1 (lane 1, A) and 428-mer duplex 2 (lane 1, B) were cleaved, respectively, into a 333-mer (lane 2, A) and a 335-mer (lane 2, B) fragment on restriction cleavage. When filaments were formed on a heterologous oligonucleotide, no protection of the restriction sites was observed (data not shown).
Fluorimetric Analysis of the Polarity of Pairing and Strand Displacement.
For the pairing assay, 3 μM RG1(−) or RG2(−) labeled at its 3′ end with fluorescein was incubated with 1.2 μM HsRad51 or ScRad51 in reaction buffer R (total volume 150 μl) to form the presynaptic filament. The mixture was incubated at 37°C for 5 min followed by the addition of RPA at 0.036 μM and further incubation for 4 min. MgCl2 concentration was increased to 30 mM, and RG1 or RG2 duplex at 3 μM was added. These duplexes were labeled with rhodamine at the 5′ end of the (+) strand (Table 1A).
Table 1.
Inhibition of homologous pairing by Rad51 in direct proportion to GC content of oligonucleotide substrates
| % GC | Homologous pairing measured by FRET (relative signal, %) |
|---|---|
| 16 | 100 |
| 29 | 72 |
| 40 | 41 |
| 71 | 04 |
The substrates were homologous single- and double-stranded 83-mer oligonucleotides with the GC base contents indicated. Secondary structure in the single-stranded oligonucleotides was minimized and reactions were constituted as described in Methods. The steady-state level of homologous pairing was assessed after incubation at 37°C for 30 min by measurement of fluorescence resonance energy transfer (FRET) as described previously (8, 21). The entries in italics were derived by interpolation from the other data.
Strand displacement reactions employed unlabeled RG1(−) or RG2(−) strand for filament formation. HsRad51, RecA, or ScRad51 was incubated with unlabeled RG1(−) or RG2(−) oligonucleotide for 5 min followed, except in the case of RecA, by addition of RPA to 0.036 μM. After 4 min, the concentration of MgCl2 was increased to 30 mM. The reaction mixture containing the filaments (40 μl) was then transferred to a reaction buffer R (110 μl) containing 3 μM homologous duplex oligonucleotides and 30 mM MgCl2. The duplex oligonucleotide was labeled with fluorescein at the 3′ end of the (−) strand and with rhodamine at the 5′ end of the (+) strand (Table 1A). The final concentrations of ssDNA, dsDNA, and proteins were 3.0, 3.0, and 1.2 μM, respectively.
In another set of experiments, reactions were done with the single-stranded RG1(+) and RG2(+) labeled with fluorescein at their 5′ rather than 3′ ends, and these were reacted with their respective duplexes in the individual assays for pairing or strand exchange (Table 1B) as described above.
For both pairing and the strand displacement reactions, the emission from fluorescein was monitored at 520 nm upon excitation at 493 nm on an SLM 8000C spectrofluorimeter (SLM–Aminco, Urbana, IL). Emission data were collected every 2 sec. For plotting purposes, the differences in fluorescence emission at the start of the reaction between different samples were normalized by reference measurements of the average fluorescein emission at 520 nm before the addition of duplex.
Protection of 5′ and 3′ Ends from Exonucleolytic Digestion.
Oligonucleotide A16(−), 32P-labeled at either the 5′ or 3′ end, was incubated at 3 μM with either 1.2 μM HsRad51 or 0.075 μM RPA, or both, for 4 min as described (total volume 10 μl). λ Exonuclease (0.05 units) or E. coli exonuclease I (2 units) was added to the reaction, and incubation was continued for 2 min. In the case of λ exonuclease, MgCl2 was increased to 5 mM before the addition of exonuclease. After 2 min, the reaction was stopped by the addition of 25 μg yeast tRNA in stop solution. Four hundred microliters of 20% TCA was added and the reaction was chilled on ice for 10 min followed by centrifugation at 8,000 × g. The supernatant (200 μl) was counted for radioactivity to quantitate the fraction of acid-soluble counts.
RESULTS
The Formation of Homologous Joints at Either End of Duplex DNA by HsRad51.
