Abstract
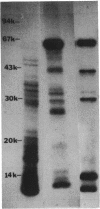
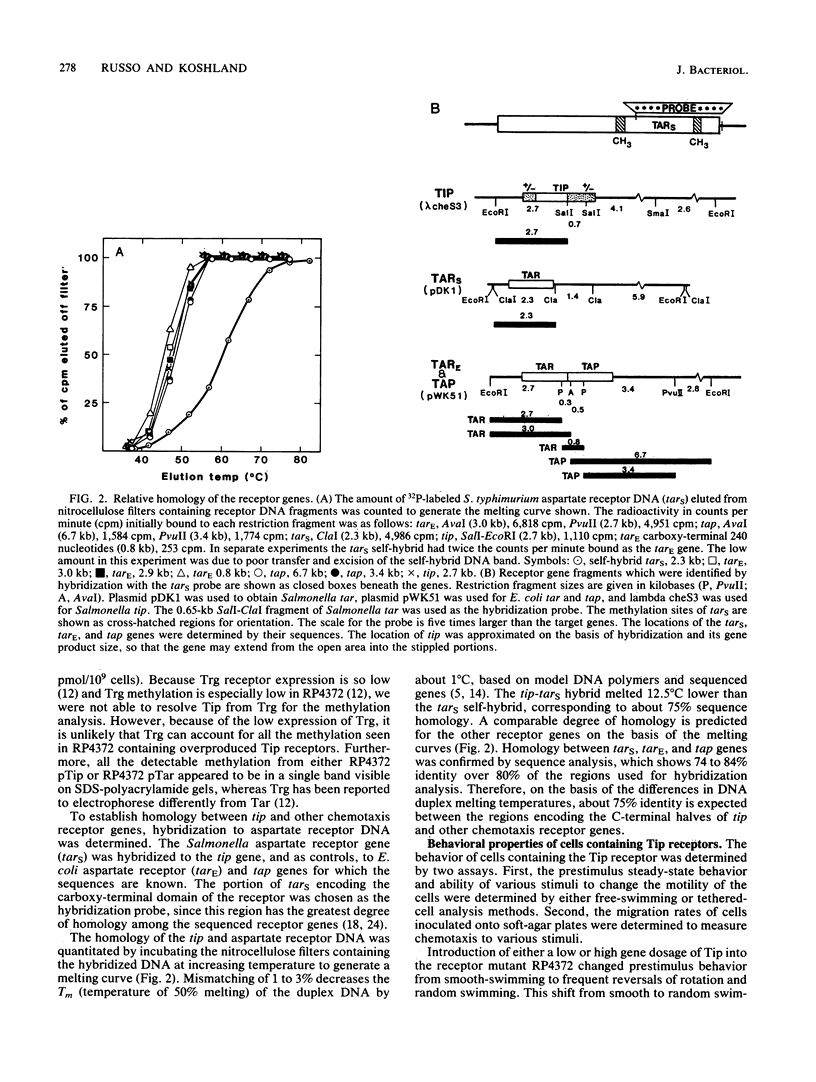
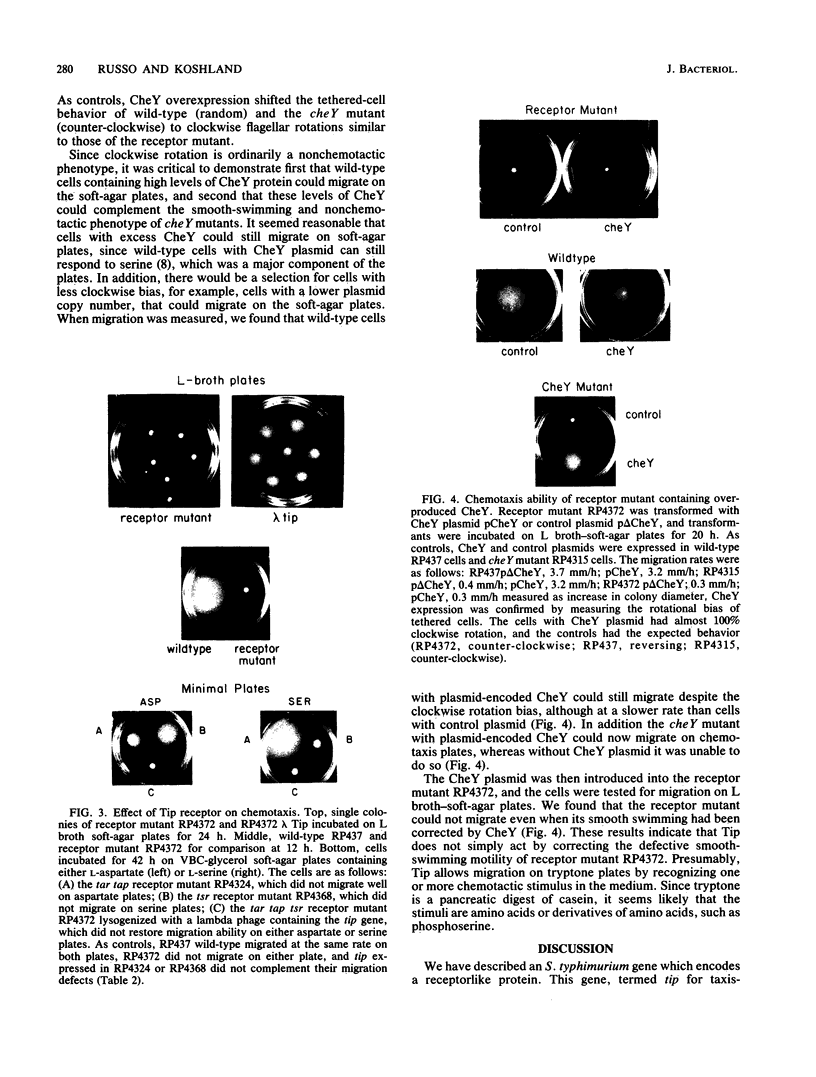
A chemotaxis gene encoding a protein with receptorlike properties has been identified in Salmonella typhimurium and termed tip for taxis-involved protein. Based on the stringency of DNA hybridization, the tip gene has about 75% homology with a region of the tar gene encoding the cytoplasmic domain of the aspartate receptor. Introduction of the tip gene into a smooth-swimming Escherichia coli receptor mutant (tar tsr tap) restored both chemotaxis ability on soft-agar-tryptone plates and a wild-type swimming phenotype. We have shown, by overexpressing the CheY protein, that shifting of the mutant swimming bias in the absence of receptors is insufficient to restore chemotaxis ability. This suggests that in addition to resetting the swimming bias, the tip gene product functions as a receptor. By functional criteria, we found that Tip is not a duplicate aspartate (Tar) or serine (Tsr) receptor gene. Based on behavioral properties, the S. typhimurium Tip receptor provides functional features similar to those of the E. coli Tap receptor.
Full text
PDF
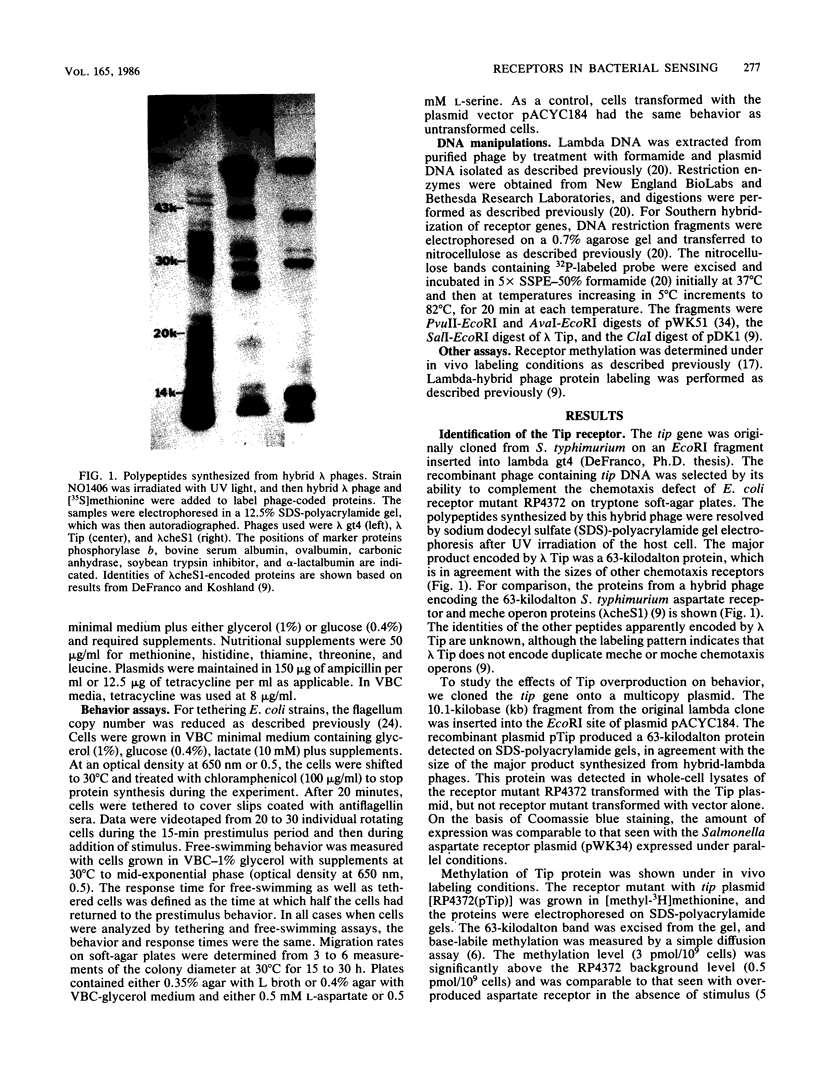



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bollinger J., Park C., Harayama S., Hazelbauer G. L. Structure of the Trg protein: Homologies with and differences from other sensory transducers of Escherichia coli. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3287–3291. doi: 10.1073/pnas.81.11.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd A., Kendall K., Simon M. I. Structure of the serine chemoreceptor in Escherichia coli. Nature. 1983 Feb 17;301(5901):623–626. doi: 10.1038/301623a0. [DOI] [PubMed] [Google Scholar]
- Boyd A., Krikos A., Simon M. Sensory transducers of E. coli are encoded by homologous genes. Cell. 1981 Nov;26(3 Pt 1):333–343. doi: 10.1016/0092-8674(81)90202-6. [DOI] [PubMed] [Google Scholar]
