Abstract
Based on the promising opioid pharmacological profile of the peptide, Tyr-Pro-Trp-Gly-NH2 (Tyr-W-MIF), Zadina et al. (1997) synthesized and screened other Gly4-substituted peptides, culminating in the synthesis of Tyr-Pro-Trp-Phe-NH2 (endomorphin-1), which displayed high affinity and selectivity for the μ opioid receptor. The amidated peptide was then isolated from bovine brain frontal cortex, as was a related peptide, Tyr-Pro-Phe-Phe-NH2 (endomorphin-2), that displayed similar high affinity and selectivity for the μ opioid receptor. The biosynthesis of the endomorphins in the brain remains obscure, since the putative precursor proteins for the peptides have not been identified. With the completion of the human genome sequencing project, we hypothesized that we should uncover the biological precursors of the peptides using a bioinformatic approach to search the entire human proteome for proteins that contained the endomorphin peptide sequences followed by Gly-Lys/Arg, the consensus sequence for peptide α-amidation and precursor cleavage. Twelve proteins were identified that contained the endomorphin-1 Tyr-Pro-Trp-Phe sequence, however none contained the Tyr-Pro-Trp-Phe-Gly sequence necessary for α-amidation. Twenty-two distinct proteins contained the endomorphin-2 tetrapeptide sequence, and two of those contained the sequence, Tyr-Pro-Phe-Phe-Gly, however, none contained the requisite peptide-Gly-Lys/Arg sequence. Western blot analysis using an endomorphin-2 antibody detected 4 prominent proteins in mouse brain, necessitating reinterpretation of previous immunocytolocalization studies in the brain. Screening of the current human proteome yielded no evidence for endomorphin precursor proteins based on accepted biochemical criteria.
Keywords: endomorphin, bioinformatics, human proteome, peptide amidation
Introduction
Humans have used opium, derived from the poppy plant, Papaver somniferum, for thousands of years for its therapeutic properties and pleasurable subjective effects. The active principle in opium was isolated and named morphine after the Greek god of dreams, Morpheus (Sertürner, 1805, 1817). The structure of morphine was predicted by Gulland and Robinson (1925), a remarkable feat for the time due to its complex ring structure, and the Robinson structure was confirmed by chemical synthesis (Gates and Tschudi, 1956). In addition to morphine and its synthetic morphinan congeners, several other distinct chemical classes, including phenylpiperidines, benzomorphans, octahydroisoquinolines, and propioanilides, have been found to exhibit morphine-like pharmacological effects (Jaffe and Martin, 1990).
Opioids are the most powerful analgesic drugs available, and are the treatment of choice for the management of moderate to severe pain (McQuay, 1999). Adverse effects, including respiratory depression, nausea, and constipation, impact their use, and protracted opioid therapy leads to drug tolerance and physical dependence. Adaptive changes at the molecular, cellular, and neural network level are induced by habitual use of opioids, however, much remains to be learned about the mechanisms underlying the plasticity (Bailey and Connor, 2005).
The discovery and characterization of opioid receptors in vertebrate brain was accomplished with the use of radioligand binding assays (Pert and Snyder, 1973; Simon et al., 1973; Terenius, 1973). Opioid receptors engaged opioid drugs in a stereoselective manner, and were found to be integral membrane proteins located predominantly in neurons. It became clear that opioid receptors displayed heterogeneous properties, and it appeared that at least three types of opioid receptors existed, classified as μ, δ, and κ receptors (Lord et al., 1977; Kosterlitz et al., 1981), and these receptors have been confirmed by molecular cloning (Evans et al., 1982; Kieffer et al., 1982; Chen et al., 1983; Yasuda et al., 1983).
Known neurotransmitters at that time did not bind to opioid receptors with high affinity, so the assumption was made that uncharacterized ligands must exist in the brain that were the physiological agonists for the receptors. Two pentapeptides were isolated from sheep brain, Met-enkephalin (Tyr-Gly-Gly-Phe-Met) and Leu-enkephalin (Tyr-Gly-Gly-Phe-Leu), which displayed high affinity for opioid receptors and had morphine-like pharmacological activity (Hughes et al., 1975). The sequence of Met-enkephalin was contained within the anterior pituitary polypeptide, β-lipotropin, and a 31-amino acid peptide derived from the carboxyl terminus of β-lipotropin, β-endorphin, was characterized as another highly active opioid peptide (Bradbury et al., 1979). Goldstein and colleagues (1981) isolated dynorphin A, a Leu-enkephalin-containing heptadecapeptide, from the posterior pituitary, which also exhibited potent opioid agonist activity. Utilizing advances in reverse-phase liquid chromatography, the Udenfriend laboratory isolated and sequenced many other biologically active opioid peptides that contained either the Met- or Leu-enkephalin sequence at their amino termini (reviewed in Howells, 1986; Chaturvedi et al., 2000). Molecular cloning revealed that all of the known opioid peptides that contained the Tyr-Gly-Gly-Phe-Met/Leu sequence at their amino termini were derived as a result of proteolytic processing of three precursor proteins, proopiomelanocortin, proenkephalin, or prodynorphin (Nakanishi et al., 1979; Noda et al., 1982; Gubler et al., 1982; Comb et al., 1982; Kakidani et al., 1982). Met- and Leu-enkephalin displayed preferential binding to the δ opioid receptor, dynorphin A and other peptides derived from prodynorphin bound preferentially to the κ receptor, and metorphamide, an amidated peptide derived from proenkephalin (Tyr-Gly-Gly-Phe-Met-Arg-Arg-Val-NH2, Weber et al., 1983), displayed the highest relative affinity for the μ receptor (see Paterson et al., 1984). Many other endogenous opioid peptides interacted with more than one receptor type with high affinity (Paterson et al., 1984).
