Abstract
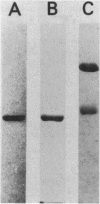
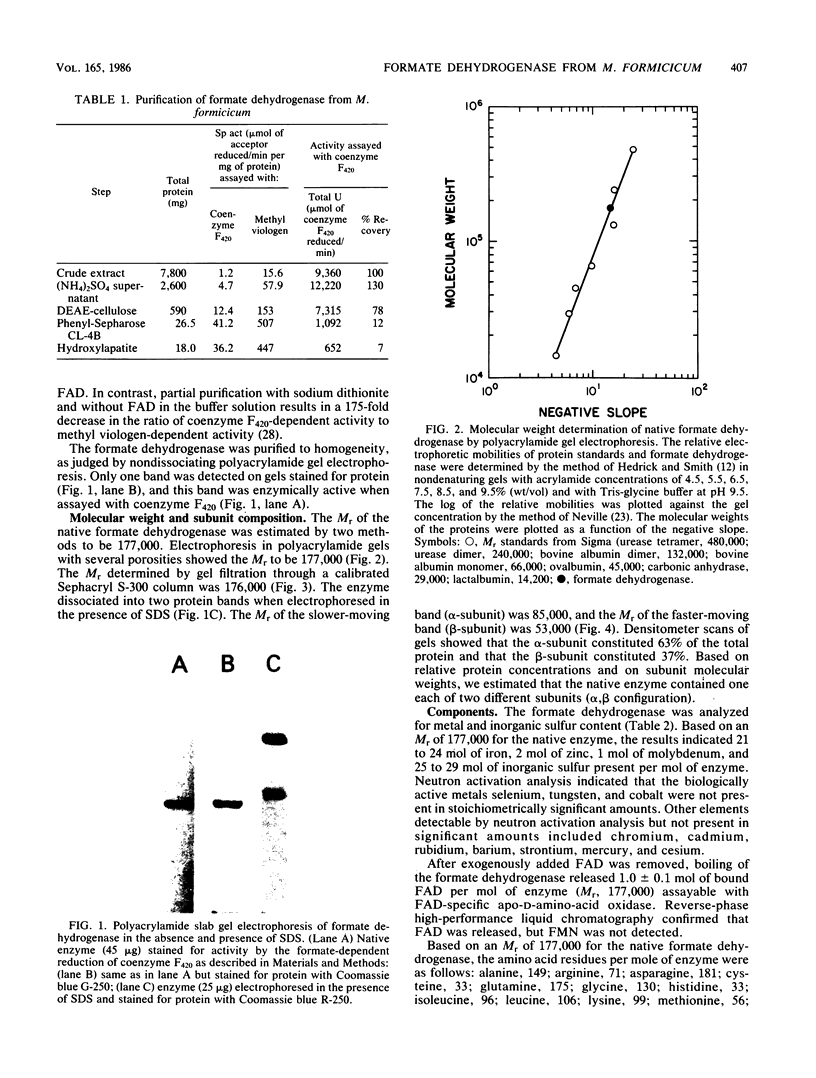
The coenzyme F420-dependent formate dehydrogenase from Methanobacterium formicicum was purified to electrophoretic homogeneity by anoxic procedures which included the addition of azide, flavin adenine dinucleotide (FAD), glycerol, and 2-mercaptoethanol to all buffer solutions to stabilize activity. The enzyme contains, in approximate molar ratios, 1 FAD molecule and 1 molybdenum, 2 zinc, 21 to 24 iron, and 25 to 29 inorganic sulfur atoms. Denaturation of the enzyme released a molybdopterin cofactor. The enzyme has a molecular weight of 177,000 and consists of one each of two different subunits, giving the composition alpha 1 beta 1. The molecular weight of the alpha-subunit is 85,000, and that of the beta-subunit is 53,000. The UV-visible spectrum is typical of nonheme iron-sulfur flavoprotein. Reduction of the enzyme facilitated dissociation of FAD, and the FAD-depleted enzyme was unable to reduce coenzyme F420. Preincubation of the FAD-depleted enzyme with FAD restored coenzyme F420-dependent activity.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barber M. J., Siegel L. M., Schauer N. L., May H. D., Ferry J. G. Formate dehydrogenase from Methanobacterium formicicum. Electron paramagnetic resonance spectroscopy of the molybdenum and iron-sulfur centers. J Biol Chem. 1983 Sep 25;258(18):10839–10845. [PubMed] [Google Scholar]
- Beinert H. Semi-micro methods for analysis of labile sulfide and of labile sulfide plus sulfane sulfur in unusually stable iron-sulfur proteins. Anal Biochem. 1983 Jun;131(2):373–378. doi: 10.1016/0003-2697(83)90186-0. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Enoch H. G., Lester R. L. The purification and properties of formate dehydrogenase and nitrate reductase from Escherichia coli. J Biol Chem. 1975 Sep 10;250(17):6693–6705. [PubMed] [Google Scholar]
- Ferry J. G., Wolfe R. S. Nutritional and biochemical characterization of Methanospirillum hungatii. Appl Environ Microbiol. 1977 Oct;34(4):371–376. doi: 10.1128/aem.34.4.371-376.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonda M. L., Anderson B. M. D-amino acid oxidase. I. Spectrophotometric studies. J Biol Chem. 1967 Sep 10;242(17):3957–3962. [PubMed] [Google Scholar]
- Fox G. E., Magrum L. J., Balch W. E., Wolfe R. S., Woese C. R. Classification of methanogenic bacteria by 16S ribosomal RNA characterization. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4537–4541. doi: 10.1073/pnas.74.10.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick J. L., Smith A. J. Size and charge isomer separation and estimation of molecular weights of proteins by disc gel electrophoresis. Arch Biochem Biophys. 1968 Jul;126(1):155–164. doi: 10.1016/0003-9861(68)90569-9. [DOI] [PubMed] [Google Scholar]
- Jacobson F. S., Daniels L., Fox J. A., Walsh C. T., Orme-Johnson W. H. Purification and properties of an 8-hydroxy-5-deazaflavin-reducing hydrogenase from Methanobacterium thermoautotrophicum. J Biol Chem. 1982 Apr 10;257(7):3385–3388. [PubMed] [Google Scholar]
- Johnson J. L., Hainline B. E., Rajagopalan K. V., Arison B. H. The pterin component of the molybdenum cofactor. Structural characterization of two fluorescent derivatives. J Biol Chem. 1984 May 10;259(9):5414–5422. [PubMed] [Google Scholar]
- Jones J. B., Stadtman T. C. Selenium-dependent and selenium-independent formate dehydrogenases of Methanococcus vannielii. Separation of the two forms and characterization of the purified selenium-independent form. J Biol Chem. 1981 Jan 25;256(2):656–663. [PubMed] [Google Scholar]