As a preliminary, we studied the formation of joints at either end of duplex DNA, because a bias in this property could confound a determination of the intrinsic directionality of strand exchange. Following common practice, we define the directionality of strand exchange, 5′ to 3′ or 3′ to 5′, with respect to the polarity of the single strand in the presynaptic nucleoprotein filament and the identical strand that is displaced from duplex DNA by the exchange. We designate the ends of duplex DNA as 5′ or 3′ in relation to that same strand; thus, a 5′ to 3′ exchange starts at the 5′ end of the common strand.
We designed two substrates with an 83-nt AT-rich sequence and an adjacent restriction site at one or the other end of the DNA to look for a bias in the formation of joints by HsRad51 at either end (Fig. 1). We used protection of the indicated restriction sites in duplex DNA to detect stable, homology-dependent interactions with HsRad51 filaments formed on A16(−), which is homologous to the 83-mer sequence engineered at either end of the duplex DNA (24).
When incubated with the respective restriction enzymes, the uniformly radiolabeled duplex DNA was converted efficiently into a ca. 340-mer DNA fragment (lane 2, Fig. 1 A and B). When the duplex substrates were incubated first with HsRad51 nucleoprotein filament formed on A16(−), the 5′ end of the duplex was 85% protected in 20 min, whereas the 3′ end was only 20% protected (Fig. 1). However, when HsRPA, the human heterotrimeric single-stranded DNA-binding protein, was added to the reaction mixture after HsRad51 had initiated filament formation on A16(−), subsequent protection of the 3′ end of the duplex DNA increased to 100%, whereas protection of the 5′ end of the duplex DNA remained at its already high level of about 85%.
Given its function as a single-stranded DNA binding protein (25), we suspected that RPA was acting on the formation of nucleoprotein filaments. To test that hypothesis, we examined the protection by HsRad51 and RPA of labeled 5′ and 3′ ends from digestion by exonucleases. We incubated terminally labeled A16(−) with HsRad51 alone, RPA alone, and both proteins together for 4 min at 37° before we tested the ability of exonucleases to make the terminal label soluble in acid. In tests that were duplicated, λ exonuclease was used to assess protection of the 5′ end, and E. coli exonuclease I was used to assess protection of the 3′ end. HsRad51 alone protected 65% of 5′ ends from digestion, whereas RPA alone or RPA and HsRad51 protected about 90% of ends. At the 3′ end, there was a bigger effect of RPA on protection. HsRad51 alone protected only 13% of ends from digestion by exonuclease I. HsRad51 together with RPA, however, protected 53% of 3′ ends, and RPA by itself protected 79% of 3′ ends. The effect of RPA on the protection of single-stranded DNA was not sequence-specific because similar results were obtained with oligonucleotide W16 (40% GC) (data not shown). These observations on protection of 5′ and 3′ ends correlate well with the observed effects of RPA on the formation of joints at 5′ vs. 3′ ends. RPA had no effect on the formation of joints at 5′ ends and little effect on protection of single-stranded 5′ ends in the HsRad51 filament from exonuclease digestion; whereas RPA had 4- to 5-fold effects on both the formation of joints at 3′ ends and the protection of 3′ ends within the HsRad51 filament. Because RPA by itself cannot form joints between homologous substrates (Fig. 1), we surmise that its effect on the formation of joints at 3′ ends is attributable at least in part to the formation of a more complete filament by HsRad51 at 3′ ends of single-stranded DNA.
Because RPA equalized the formation of joints at both ends of duplex DNA, it became a useful reagent to nullify a possible source of bias in the determination of directionality of strand exchange.
Strategy for Determination of the Progression of Strand Exchange from One End to the Other of Oligonucleotide Substrates.