Other atypical peptides with opioid-like activity that were not derived from proopiomelanocortin, proenkephalin or prodynorphin, have been generated by proteolytic fragmentation of the milk protein, casein (the β-casomorphins, Brantl et al., 1979) or the blood protein, hemoglobin (the hemorphins, Brantl et al., 1986; Nyberg et al., 1997) (reviewed in Janecka et al., 2004). In addition, atypical peptides have been isolated from frog skin that are mu receptor-selective (the dermorphins, Broccardo et al., 1981) or delta receptor-selective (the deltorphins, Erspamer et al., 1989). The distinctive feature of the amphibian peptides with opioid-like activity is the presence of D-amino acids at the second position of their amino termini, Tyr-D-Xaa-Phe (reviewed in Negri et al., 2000). The presence of the D-enantiomer is due, presumably, to a post-translational modification of the precursor protein involving epimerization of the naturally occurring L-amino acid (Mor et al., 1992; Heck et al., 1996).
In 1993, Kastin and colleagues reported the isolation of Tyr-Pro-Trp-Gly-NH2 (Tyr-W-MIF-1) from human and bovine brain (Erchegyi et al., 1992; Hackler et al., 1993). This peptide was related in sequence to Tyr-Pro-Leu-Gly-NH2 (Tyr-MIF-1) and Pro-Leu-Gly-NH2 (MIF-1, melanotropin release inhibiting factor) that had been isolated previously from brain (Nair et al., 1971; Horvath and Kastin, 1989). Tyr-W-MIF-1 displayed high affinity (Ki = 70 nM) and selectivity (>200-fold) for the μ opioid receptor (Zadina et al., 1994).
Based on the promising affinity and selectivity of Tyr-W-MIF-1 for the μ opioid receptor, all possible amino acid substitutions at position 4 of the peptide were synthesized and screened for their affinity for the μ receptor. The Phe-substituted peptide, Tyr-Pro-Trp-Phe-NH2, had a Ki = 0.4 nM, and μ/δ and μ/κ selectivity ratios of 4000 and 15000, respectively. This peptide, named endomorphin-1, possessed potent, naloxone-reversible agonist activity in the guinea-pig ileum bioassay and analgesic activity in the mouse tail-flick assay. An antibody to endomorphin-1 was generated, and endomorphin-1 was isolated from bovine and human brain. A related peptide, Tyr-Pro-Phe-Phe-NH2 (named endomorphin-2) was also isolated and sequenced, and it displayed a similar pharmacological profile (Zadina et al., 1997; Hackler et al., 1997).
The goal of this study was to use bioinformatics to search the current human proteome for putative endomorphin precursor proteins, which has become feasible with the completion of the human genome sequencing project.
Materials and Methods
Bioinformatic screening of the human proteome for endomorphin-1 and endomorphin-2
Several protein databases were searched in this study, including NCBI RefSeq database (September 2006 version) (Pruitt et al., 2007), Swiss-Prot database (October 2006 version), and TrEBML database (October 2006 version) (Wu et al., 2006). These databases constitute a comprehensive collection of all known (RefSeq and Swiss-Prot) and predicted (RefSeq and TrEMBL) protein sequences.
A PERL (Practical Extraction and Report Language) program was written to search for sequences with various patterns: YPWF (Tyr-Pro-Trp-Phe) or YPFF (Tyr-Pro-Phe-Phe) alone or including G (Gly) at the carboxyl terminus, and flanked by either K (Lys) or R (Arg) with and without G. For example, endomorphin-2 patterns with YPFF, YPFFG, YPFF K/R, YPFFG K/R, K/R YPFF, K/R YPFFG, K/R YPFF K/R, and K/R YPFFG K/R would be extracted, and a similar strategy was employed to search for endomorphin-1 sequences. Human proteins were extracted from Swiss-Prot and TrEMBL databases and records overlapping with RefSeq were removed.
Cell culture
Human embryonic kidney 293 cells stably expressing the FLAG-tagged δ opioid receptor (Yadav et al., 2007) were maintained at 37°C in a humidified atmosphere containing 5% CO2 in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin sulfate and 0.25 mg/ml G418. Human SH-SY5Y neuroblastoma cells (American Type Culture Collection, Manassas, VA) were grown in Dulbecco's modified Eagle's medium/Ham's F12 medium containing 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin sulfate. Mouse neuroblastoma × rat glioma hybrid NG108-15 cells (American Type Culture Collection, Manassas, VA) were cultured in Dulbecco's modified Eagle's medium containing 5% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin sulfate.