- Boyd A., Simon M. Bacterial chemotaxis. Annu Rev Physiol. 1982;44:501–517. doi: 10.1146/annurev.ph.44.030182.002441. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Graham D. E., Neufeld B. R. Analysis of repeating DNA sequences by reassociation. Methods Enzymol. 1974;29:363–418. doi: 10.1016/0076-6879(74)29033-5. [DOI] [PubMed] [Google Scholar]
- Chelsky D., Gutterson N. I., Koshland D. E., Jr A diffusion assay for detection and quantitation of methyl-esterified proteins on polyacrylamide gels. Anal Biochem. 1984 Aug 15;141(1):143–148. doi: 10.1016/0003-2697(84)90437-8. [DOI] [PubMed] [Google Scholar]
- Clarke S., Koshland D. E., Jr Membrane receptors for aspartate and serine in bacterial chemotaxis. J Biol Chem. 1979 Oct 10;254(19):9695–9702. [PubMed] [Google Scholar]
- Clegg D. O., Koshland D. E., Jr The role of a signaling protein in bacterial sensing: behavioral effects of increased gene expression. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5056–5060. doi: 10.1073/pnas.81.16.5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFranco A. L., Koshland D. E., Jr Molecular cloning of chemotaxis genes and overproduction of gene products in the bacterial sensing system. J Bacteriol. 1981 Aug;147(2):390–400. doi: 10.1128/jb.147.2.390-400.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFranco A. L., Parkinson J. S., Koshland D. E., Jr Functional homology of chemotaxis genes in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1979 Jul;139(1):107–114. doi: 10.1128/jb.139.1.107-114.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer G. L., Engström P., Harayama S. Methyl-accepting chemotaxis protein III and transducer gene trg. J Bacteriol. 1981 Jan;145(1):43–49. doi: 10.1128/jb.145.1.43-49.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer G. L., Engström P. Parallel pathways for transduction of chemotactic signals in Escherichia coli. Nature. 1980 Jan 3;283(5742):98–100. doi: 10.1038/283098a0. [DOI] [PubMed] [Google Scholar]
- Ingolia T. D., Koshland D. E., Jr Response to a metal ion-citrate complex in bacterial sensing. J Bacteriol. 1979 Dec;140(3):798–804. doi: 10.1128/jb.140.3.798-804.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. W., Rosenthal N., Rodakis G. C., Kafatos F. C. Evolution of two major chorion multigene families as inferred from cloned cDNA and protein sequences. Cell. 1979 Dec;18(4):1317–1332. doi: 10.1016/0092-8674(79)90242-3. [DOI] [PubMed] [Google Scholar]