- Kröger A., Winkler E., Innerhofer A., Hackenberg H., Schägger H. The formate dehydrogenase involved in electron transport from formate to fumarate in Vibrio succinogenes. Eur J Biochem. 1979 Mar;94(2):465–475. doi: 10.1111/j.1432-1033.1979.tb12914.x. [DOI] [PubMed] [Google Scholar]
- Light D. R., Walsh C., Marletta M. A. Analytical and preparative high-performance liquid chromatography separation of flavin and flavin analog coenzymes. Anal Biochem. 1980 Nov 15;109(1):87–93. doi: 10.1016/0003-2697(80)90014-7. [DOI] [PubMed] [Google Scholar]
- Liu C. L., Mortenson L. E. Formate dehydrogenase of Clostridium pasteurianum. J Bacteriol. 1984 Jul;159(1):375–380. doi: 10.1128/jb.159.1.375-380.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey V., Curti B. A new method of preparation of D-amino acid oxidase apoprotein and a conformational change after its combination with flavin adenine dinucleotide. J Biol Chem. 1966 Jul 25;241(14):3417–3423. [PubMed] [Google Scholar]
- Müller U., Willnow P., Ruschig U., Höpner T. Formate dehydrogenase from Pseudomonas oxalaticus. Eur J Biochem. 1978 Feb;83(2):485–498. doi: 10.1111/j.1432-1033.1978.tb12115.x. [DOI] [PubMed] [Google Scholar]
- Nelson M. J., Brown D. P., Ferry J. G. FAD requirement for the reduction of coenzyme F420 by hydrogenase from Methanobacterium formicicum. Biochem Biophys Res Commun. 1984 May 16;120(3):775–781. doi: 10.1016/s0006-291x(84)80174-6. [DOI] [PubMed] [Google Scholar]
- Neville D. M., Jr Molecular weight determination of protein-dodecyl sulfate complexes by gel electrophoresis in a discontinuous buffer system. J Biol Chem. 1971 Oct 25;246(20):6328–6334. [PubMed] [Google Scholar]
- Olson S. T., Massey V. Purification and properties of the flavoenzyme D-lactate dehydrogenase from Megasphaera elsdenii. Biochemistry. 1979 Oct 16;18(21):4714–4724. doi: 10.1021/bi00588a036. [DOI] [PubMed] [Google Scholar]
- Reisner A. H., Nemes P., Bucholtz C. The use of Coomassie Brilliant Blue G250 perchloric acid solution for staining in electrophoresis and isoelectric focusing on polyacrylamide gels. Anal Biochem. 1975 Apr;64(2):509–516. doi: 10.1016/0003-2697(75)90461-3. [DOI] [PubMed] [Google Scholar]
- Schauer N. L., Ferry J. G. FAD requirement for the reduction of coenzyme F420 by formate dehydrogenase from Methanobacterium formicicum. J Bacteriol. 1983 Aug;155(2):467–472. doi: 10.1128/jb.155.2.467-472.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N. L., Ferry J. G. Metabolism of formate in Methanobacterium formicicum. J Bacteriol. 1980 Jun;142(3):800–807. doi: 10.1128/jb.142.3.800-807.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N. L., Ferry J. G. Properties of formate dehydrogenase in Methanobacterium formicicum. J Bacteriol. 1982 Apr;150(1):1–7. doi: 10.1128/jb.150.1.1-7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzing S. F., Bryant M. P., Wolfe R. S. Factor 420-dependent pyridine nucleotide-linked formate metabolism of Methanobacterium ruminantium. J Bacteriol. 1975 Jan;121(1):192–196. doi: 10.1128/jb.121.1.192-196.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T. Purification and properties of cytochrome c-553, an electron acceptor for formate dehydrogenase of Desulfovibrio vulgaris, Miyazaki. Biochim Biophys Acta. 1979 Oct 10;548(1):96–105. doi: 10.1016/0005-2728(79)90190-7. [DOI] [PubMed] [Google Scholar]
- Yamamoto I., Saiki T., Liu S. M., Ljungdahl L. G. Purification and properties of NADP-dependent formate dehydrogenase from Clostridium thermoaceticum, a tungsten-selenium-iron protein. J Biol Chem. 1983 Feb 10;258(3):1826–1832. [PubMed] [Google Scholar]