Immunoblot analysis
Brains were removed from C57BL6 mice and homogenized in 50 mM Tris HCl, pH 7.5 with a Tekmar tissuemizer (Cincinnati, OH). Protein lysates were prepared by solubilization of the tissue in 5 volumes of 1% n-dodecyl-β-D-maltoside, 50 mM Tris HCl, pH 7.5, 150 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 10% glycerol and a protease inhibitor cocktail (1:1000, Sigma, St. Louis, MO). Lysates were similarly prepared from human embryonic kidney 293, human SH-SY5Y neuroblastoma, and mouse neuroblastoma × rat glioma NG108-15 cell lines. Detergent lysates were centrifuged at 16,000 × g for 20 min, and the supernatants were recovered for immunoblot analysis. Protein concentrations in the supernatants were determined using the BioRad DC assay with bovine serum albumin as standard. Cell extracts containing approximately 75 μg of protein were mixed with 5X SDS-PAGE gel loading buffer and heated at 50°C for 5 min. Proteins were resolved using 12% SDS-PAGE and transferred to PVDF membranes (Immobilon P, Millipore, Billarica, MA). Membranes were blocked for 1 h in 3% dried milk, 50 mM Tris HCl, pH 7.5, 150 mM NaCl, 1 mM CaCl2, 10% glycerol and 0.1% Tween 20, followed by overnight incubation at 4°C with a 1:2000 dilution of the anti-endomorphin polyclonal antibody (AB 5106, Upstate/Chemicon, Temecula, CA) for detection of endomorphin-like immunoreactivity. The AB 5106 antibody was raised in rabbits against synthetic endomorphin-2 (Tyr-Pro-Phe-Phe-NH2) conjugated to bovine serum albumin, and has been used by Evans et al. (2007) to study endomorphin-2 immunoreactivity in the rat insular cortex using immunohistochemical methods. The vendor's specifications assert that the polyclonal antiserum cross-reacts 100% with endomorphin-2, 70% with endomorphin-1, <0.03% with Met-enkephalin, and <0.01% with Leu-enkephalin and β-endorphin. Membranes were then washed and incubated with goat anti-rabbit IgG conjugated with horse radish peroxidase (Santa Cruz Biotechnology, Santa Cruz, CA) for 1 h at room temperature and developed using CDP-Star western blot chemiluminescence reagent (PerkinElmer Life Sciences, Boston, MA). The apparent molecular masses of the endomorphin-like immunoreactive proteins were estimated by plotting the migration distance of six protein standards versus the logarithm of their molecular mass. A straight line was obtained (r2 = 0.987), and the molecular masses of the immunoreactive proteins were obtained by interpolation based on their relative migration.
Results
Following the discovery of endomorphin-1 and endomorphin-2 (Zadina et al., 1997; Hackler et al., 1997), many research groups have attempted to characterize the biosynthetic precursor(s) of the peptides using molecular cloning methodology. These efforts have failed thus far. With the complete sequencing of the human genome accomplished, coupled with in silico translation of all predicted open reading frames, it has become possible to search the entire human proteome for putative endomorphin precursors using a bioinformatic approach.
The precursors of all α-amidated peptides contain a Gly residue on the carboxyl-terminal side of the amino acid that becomes amidated (Eipper et al., 1992), and the Gly is followed by a basic amino acid that serves as a site for endoproteolytic cleavage, unless the amidated residue-Gly sequence is at the extreme carboxyl terminus of the protein (Table 1). We therefore searched the human proteome for 1) the endomorphin-1 and endomorphin-2 tetrapeptide sequences, 2) the endomorphin-1 and endomorphin-2 tetrapeptide sequences followed by a Gly at the carboxyl terminus, and 3) the YPWFG and YPFFG sequences flanked by basic (Lys or Arg) amino acids. The results of the search for the endomorphin-1 precursor protein are shown in Table 2. We found the endomorphin-1 tetrapeptide sequence, Tyr-Pro-Trp-Phe, within 12 proteins in the protein databases. Three of these proteins were α-subunits of the guanine nucleotide binding protein family, Gα11, Gαq, and Gα14, with accession annotations of NP_002058, NP_002063, and NP_004288, respectively. Four SH2 domain-containing proteins also had the Tyr-Pro-Trp-Phe sequence. They were the Vav 2 oncogene (NP_003362), lymphocyte adaptor (NP_005466), SH2-B homolog (NP_056318), and SH2 adaptor protein (NP_066189). Five members of the G protein-coupled receptor family also contained the tetrapeptide Tyr-Pro-Trp-Phe sequence, including an olfactory receptor (NP_001004760), and four trace amine receptors, R1 (NP_612200), R2 (NP_055441), R5 (NP_444508), and R6 (NP_778237). None of these proteins, or any other proteins in the databases, contained Tyr-Pro-Trp-Phe-Gly, which is a necessary prerequisite for α-amidation of the endormorphin-1 tetrapeptide sequence.
Table 1.