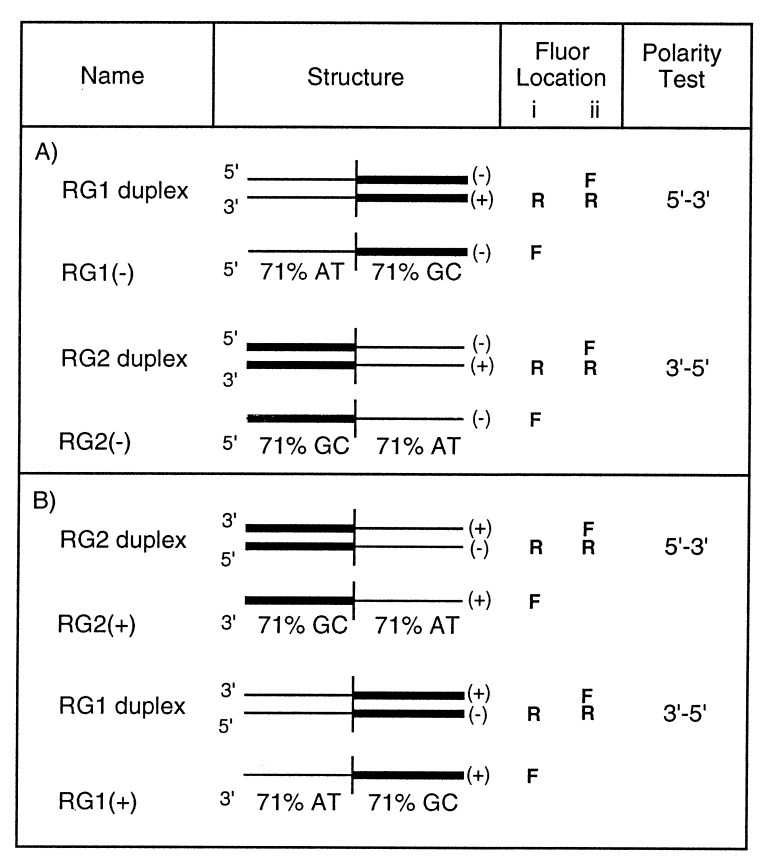
In experiments that will be described in detail elsewhere, we found that homologous pairing by Rad51 (as well as by RecA protein) is inhibited in direct proportion to the GC content of the oligonucleotide substrates (Table 1). The efficiency of formation of a homologous joint in a molecule that had 29% GC was more than 18 times greater than in a molecule that had 71% GC (Table 1). We further determined that even segments of only 12 GC base pairs located at both ends of an otherwise AT-rich 83-mer could completely inhibit strand exchange as measured by gel electrophoresis (data not shown). Therefore, we constructed oligonucleotide substrates in which half of the molecule was AT-rich and half was GC-rich (Fig. 2). By this means, we could strongly bias the formation of joints to either a 5′ or 3′ AT-rich end and ask whether strand exchange could be propagated subsequently through the GC-rich sequence at the other end.
Figure 2.
Chimeric substrates for detection of the polarity of strand exchange. The substrates used for fluorimetric assays of pairing and strand-displacement reactions. Thick and thin lines represent the GC-rich and AT-rich segments, respectively. Fluor location i and ii represent the placements of fluorescein (F) and rhodamine (R) on different strands for pairing and strand displacement, respectively. The dyes were placed at the right-hand ends of the symbolized strands: in the substrates shown in A, fluorescein was placed at the 3′ end of the single strand in the assay for pairing and at the 3′ end of the displaced strand in the assay for strand exchange; rhodamine was placed at the 5′ end of the plus strand in either assay. In B, the single-stranded oligonucleotides were the complements of those used in A, and fluorescein was located at the 5′ end of the single strand in the assay for pairing and at the 5′ end of the displaced strand in the assay for strand exchange, whereas rhodamine was at the 3′ end of the minus strand in either assay.
To distinguish effects on homologous pairing from effects on subsequent strand exchange, we used recently designed assays that are based on FRET (21). Depending on the placement of fluorescent dyes at the ends of strands of oligonucleotide substrates, these assays detect the conjunction of strands that occurs upon homologous pairing or the subsequent separation of homologous strands that occurs upon strand exchange.
The Preferential Propagation of Strand Exchange Through a Region That Lies 3′ to a Joint.
