Abstract
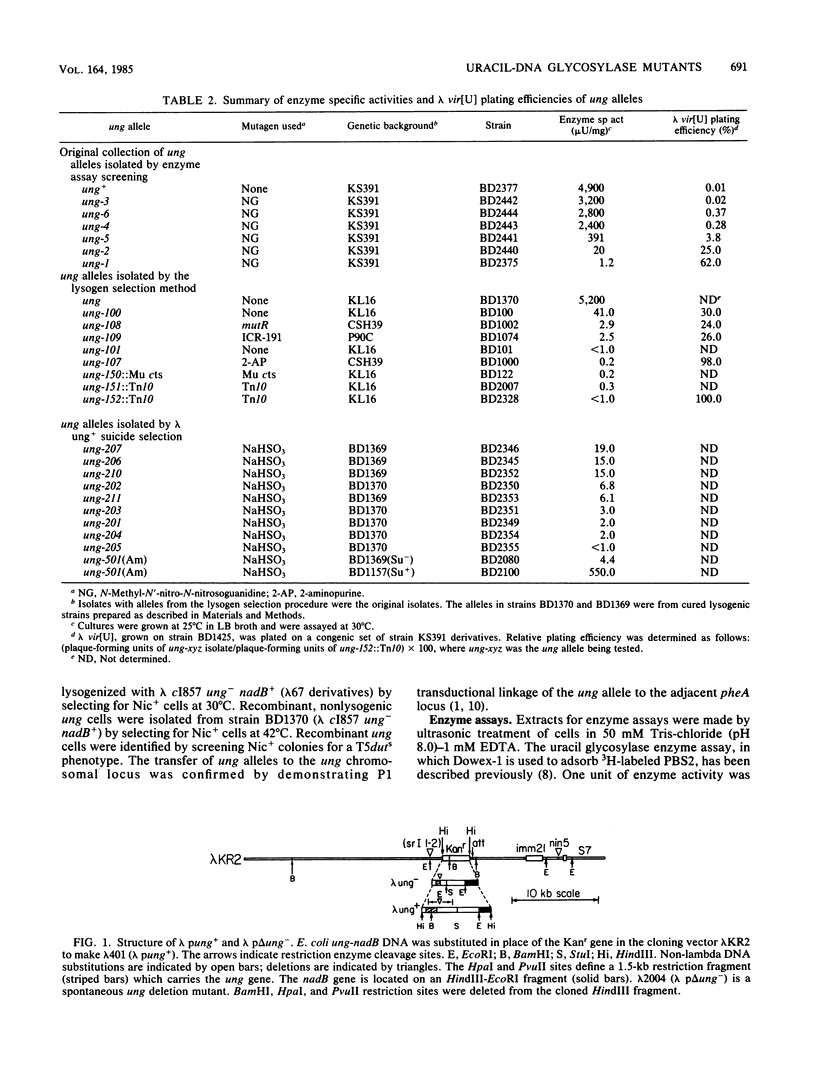
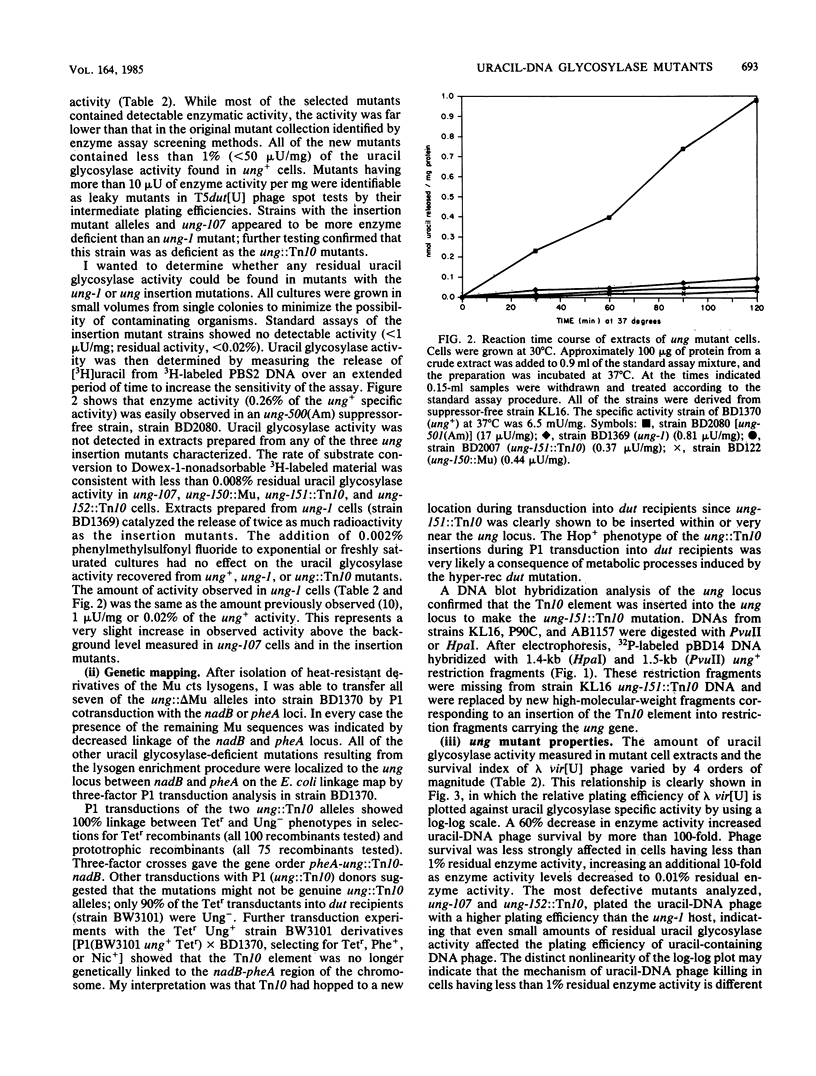
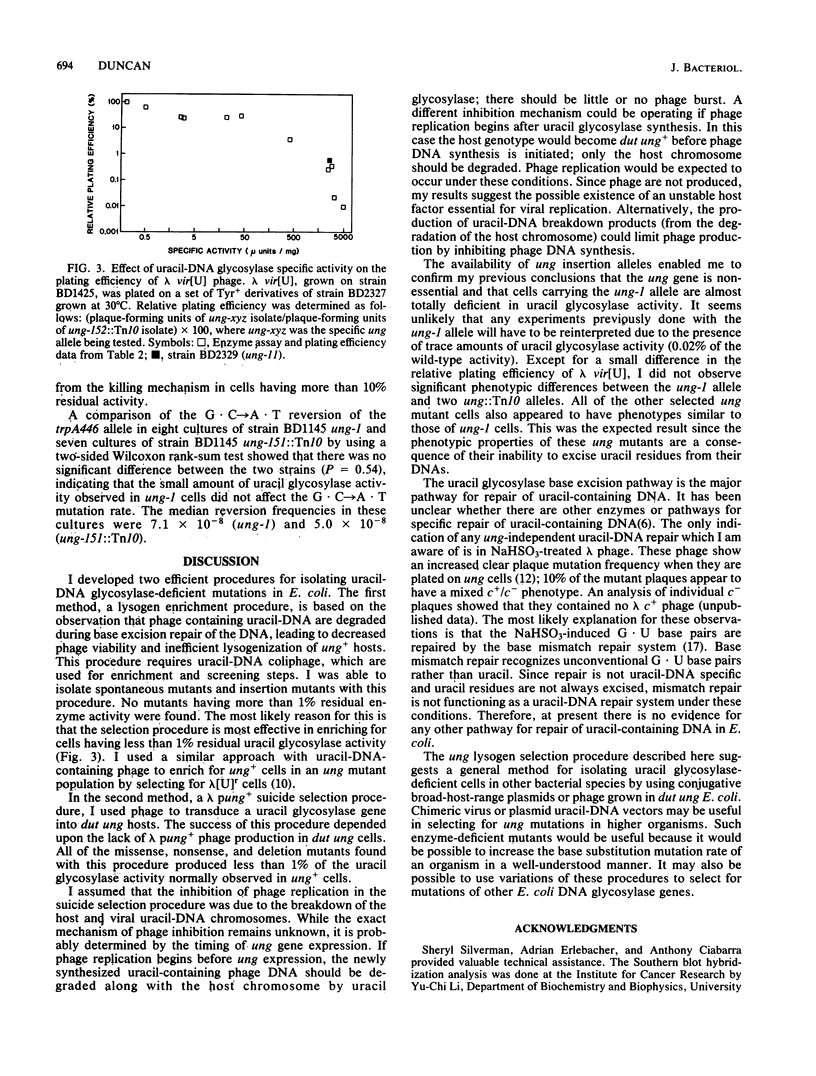
Two uracil-DNA glycosylase (ung) mutation selection procedures based upon the ability of uracil glycosylase to degrade the chromosomes of organisms containing uracil-DNA were devised to obtain a collection of well-defined ung alleles. In an enrichment procedure, lysogens were selected from Escherichia coli cultures infected with lambda pKanr phage containing uracil in their DNA. (These uracil-DNA phage were prepared by growth on host cells deficient in both dUTPase and uracil-DNA glycosylase.) The lysogenic Kanr population was enriched for uracil glycosylase-deficient mutants by a factor of 10(4). In a phage suicide selection procedure, lambda pung+ phage were unable to form plaques on dut ung cells containing uracil-DNA in their chromosomes, and all of the progeny were lambda pung-. Deletion, insertion (ung::Mu and ung::Tn10), nonsense, and missense mutants were isolated by using these procedures. Extracts of three insertion mutants contained no detectable enzyme activity. All of the other mutant isolates had less than 1% of the normal uracil glycosylase specific activity. The previously studied ung-1 allele, which was derived by N-methyl-N'-nitro-N-nitrosoguanidine mutagenesis, produced about 0.02% of the normal amount of uracil glycosylase activity. No significant phenotypic differences between ung-1 and ung::Tn10 alleles were observed. Variations of the lysogen selection procedure may be helpful for isolating other DNA glycosylase mutations in E. coli and other organisms.