Human α-amidated peptides and sequences in the precursor proteins that flank the bioactive amidated peptides (underlined).
| Amidated Peptide (Annotation) | Context of the Amidated Peptide Sequence in its Precursor Protein |
|---|---|
| Amylin
(NP_000406) |
KRKCNTATCATQRLANFLIRSSNNLGAILSPTNVGSNTYGKR |
| Calcitonin
(AAA58403) |
KRCGNLSTCMLGTYTQDFNKFHTFPQTAIGVGAPGKKR |
| Calcitonin Gene Related Peptide
(CAA26190) |
KRACDTATCVTHRLAGLLSRSGGVVKNNFVPTNVGSKAFGRRRR |
| Corticotropin Releasing Factor
(CAA2383) |
RRSEEPPISLDLTFHLLREVLEMARAEQLAQQAHSNRKLMEIIGK |
| Big Endothelin-3
(NP_996917) |
RRCTCPTYKDKECVYYCHLDIIWINTPEQTVPYGLSNYRGSFRGKR |
| Growth hormone releasing factor
(NP_066567) |
RRYADAIFTNSYRKVLGQLSARKLLQDIMSRQQGESNQERGARARLGR |
| Luteinizing hormone-releasing hormone
(NP_00081) |
pEHWSYGLRPGGKR |
| α-Melanocyte stimulating hormone
(NP_001030333) |
KRSYSMEHFRWGKPVGKKRR |
| Metorphamide
(CAA23767) |
KRYGGFMRRVGR |
| Neurokinin A
(NP_003173) |
KRHKTDSFVGLMGKR |
| Neuropeptide Y
(NP_00089) |
YPSKPDNPGEDAPAEDMARYYSALRHYINLITRQRYGRR |
| Oxytocin
(NP_000906) |
CYIQNCPLGGKR |
| Pituitary adenylate cyclase activating polypeptide 27
(AAB21470) |
KRHSDGIFTDSYSRYRKQMAVKKYLAAVLGKR |
| Pancreastatin
(NP_001266) |
RKGESRSEALAVDGAGKPGAEEAQDPEGKGEQEHSQQKEEEEEMAVVPQGLFRGGK |
| Pancreatic polypeptide
(EAW51649) |
APLEPVYPGDNATPEQMAQYAADLRRYINMLTRPRYGKR |
| Peptide YY
(NP_004151) |
YPIKPEAPGEDASPEELNRYYASLRHYLNLVTRQRYGKR |
| PHM-27
(NP_919416) |
RHADGVFTSDFSKLLGQLSAKKYLESLMGKR |
| Secretin
(NP_068739) |
RRHSDGTFTSELSRLREGARLQRLLQGLVGKR |
| Substance P
(NP_003173) |
RRPKPQQFFGLMGKR |
| Thyrotropin releasing hormone, repeat 1
(NP_009048) |
KRQHPGKR |
| Thyrotropin releasing hormone, repeat 2
(NP_009048) |
KRQHPGRR |
| Thyrotropin releasing hormone, repeat 3
(NP_009048) |
KRQHPGRR |
| Thyrotropin releasing hormone, repeat 4
(NP_009048) |
KRQHPGRR |
| Thyrotropin releasing hormone, repeat 5
(NP_009048) |
KRQHPGKR |
| Thyrotropin releasing hormone, repeat 6
(NP_009048) |
KRQHPGRR |
| Vasoactive intestinal polypeptide
(AAA61289) |
KRHSDAVFTDNYTRLRLQMAVKKYLNSILNGKR |
| Vasopressin
(AAA61291) |
CYFQNCPRGGKR |
Table 2.
Search for endomorphin-1 in human protein database.
| PEPTIDE | PROTEIN FAMILY | PROTEIN (ANNOTATION) | AA NUMBER* (MOL. WT.) | SEQUENCE CONTEXT† |
|---|---|---|---|---|
| YPWF
(endomorphin-1 lacking the C-terminal amide) |
GTP-binding protein | G alpha 11
(NP_002058) |
261
(41992) |
RTIITYPWFQNSSV |
| G alpha q
(NP_002063) |
261
(42011) |
RTIITYPWFQNSSV | ||
| G alpha 14
(NP_004288) |
257
(41440) |
KTIITYPWFLNSSV | ||
| SH2 domain proteins | Vav 2 oncogene
(NP_003362) |
661
(96902) |
IDYTAYPWFAGNME | |
| Lymphocyte adaptor
(NP_005466) |
362
(63094) |
HFLSCYPWFHGPIS | ||
| SH2-B homolog
(NP_056318) |
525
(70620) |
QPLSGYPWFHGMLS | ||
| SH2 adaptor
(NP-066189) |
415
(67607) |
LELSDYPWFHGTLS | ||
| G protein-coupled receptor | Trace amine R1
(NP_612200) |
308
(38961) |
VYAFFYPWFRKALK | |
| Trace amine R2
(NP_055441) |
274
(34822) |
IYGFFYPWFRRALK | ||
| Trace amine R5
(NP_444508) |
314
(37898) |
IYALFYPWFRKAIK | ||
| Trace amine R6
(NP_778237) |
315
(38320) |
IYALFYPWFRKAIK | ||
| Olfactory receptor
(NP_001004760) |
33
(36751) |
GMEQQYPWFSIPFS | ||
| YPWFG
(precursor required for amidation) |
No hits | |||
Designates amino acid number of the Y at the N-terminus of endomorphin 2; Mol. Wt., predicted molecular weight of the mature full-length protein.
Sequence context, amino acid sequence of the protein flanking the endomorphin sequence (underlined).