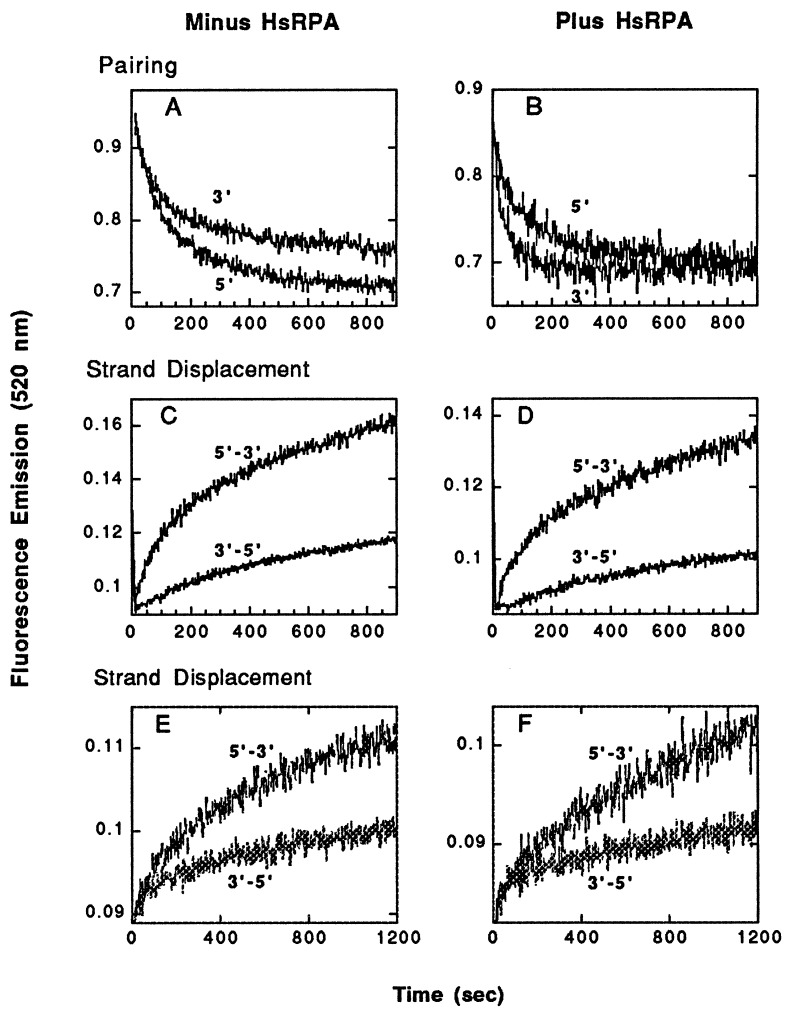
In a first set of experiments, we used the fluorescence assay to assess the formation of joints when the substrates in Fig. 2 had AT-rich sequences at 5′ vs. 3′ ends. The presynaptic filament was formed on RG1(−)F or RG2(−)F single-stranded oligonucleotides and was paired with the corresponding duplex (Fig. 2A). Quenching in fluorescence emission was recorded every 2 sec after the addition of duplex to an otherwise complete reaction mixture at 37°C. In the absence of RPA, the yield of joint molecules was about 30% greater when AT-rich sequences were located at the 5′ vs. the 3′ end of paired substrates (Fig. 3A). Addition of RPA eliminated the relative deficit seen when 3′ ends were AT-rich, and indeed the apparent rate of formation of joints was three times greater when AT-rich DNA was present at the 3′ end (Fig. 3B; rates were estimated from apparent first-order rate constants provided by a curve-fitting program). These observations are consistent with the experiments depicted in Fig. 1, which showed that HsRad51 alone forms joints better at 5′ than 3′ ends, but that addition of RPA corrects the deficit at 3′ ends.
Figure 3.
The polarity of strand exchange promoted by HsRad51, as determined by the fluorimetric assay. In A–D, the substrates used were those shown in Fig. 2A. (A and B) Pairing promoted by HsRad51 without and with RPA, respectively. The labels 3′ and 5′ refer to the end of the single-stranded oligonucleotide, either RG1(−) or RG2(−), at which AT-rich sequences were located. (C and D) Strand exchange promoted by HsRad51 without and with RPA, respectively. In E and F, the substrates used were those shown in Fig. 2B in which there was an alternative arrangement of the fluorescent dyes. (E) Strand exchange in the absence of RPA. (F) Strand exchange in the presence of RPA.
With the fluorescent dyes located on the substrates to detect strand exchange, we then examined the ability of HsRad51 to propagate strand exchange through a GC-rich region lying either 5′ or 3′ to the joints formed in AT-rich DNA. Exchange indeed was propagated through GC-rich regions but more effectively when GC-rich DNA was located 3′ to the AT-rich sequences (Fig. 3C). This bias in the propagation of strand exchange was unaffected by the addition of RPA (Fig. 3D). The observed preference for propagation of exchange through a GC region lying 3′ to an AT-rich region is consistent with preferential strand exchange in the 5′ to 3′ direction.