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Csonka L. N., Clark A. J. Construction of an Hfr strain useful for transferring recA mutations between Escherichia coli strains. J Bacteriol. 1980 Jul;143(1):529–530. doi: 10.1128/jb.143.1.529-530.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demple B., Linn S. On the recognition and cleavage mechanism of Escherichia coli endodeoxyribonuclease V, a possible DNA repair enzyme. J Biol Chem. 1982 Mar 25;257(6):2848–2855. [PubMed] [Google Scholar]
- Duncan B. K., Chambers J. A. The cloning and overproduction of Escherichia coli uracil-DNA glycosylase. Gene. 1984 May;28(2):211–219. doi: 10.1016/0378-1119(84)90258-0. [DOI] [PubMed] [Google Scholar]
- Duncan B. K., Miller J. H. Mutagenic deamination of cytosine residues in DNA. Nature. 1980 Oct 9;287(5782):560–561. doi: 10.1038/287560a0. [DOI] [PubMed] [Google Scholar]
- Duncan B. K., Rockstroh P. A., Warner H. R. Escherichia coli K-12 mutants deficient in uracil-DNA glycosylase. J Bacteriol. 1978 Jun;134(3):1039–1045. doi: 10.1128/jb.134.3.1039-1045.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan B. K., Warner H. R. Metabolism of uracil-containing DNA: degradation of bacteriophage PBS2 DNA in Bacillus subtilis. J Virol. 1977 Jun;22(3):835–838. doi: 10.1128/jvi.22.3.835-838.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan B. K., Weiss B. Specific mutator effects of ung (uracil-DNA glycosylase) mutations in Escherichia coli. J Bacteriol. 1982 Aug;151(2):750–755. doi: 10.1128/jb.151.2.750-755.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karran P., Lindahl T., Ofsteng I., Evensen G. B., Seeberg E. Escherichia coli mutants deficient in 3-methyladenine-DNA glycosylase. J Mol Biol. 1980 Jun 15;140(1):101–127. doi: 10.1016/0022-2836(80)90358-7. [DOI] [PubMed] [Google Scholar]