The results of the search for the endomorphin-2 precursor protein are displayed in Table 3. We identified 22 different known or putative proteins that contained the endomorphin-2 tetrapeptide sequence, Tyr-Pro-Phe-Phe. These proteins included galactocerebrosidase (NP_000144), ataxin 1 (NP_000323), insulin-like growth factor receptor 1 precursor (NP_000866), choline kinase α isoforms a (NP_001268) and b (NP_997634), mycotubularin-related protein 9 (NP_056273), isoforms a through g of MAP kinase-activating death domain-containing protein (NP_569832, NP_569827, NP_569828, NP-003676, NP_569829, NP_569830, and NP_569831, respectively), the D123 gene product (NP_006014), integrin α11 (NP_036343), a hypothetical protein (NP_067008), uncharacterized protein C19orf15 precursor (CS015_HUMAN), DENN (Q15741), 1-acylglycerol-3-phosphate O-acyltransferase 4 (NP_064518), longevity assurance gene 1 (NP_067090), secreted frizzled-related protein 5 (NP_003006), testis-specific protein disulfide isomerase-like protein (NP_777584), tripartite motif-containing 72 (NP_001008275) and putative human proteins FUSIP1 (Q61Q42), AGPAT4 (Q6PJN9), unknown protein (Q9P1N2) and unknown protein (Q5JWK2). Two proteins in the database, a sodium/sulfate symporter (NP_036582) and a coiled-coil domain-containing protein (NP_694955) contained the pentapeptide sequence, Tyr-Pro-Phe-Phe-Gly, which is a requirement for carboxyl terminal α-amidation of endomorphin-2, however, neither sequence was flanked by basic amino acids that serve as signals for proteases involved in processing bioactive peptides (Table 1).
Table 3.
Search for endomorphin-2 in human protein database.
| PEPTIDE | PROTEIN FAMILY | PROTEIN (ANNOTATION) | AA NUMBER* (MOL. WT.) | SEQUENCE CONTEXT† |
|---|---|---|---|---|
| YPFF
(endomorphin-2 lacking the C-terminal amide) |
Lysosomal hydrolase | Galacto-Cerebrosidase
(NP_000144) |
498
(72872) |
DFNVDYPFFSEAPN |
| PolyQ-tract protein | Ataxin 1
(NP_000323) |
648
(86792) |
EVLVEYPFFVFGQG | |
| Growth factor receptor | Insulin-like growth factor receptor 1 precursor
(NP_000866) |
775
(151864) |
ELETEYPFFESRVD | |
| Kinase | Choline kinase alpha isoform a
(NP_001268) |
359
(52118) |
YSYEKYPFFRANIR | |
| Choline kinase alpha isoform b
(NP_997634) |
341
(50024) |
YSYEKYPFFRANIR | ||
| Phosphatase cofactor | Mycotubularin-related protein 9
(NP_056273) |
109
(63331) |
SITLMYPFFYRPMF | |
| Proapoptotic signal transduction | MAP kinase-Activating death domain-Containing protein isoform a
(NP_569832) |
183
(176424) |
CVLSHYPFFSTFRE | |
| MAP kinase-Activating death domain-Containing protein isoform b
(NP_569827) |
183
(173297) |
CVLSHYPFFSTFRE | ||
| MAP kinase-Activating death domain-Containing protein isoform c
(NP_569828) |
183
(171294) |
CVLSHYPFFSTFR | ||
| MAP kinase-Activating death domain-Containing protein isoform d
(NP_003673) |
183
(183205) |
CVLSHYPFFSTFRE | ||
| MAP kinase-Activating death domain-Containing protein isoform e
(NP_569829) |
183
(178460) |
CVLSHYPFFSTFRE | ||
| MAP kinase-Activating death domain-Containing protein isoform f
(NP_569830) |
183
(163907) |
CVLSHYPFFSTFRE | ||
| MAP kinase-Activating death domain-Containing protein isoform g
(NP_569831) |
183
(175786) |
CVLSHYPFFSTFRE | ||
| Cell cycle control | D123 gene product
(NP_006014) |
15
(39057) |
QFSAWYPFFRGVTI | |
| Membrane receptor | Integrin alpha 11
(NP_036343) |
897
(131043) |
VCNVSYPFFRAKAK | |
| Membrane protein | Hypothetical protein
(NP_067008) |
566
(91653) |
FRDIFYPFFLIQDL | |
| Uncharacterized protein C19orf15 precursor
(CS015_HUMAN) |
480
(129175) |
FRDIFYPFFLIQDL | ||
| DENN
(Q15741) |
183
(142500) |
CVLSHYPFFSTFRE | ||
| Transferase | 1-acylglycerol-3-phosphate O-acyltransferase 4
(NP_064518) |
320
(43890) |
ASLVLYPFFQFLVS | |
| Signaling factor | Longevity assurance gene 1
(NP_067090) |
125
(39405) |
LFGTDYPFFHDPPS | |
| Secreted Frizzled-related protein 5
(NP_003006) |
307
(35446) |
PCSLYYPFFYGAAE | ||
| Isomerase | Testis-specific protein disulfide isomerase-like protein
(NP_777584) |
460
(66526) |
LMYLDRYPFFRLFPS | |
| RING-finger | Tripartite motif-containing 72
(NP_001008275) |
450
(52547) |
LPRPVYPFFDVCWH | |
| unknown | FUSIP1
(Q6IQ42) |
58
(8330) |
FAYVQYPFFRWLIS | |
| unknown | AGPAT4
(Q6PJN9) |
15
(8257) |
ASLVLYPFFQFLVS | |
| unknown | (Q9P1N2) | 44
(8592) |
PLIYPYPFFEHYLT | |
| unknown | (Q5JWK2) | 142
(24175) |
VRKQLYPFFKELQK | |
| YPFFG
(precursor required for C-terminal amidation) |
Transporter | SUT-1 Sodium/sulfate symporter
(NP_036582) |
66
(69228) |
VPAFLYPFFGVLRSN |
| ATPase | Coiled-coil domain containing 100
(NP_694955) |
480
(112510) |
ILRYSYPFFGSAAPI |
Designates amino acid number of the Y at the N-terminus of endomorphin 2; Mol. Wt., predicted molecular weight of the mature full-length protein.