In the experiments shown in Fig. 3 A–D, fluorescein was located at the 3′ ends of the single strands in the presynaptic filaments, which governed the locations of fluorescein and rhodamine in the duplex oligonucleotides as indicated in Fig. 2A both for pairing and strand-displacement assays. To determine whether the placement of the reporter dyes biased the outcome of either pairing or strand-exchange experiments, we prepared a set of substrates based on the location of fluorescein at the 5′ end of the strand in the presynaptic filament (Fig. 2B), which in this case was the complement of the single strand diagrammed in Fig. 2A. We observed a similar bias in strand exchange with or without the presence of RPA; strand exchange was faster when the substrate was 5′-(AT)42(GC)42-3′ rather than the inverse orientation (Fig. 3 E and F). The results of pairing assays also were similar to those shown in Fig. 3 A and B (data not shown). From these experiments, we conclude that the bias in strand exchange was not determined by the location of the reporter dyes but rather by the relative orientation of the AT-rich and GC-rich sequences. We also note that in this second set of experiments the oligonucleotide in the filament had a different sequence than that in the first set, indicating that nucleotide sequence is not a unique determinant.
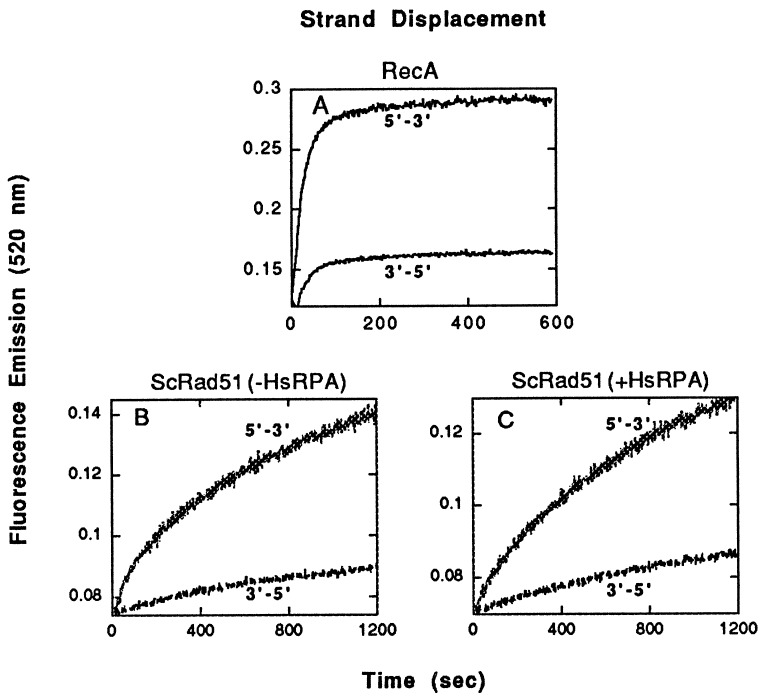
As a control, we used the fluorimetric assay to examine the action of RecA protein, which is known to promote exchange in the 5′ to 3′ direction (10–12). The RecA reaction also was clearly biased in favor of the substrate in which strand exchange was required to move 5′ to 3′ through a GC-rich region that lay 3′ to AT-rich DNA (Fig. 4A).
Figure 4.
The polarity of strand exchange promoted by RecA and ScRad51, as determined by the fluorimetric assay. The substrates were those shown in Fig. 2A. (A) Strand exchange by RecA. (B and C) Strand exchange by ScRad51 without and with human RPA, respectively.
Because the apparent polarity of strand exchange is the same for RecA protein from E. coli and Rad51 from H. sapiens, we were curious about the polarity of Rad51 from S. cerevisiae. Applying the same tests, we found that yeast Rad51 also propagated strand exchange preferentially through a GC-rich region that lay 3′ to AT-rich sequences rather than 5′ (Fig. 4 B and C). We also observed that human RPA stimulated the formation of 3′ joints by yeast Rad51 protein (data not shown).
DISCUSSION
The Polarity of Strand Exchange Promoted by Human Rad51 Protein.