- Kihara M., Macnab R. M. Chemotaxis of Salmonella typhimurium toward citrate. J Bacteriol. 1979 Oct;140(1):297–300. doi: 10.1128/jb.140.1.297-300.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D. E., Jr Biochemistry of sensing and adaptation in a simple bacterial system. Annu Rev Biochem. 1981;50:765–782. doi: 10.1146/annurev.bi.50.070181.004001. [DOI] [PubMed] [Google Scholar]
- Koshland D. E., Jr, Russo A. F., Gutterson N. I. Information processing in a sensory system. Cold Spring Harb Symp Quant Biol. 1983;48(Pt 2):805–810. doi: 10.1101/sqb.1983.048.01.083. [DOI] [PubMed] [Google Scholar]
- Krikos A., Mutoh N., Boyd A., Simon M. I. Sensory transducers of E. coli are composed of discrete structural and functional domains. Cell. 1983 Jun;33(2):615–622. doi: 10.1016/0092-8674(83)90442-7. [DOI] [PubMed] [Google Scholar]
- Macnab R. M., Koshland D. E., Jr The gradient-sensing mechanism in bacterial chemotaxis. Proc Natl Acad Sci U S A. 1972 Sep;69(9):2509–2512. doi: 10.1073/pnas.69.9.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S. Complementation analysis and deletion mapping of Escherichia coli mutants defective in chemotaxis. J Bacteriol. 1978 Jul;135(1):45–53. doi: 10.1128/jb.135.1.45-53.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S. Novel mutations affecting a signaling component for chemotaxis of Escherichia coli. J Bacteriol. 1980 Jun;142(3):953–961. doi: 10.1128/jb.142.3.953-961.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reader R. W., Tso W. W., Springer M. S., Goy M. F., Adler J. Pleiotropic aspartate taxis and serine taxis mutants of Escherichia coli. J Gen Microbiol. 1979 Apr;111(2):363–374. doi: 10.1099/00221287-111-2-363. [DOI] [PubMed] [Google Scholar]
- Russo A. F., Koshland D. E., Jr Separation of signal transduction and adaptation functions of the aspartate receptor in bacterial sensing. Science. 1983 Jun 3;220(4601):1016–1020. doi: 10.1126/science.6302843. [DOI] [PubMed] [Google Scholar]