Sequence context, amino acid sequence of the protein flanking the endomorphin sequence (underlined).
Several groups have studied the immunocytochemical localization of endomorphin-like immunoreactivity in the spinal cord and brain (Martin-Schild et al., 1997, 1998, 1999; Pierce et al., 1998; Schreff et al., 1998; Pierce and Wessendorf, 2000, Wang et al., 2002; Sanderson Nydahl et al., 2004; Evans et al., 2007), however, the peptide/protein species that were recognized by the endomorphin antibodies were not characterized biochemically. Based on our bioinformatic search of the human proteome, it appeared likely that many proteins would be recognized by the endomorphin antibodies due to the presence of the endomorphin tetrapeptide sequence within them. We therefore used western blotting to characterize the endomorphin-like immunoreactivity in mouse brain, and several other murine and human tissue samples, using a commercial polyclonal antibody generated against synthetic endomorphin-2 for detection. This is the same antibody that was used recently by Evans et al. (2007) for immunohistochemical studies in rat brain. Mouse brain lysates contained four major immunoreactive protein bands with apparent molecular masses of 25, 28, 78, and 117 kilodaltons (kDa). Murine NG108-15 cells also displayed the 28 kDa species prominently, along with another protein of 48 kDa, in addition to other lower intensity bands. Three major immunoreactive protein bands were evident in the human cell lines, which had apparent molecular masses of approximately 48, 64 and 78 kDa (Fig. 1).
Figure 1. Western blot analysis of endomorphin-like immunoreactivity in mouse brain and human and murine cell lines.
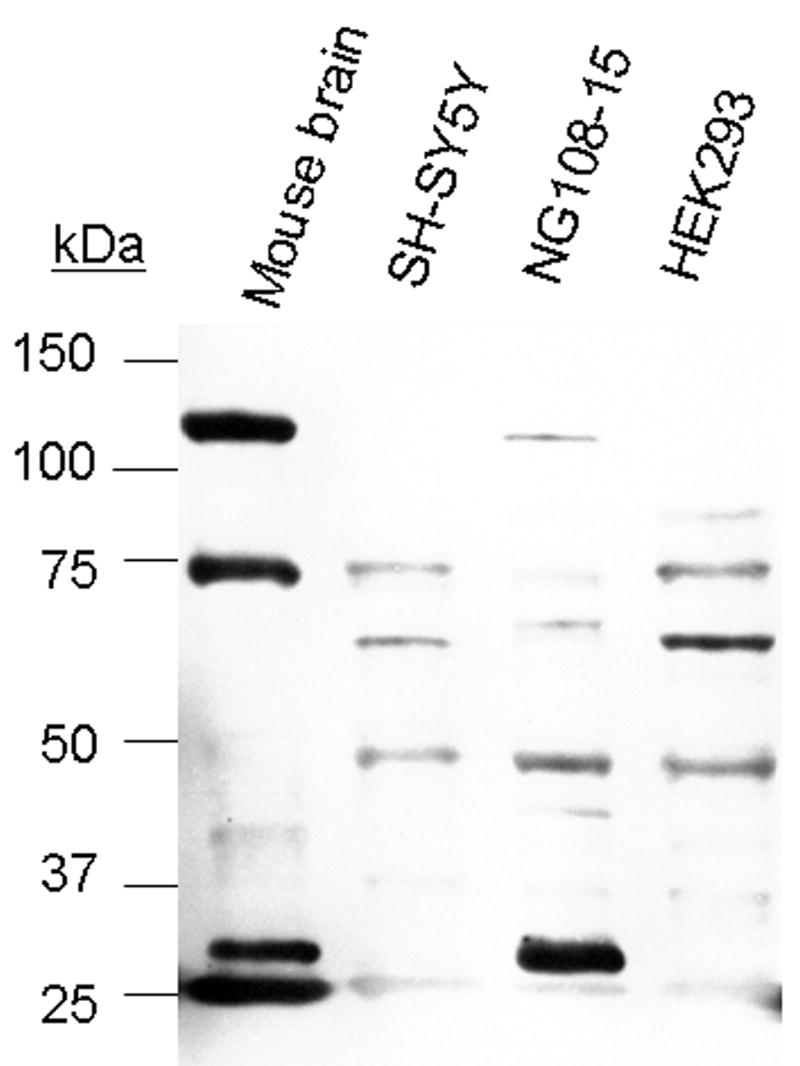
Detergent extracts were centrifuged at 16,000 × g for 20 min, and the supernatants were recovered for immunoblot analysis. Protein extracts were resolved using 12% SDS-PAGE and transferred to PVDF membranes. Following overnight incubation at 4°C with a 1:2000 dilution of the anti-endomorphin antibody, membranes were incubated with secondary antibody and developed. Left: Migration position of protein standards. Lanes left to right, endomorphin-like immunoreactivity in mouse brain, human SH-SY5Y neuroblastoma cells, mouse neuroblastoma × rat glioma NG 108-15 cells, and human embryonic kidney 293 cells, respectively. The western blot analysis was repeated three times with similar results.