Although the formation of homologous joints at one end or the other of duplex DNA may provide clues on the polarity of strand exchange promoted by recombination filaments, the determination of directionality clearly requires the demonstration that exchange progresses with time from one end to the other. In the present study, two tools enabled us to do so. The first was provided by the observation that the yield of homologous joints by HsRad51 or RecA protein is inversely proportional to GC content (Table 1 and unpublished observations). This made it possible to bias the formation of joints to either end of a duplex oligonucleotide by putting AT-rich DNA at one end and GC-rich DNA at the other (Fig. 2). The second tool was provided by recently developed fluorimetric assays, which distinguish homologous pairing from subsequent strand exchange by a nondisruptive spectroscopic method (21). When we examined the preferential formation of joints by HsRad51 in AT-rich regions at 5′ vs. 3′ ends of oligonucleotides we found a preference for 5′ ends in the absence of RPA, and a smaller preference for 3′ ends in the presence of RPA. These preferences, however, had no observable effect on the bias of subsequent strand exchange to proceed in the direction, 5′ to 3′ (Fig. 3). Thus, we infer that the observed directionality of strand exchange probably did not reflect a functional bias imposed before strand exchange.
In the present and previous studies of human Rad51 involving oligonucleotides or polynucleotides as substrates, blunt ends in the DNA did not completely inhibit homologous pairing and strand exchange (8), as was reported for yeast Rad51 acting on circular single-stranded DNA and linear duplex DNA (20). We have observed, however, that overhanging complementary single-stranded ends stimulate the reactions of human Rad51 (data not shown).
A Uniform Polarity of Strand Exchange Promoted by Nucleoprotein Filaments.
The findings reported here show that in short-range interactions involving the exchange of a few dozen base pairs, proteins of the RecA family, including one from a prokaryote, one from a lower eukaryote, and one from a higher eukaryote all promote strand exchange preferentially in the 5′ to 3′ direction. All of these proteins produce structurally similar nucleoprotein filaments, and it is easier to comprehend an intrinsic and uniform polarity, rather than a nonuniform polarity, associated with a structure that has been conserved through evolution. It is conceivable, however, that other factors influence the apparent directionality of exchange when the substrates are long DNA molecules. For example, the separation of kilobase lengths of a displaced single strand from the heteroduplex product could be governed by factors that reverse the apparent polarity.
The Polarity Paradox.
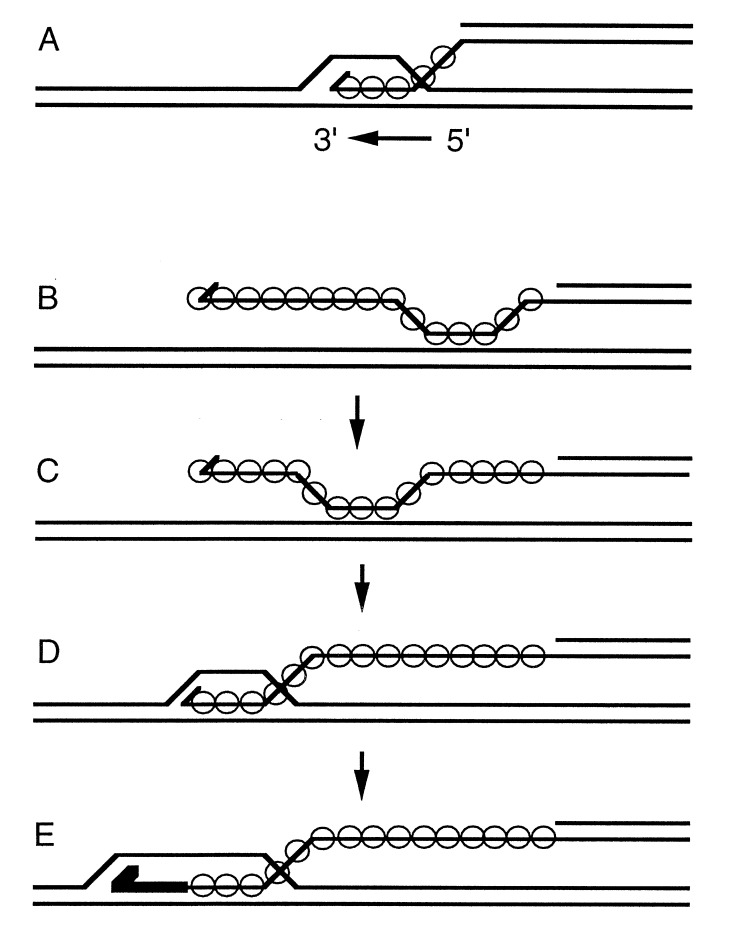