- Kleckner N., Barker D. F., Ross D. G., Botstein D. Properties of the translocatable tetracycline-resistance element Tn10 in Escherichia coli and bacteriophage lambda. Genetics. 1978 Nov;90(3):427–461. doi: 10.1093/genetics/90.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A. L., Clark S., Modrich P. Methyl-directed repair of DNA base-pair mismatches in vitro. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4639–4643. doi: 10.1073/pnas.80.15.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai F., Hawryluk I., Shapiro R. The mutagenic specificity of sodium bisulfite. Biochem Biophys Res Commun. 1970 Jun 5;39(5):983–988. doi: 10.1016/0006-291x(70)90421-3. [DOI] [PubMed] [Google Scholar]
- Peden K. W., Nathans D. Local mutagenesis within deletion loops of DNA heteroduplexes. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7214–7217. doi: 10.1073/pnas.79.23.7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piechaczyk M., Jeanteur P., Louarn J. M. An easy method for the selection of restriction- and modification-deficient mutants of Escherichia coli K-12. Gene. 1980 Oct;11(1-2):173–175. doi: 10.1016/0378-1119(80)90098-0. [DOI] [PubMed] [Google Scholar]
- Riazuddin S., Lindahl T. Properties of 3-methyladenine-DNA glycosylase from Escherichia coli. Biochemistry. 1978 May 30;17(11):2110–2118. doi: 10.1021/bi00604a014. [DOI] [PubMed] [Google Scholar]