Discussion
The bioinformatic search of the human proteome uncovered 12 different proteins that contain the endomorphin-1 tetrapeptide sequence, Tyr-Pro-Trp-Phe (Table 2), however, none of these proteins contained a Gly residue on the carboxyl-terminal side of the sequence that is required for peptide α-amidation (Eipper et al., 1992).
Twenty-two different known and putative proteins were found to contain the endomorphin-2 tetrapeptide sequence, Tyr-Pro-Phe-Phe, and two of these, the Na+-coupled sulfate transporter, SUT-1 (NP_036582) and a coiled-coil domain-containing putative ATPase (NP_694955) contain Gly on the carboxyl terminal side adjacent to the endomorphin-2 sequence (Table 3). The Tyr-Pro-Phe-Phe-Gly sequence in the sulfate transporter, SUT-1, beginning at amino acid residue 66 of the protein, is located within the second transmembrane domain of the 12-transmembrane domain-containing transporter protein (Girard et al., 1999). The mRNA encoding this protein exhibits a highly restricted human tissue distribution, and is abundant in the placenta, with significantly lower levels in the testis and heart, and is absent in the brain and 13 other tissues (Girard et al., 1999). Information regarding the structure of the domain containing the Tyr-Pro-Phe-Phe-Gly sequence in the putative ATPase protein is lacking. Another amino acid sequence constraint of substrates subject to α-amidation, however, is that they contain either Lys or Arg adjacent to the carboxyl terminal side of the Gly (Eipper et al., 1992), and the SUT-1 sodium sulfate transporter contains Val at this position, while the putative ATPase contains Ser adjacent to the Gly. It is possible, although unlikely, that endomorphin-2 is cleaved at the Gly-Val or Gly-Ser sequence by a novel processing enzyme. Support for the necessity of the peptide-Gly-Lys/Arg sequence for α-amidation is provided in Table 1, which contains a list of endogenous human bioactive peptides that have been confirmed to be α-amidated. In all cases, the peptide-Gly-Lys/Arg sequence is present.
Endomorphin-1 and endomorphin-2 were both reported to be amidated at the carboxyl terminus (Zadina et al., 1997; Hackler et al., 1997). Synthetic endomorphin-1-OH and endomorphin-2-OH, that lack the carboxyl-terminal amide group, display a 555-fold and 290-fold lower affinity, respectively, for the μ opioid receptor, and display a 400-fold and >1700-fold decrease, respectively, in agonist activity in the guinea pig ileum bioassay (In et al., 2005). Thus, to be considered as relevant endogenous agonists for the μ opioid receptor, the endomorphin peptides would have to be amidated at the carboxyl terminus.
Following the proposal that endomorphin-1 and endomorphin-2 were the endogenous ligands for the μ opioid receptor (Zadina et al., 1997), several laboratories investigated the distribution of endomorphin-like immunoreactivity in the central nervous system, with particular regard to whether the endomorphin-like immunoreactivity was co-localized with the μ receptor. In the spinal cord, it was reported that superficial layers of the dorsal horn contained endomorphin-2-like immunoreactivity, with the source being afferent fibers emanating from the dorsal root ganglion (Martin-Schild et al., 1997, 1998; Pierce et al., 1998, Schreff et al., 1998, Sanderson Nydahl et al., 2004). Evidence for release of endomorphin-2-like substances, defined by their ability to displace [125I]endomorphin-2 from an antibody that recognized the peptide, from rat spinal cord has also been reported (Williams et al., 1999; Gupta et al., 2007). In the brain, endomorphin-2-like immunoreactivity was largely confined to varicose fibers that were enriched in many areas that contained relatively high levels of the μ opioid receptor, but the immunoreactivity was noticeably absent in other regions, such as the striatum, olfactory bulb, hippocampus, cortex and nucleus of the solitary tract, in which the μ opioid receptor is abundant (Schreff et al., 1998). Cell bodies containing endomorphin-2-like immunoreactivity were only observed in the dorsomedial hypothalamus of the mouse. Pierce and Wessendorf (2000) also reported endomorphin-1-like immunoreactivity in many rat brain areas, some of which, but not all, were also enriched in the μ opioid receptor, however, in contrast to the report by Schreff et al. (1998), they found that the region displaying the greatest number of immunoreactive cell bodies was the nucleus of the solitary tract. Based on our western blot results, there are certainly many proteins that would potentially cross-react with the endomorphin-2 antibodies used in these immunocytochemical localization studies. Our western blot analysis revealed four major immunoreactive proteins in mouse brain. Attempts to identify each of these endomorphin-2 immunoreactive bands observed in one-dimensional western blots based on the theoretical molecular masses of the proteins uncovered by our bioinformatic search of the human proteome would be fraught with error and uncertainty, so we will make no such attempts. Chromatographic enrichment of the endomorphin-2-like immunoreactive proteins will probably be necessary to identify them using mass spectrometry, however, that project is beyond the scope of the current study. It is worthwhile pointing out that, although the biosynthetic precursors of the dermorphins and deltorphins are well established based on cDNA cloning of transcripts from frog skin, immunohistochemical studies of these peptides have been described in rodent brain (Mor et al., 1989; Tooyama et al., 1993; Yu et al., 2000). To our knowledge, however, there is no evidence for the synthesis of dermorphins and deltorphins in rodent brain, highlighting the difficulties that arise when interpreting immunohistochemical studies with antibodies that may cross-react with unknown antigens. One of the most powerful negative controls to analyze antibody specificity is to look for cross-reacting proteins in transgenic animals in which the relevant gene encoding the antigen has been knocked out; this is obviously not possible for the endomorphins, or for the dermorphins and deltorphins, in mammalian brain.