Genetic and biochemical studies in E. coli and in S. cerevisiae have shown that invasion of duplex DNA by a 3′ single-stranded end commonly initiates homologous recombination (27–30). Paradoxically, however, strand exchange in the 5′ to 3′ direction would undo joints that are made by 3′ invasion (Fig. 5A) (31). Previous observations of apparent 3′ to 5′ polarity of eukaryotic homologs of RecA suggested that the paradox might be limited to prokaryotes. If polarity is uniform, the paradox may be general. One way to resolve the quandary is to suppose that the intrinsic polarity that we have associated with exchange over short distances in vitro has nothing to do with the extensive processing of a plectonemic joint that produces long heteroduplex regions, which is likely to be the function of other proteins (32). Instead, the recombination filament may govern the 5′ to 3′ polarity of migration of precursor paranemic joints with the result that 3′ ends are aligned rapidly at homologous sites where they can “invade” duplex DNA, leading to further stabilizing events, such as the priming of new DNA synthesis (Fig. 5 B–E).¶ In eukaryotes, the role of RPA is readily reconciled with that scenario. In view of its vital functions in replication, repair, and recombination (25, 33–35), its physical interaction with Rad52 (36) and Rad51 (E.I.G., R.C.G., T. Haaf, M.S.W., and C.M.R., unpublished observations), and its ability to stimulate the formation of joints at 3′ ends, RPA might play a role in ensuring 3′ invasion and then mediating further interactions with enzymes that are required to process the initial D-loop intermediate.
Figure 5.
The polarity paradox. Circles indicate the location of RecA protein or one of its homologs. A spur at the end of a line designates a 3′ end. The heavy line in E represents new DNA synthesis. (A) Polarity, 5′ to 3′ with respect to the single strand on which the nucleoprotein filament had formed, would dissociate all joints. (B–E) Another view of the role of polarity, according to which the polarity intrinsic to the recombination filament is only manifested in the migration of paranemic joints. Before strand invasion (B and C), 5′ to 3′ polarity would serve to drive a paranemic joint toward the 3′ end of the single strand where strand invasion (D) can occur. As in the double-strand break model for recombination, new DNA synthesis would extend and stabilize the joint (E). The subsequent action of enzymes like RuvA, B, and C can extend the region of heteroduplex DNA and finally resolve the joints into recombined products. According to this hypothesis, the polarity intrinsic to the filament only plays a biological role before strand invasion, after which the actions of other enzymes supervene.
Acknowledgments
This research was supported by Grant No R37-GM33504 from the National Institutes of Health. We are grateful to Oleg Kovalenko, Andrej Stasiak, Patrick Sung, and Michael Liskay for helpful comments on the manuscript, to Jan Zulkeski for data processing, and to Zhufang Li for technical assistance.
ABBREVIATION
- FRET
fluorescence resonance energy transfer
Footnotes
A related but different proposal was made previously suggesting that RecA protein in vitro finds DNA ends with 3′ to 5′ polarity and subsequently extends heteroduplex formation in the 5′ to 3′ direction (26).