- Slocum M. K., Parkinson J. S. Genetics of methyl-accepting chemotaxis proteins in Escherichia coli: organization of the tar region. J Bacteriol. 1983 Aug;155(2):565–577. doi: 10.1128/jb.155.2.565-577.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M. S., Goy M. F., Adler J. Protein methylation in behavioural control mechanisms and in signal transduction. Nature. 1979 Jul 26;280(5720):279–284. doi: 10.1038/280279a0. [DOI] [PubMed] [Google Scholar]
- Springer M. S., Goy M. F., Adler J. Sensory transduction in Escherichia coli: two complementary pathways of information processing that involve methylated proteins. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3312–3316. doi: 10.1073/pnas.74.8.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer W. R., Koshland D. E., Jr Identification of a protein methyltransferase as the cheR gene product in the bacterial sensing system. Proc Natl Acad Sci U S A. 1977 Feb;74(2):533–537. doi: 10.1073/pnas.74.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J. L., Koshland D. E., Jr Quantitation of the sensory response in bacterial chemotaxis. Proc Natl Acad Sci U S A. 1975 Feb;72(2):710–713. doi: 10.1073/pnas.72.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J. B., Koshland D. E., Jr A protein methylesterase involved in bacterial sensing. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3659–3663. doi: 10.1073/pnas.75.8.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tso W. W., Adler J. Negative chemotaxis in Escherichia coli. J Bacteriol. 1974 May;118(2):560–576. doi: 10.1128/jb.118.2.560-576.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Wang E. A., Koshland D. E., Jr Receptor structure in the bacterial sensing system. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7157–7161. doi: 10.1073/pnas.77.12.7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E. A., Mowry K. L., Clegg D. O., Koshland D. E., Jr Tandem duplication and multiple functions of a receptor gene in bacterial chemotaxis. J Biol Chem. 1982 May 10;257(9):4673–4676. [PubMed] [Google Scholar]