- Shortle D., Nathans D. Local mutagenesis: a method for generating viral mutants with base substitutions in preselected regions of the viral genome. Proc Natl Acad Sci U S A. 1978 May;75(5):2170–2174. doi: 10.1073/pnas.75.5.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner H. R., Demple B. F., Deutsch W. A., Kane C. M., Linn S. Apurinic/apyrimidinic endonucleases in repair of pyrimidine dimers and other lesions in DNA. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4602–4606. doi: 10.1073/pnas.77.8.4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner H. R., Duncan B. K., Garrett C., Neuhard J. Synthesis and metabolism of uracil-containing deoxyribonucleic acid in Escherichia coli. J Bacteriol. 1981 Feb;145(2):687–695. doi: 10.1128/jb.145.2.687-695.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner H. R., Duncan B. K. In vivo synthesis and properties of uracil-containing DNA. Nature. 1978 Mar 2;272(5648):32–34. doi: 10.1038/272032a0. [DOI] [PubMed] [Google Scholar]
- Warner H. R., Thompson R. B., Mozer T. J., Duncan B. K. The properties of a bacteriophage T5 mutant unable to induce deoxyuridine 5'-triphosphate nucleotidohydrolase. Synthesis of uracil-containing T5 deoxyribonucleic acid. J Biol Chem. 1979 Aug 25;254(16):7534–7539. [PubMed] [Google Scholar]
- Wood W. B. Host specificity of DNA produced by Escherichia coli: bacterial mutations affecting the restriction and modification of DNA. J Mol Biol. 1966 Mar;16(1):118–133. doi: 10.1016/s0022-2836(66)80267-x. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y., Katsuki M., Sekiguchi M., Otsuji N. Escherichia coli gene that controls sensitivity to alkylating agents. J Bacteriol. 1978 Jul;135(1):144–152. doi: 10.1128/jb.135.1.144-152.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]