References
- 1.Shinohara A, Ogawa H, Matsuda Y, Ushio N, Ikeo K, Ogawa T. Nat Genet. 1993;4:239–243. doi: 10.1038/ng0793-239. [DOI] [PubMed] [Google Scholar]
- 2.Kowalczykowski S C, Eggleston A K. Annu Rev Biochem. 1994;63:991–1043. doi: 10.1146/annurev.bi.63.070194.005015. [DOI] [PubMed] [Google Scholar]
- 3.Shinohara A, Ogawa H, Ogawa T. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 4.Bishop D K, Park D, Xu L, Kleckner N. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 5.Li M-J, Peakman M-C, Golub E I, Reddy G, Ward D C, Radding C M, Maizels N. Proc Natl Acad Sci USA. 1996;93:10222–10227. doi: 10.1073/pnas.93.19.10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sung P. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 7.Baumann P, Benson F, West S C. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 8.Gupta R, Bazemore L R, Golub E I, Radding C M. Proc Natl Acad Sci USA. 1997;94:463–468. doi: 10.1073/pnas.94.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, Golub E I, Gupta R, Radding C M. Proc Natl Acad Sci USA. 1997;94:11221–11226. doi: 10.1073/pnas.94.21.11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox M M, Lehman I R. Proc Natl Acad Sci USA. 1981;78:6018–6022. doi: 10.1073/pnas.78.10.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kahn R, Cunningham R P, DasGupta C, Radding C M. Proc Natl Acad Sci USA. 1981;78:4786–4790. doi: 10.1073/pnas.78.8.4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.West S C, Cassuto E, Howard-Flanders P. Proc Natl Acad Sci USA. 1981;78:6149–6153. doi: 10.1073/pnas.78.10.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Register J C, Griffith J. J Biol Chem. 1985;260:12308–12312. [PubMed] [Google Scholar]
- 14.Stasiak A, Egelman E H, Howard-Flanders P. J Mol Biol. 1988;202:659–662. doi: 10.1016/0022-2836(88)90293-8. [DOI] [PubMed] [Google Scholar]
- 15.Ogawa T, Yu X, Shinohara A, Egelman E H. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- 16.Benson F E, Stasiak A, West S C. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kmiec E B, Holloman W K. Cell. 1983;33:857–864. doi: 10.1016/0092-8674(83)90028-4. [DOI] [PubMed] [Google Scholar]
- 18.Sung P, Robberson D L. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 19.Baumann P, West S C. EMBO J. 1997;16:5198–5206. doi: 10.1093/emboj/16.17.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Namsaraev E, Berg P. Mol Cell Biol. 1997;17:5359–5368. doi: 10.1128/mcb.17.9.5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bazemore L R, Takahashi M, Radding C M. J Biol Chem. 1997;272:14672–14682. doi: 10.1074/jbc.272.23.14672. [DOI] [PubMed] [Google Scholar]
- 22.Bazemore L R, Folta-Stogniew E, Takahashi M, Radding C M. Proc Natl Acad Sci USA. 1997;94:11863–11868. doi: 10.1073/pnas.94.22.11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henricksen L A, Umbricht C B, Wold M S. J Biol Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 24.Hsieh P, Camerini-Otero C S, Camerini-Otero R D. Proc Natl Acad Sci USA. 1992;89:6492–6496. doi: 10.1073/pnas.89.14.6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wold M S. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 26.Morel P, Stasiak A, Ehrlich S D, Cassuto E. J Biol Chem. 1994;269:19830–19835. [PubMed] [Google Scholar]
- 27.Smith G R. Annu Rev Genet. 1987;21:179–201. doi: 10.1146/annurev.ge.21.120187.001143. [DOI] [PubMed] [Google Scholar]
- 28.Sun H, Treco D, Szostak J. Cell. 1991;64:1155–1161. doi: 10.1016/0092-8674(91)90270-9. [DOI] [PubMed] [Google Scholar]
- 29.Silberstein Z, Shalit M, Cohen A. Genetics. 1993;133:439–448. doi: 10.1093/genetics/133.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friedman-Ohana R, Cohen A. Proc Natl Acad Sci USA. 1998;95:6909–6914. doi: 10.1073/pnas.95.12.6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dutreix M, Rao B J, Radding C M. J Mol Biol. 1991;219:645–654. doi: 10.1016/0022-2836(91)90661-o. [DOI] [PubMed] [Google Scholar]
- 32.Eggleston A K, Mitchell A H, West S C. Cell. 1997;89:607–617. doi: 10.1016/s0092-8674(00)80242-1. [DOI] [PubMed] [Google Scholar]
- 33.Longhese M P, Plevani P, Lucchini G. Mol Cell Biol. 1994;14:7884–7890. doi: 10.1128/mcb.14.12.7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Firmenich A A, Elias-Arnanz M, Berg P. Mol Cell Biol. 1995;15:1620–1631. doi: 10.1128/mcb.15.3.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith J, Rothstein R. Mol Cell Biol. 1995;15:1632–1641. doi: 10.1128/mcb.15.3.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park M S, Ludwig D L, Stigger E, Lee S-H. J Biol Chem. 1996;271:18996–19000. doi: 10.1074/jbc.271.31.18996. [DOI] [PubMed] [Google Scholar]