Very recently, Wolfe et al. (2007) constructed an artificial synthetic gene that directed production, cleavage and amidation of endomorphin-2 in a Herpes simplex virus vector. When primary dorsal root ganglion cultures were transduced with the virus, amidated endomorphin-2 was detected in the culture medium by radioimmunoassay, but it was not detectable in non-transduced control cultures. Subcutaneous inoculation of the viral vector provided an analgesic effect in a spinal nerve ligation model of neuropathic pain, and the analgesic effect was blocked by the μ-receptor-selective antagonist, CTOP (Wolfe et al., 2007).
We do not know with any certainty the source of the endomorphin-1 and endomorphin-2 that were initially isolated and sequenced from bovine and human cortex by Kastin and colleagues (Zadina et al., 1997; Hackler et al., 1997). It is conceivable that the endomorphins are synthesized de novo through a non-ribosomal enzymatic pathway, as is the case for the tripeptide, glutathione (γ-glutamylcysteinylglycine), the synthesis of which is catalyzed by γ-glutamylcysteine synthetase and glutathione synthetase. Similarly, the dipeptides, carnosine (β-alanylhistidine) and homocarnosine (γ-aminobutyrylhistidine), are synthesized in an ATP-dependent manner by the enzyme carnosine synthetase, however, both of these peptides contain amino acids not present in proteins (β-alanine and γ-aminobutyric acid, respectively). The Drosophila protein, Ebony, is a non-ribosomal peptide synthetase for the conjugation of β-alanine with biogenic amines (Richardt et al., 2003). Ronai et al. (2006) proposed that endomorphins may be produced by a de novo synthetic route involving reversal of peptidehydrolase catalytic activities, based on incorporation in rat brain of intracerebroventricularly injected [3H]-Tyr-Pro, however, a radioactive peptide with the HPLC retention time of endomorphin-1 was never observed in 15 experimental samples, and a peptide with the retention time of endomorphin-2 was observed in only 1 of 15 experimental samples. Clearly, this proposed endomorphin de novo biosynthetic pathway requires more robust experimental support, including chemical identity of the peptides with the incorporated label. Certain bacteria and fungi are capable of synthesizing a diverse set of peptides using multienzyme complexes (the non-ribosomal peptide synthetases) and some of these peptides, for example, vancomycin, are therapeutically important (reviewed in Grunewald and Marahiel, 2006). Like carnosine, these non-ribosomally synthesized peptides generally contain building blocks in addition to the 20 common amino acids found in proteins, such as D-amino acids, N-acyl groups, methylated residues, oxidized groups, glycosylated side chains, and heterocyclic moieties. In addition, these peptides are often cyclic. It is possible, though not very probable, that the endomorphins isolated from bovine and human brain were synthesized by microorganisms in the gut, or arose from dietary sources, however, the amidated tetrapeptides would have to cross the intestinal epithelia and blood brain barrier in order to gain entry into the brain. It also appears unlikely that the endomorphins would be synthesized through a novel non-ribosomal mechanism, based on comparison with all of the other human amidated peptides listed in Table 1, but this possibility cannot be ruled out.
It is conceivable that the genes encoding putative endomorphin-1 and endomorphin-2 precursors are present in the human genome, but have not yet been sequenced, hence their open reading frames would not have been translated and would not be found in our search. This scenario is also unlikely due to the extent of completion of the human genome sequencing project. We cannot rule out the possibility that endomorphin transcripts are produced by alternative splicing within the regions encoding the peptides, although there is no evidence from sequencing brain cDNA libraries to support this notion at this time. Several large human brain transcriptome projects are underway, and we will continue to analyze that data as it becomes available. It is also conceivable that the original isolation of the endomorphins from brain was an artifact, possibly due to contamination of the tissue samples with the synthetic peptides.
Conclusion
Based on our bioinformatic search of the human proteome, the assertion that endomorphin-1 and endomorphin-2 are the endogenous ligands for the μ opioid receptor should be questioned. In addition, reports in the literature describing the anatomical distribution of endomorphin-like immunoreactivity with antibodies raised against the synthetic peptides must also be interpreted with caution based on the western blot data reported in this study. To our knowledge, our report is the first to describe results of western blotting using endomorphin antibodies that have been used previously for immunohistochemical studies. Endomorphin-1 and endomorphin-2 are useful as highly selective and potent μ opioid receptor agonists, however, we have found no evidence for endogenous precursor proteins that are encoded by the human genome.
Acknowledgments
This investigation was supported by Grant DA09113 from the National Institute on Drug Abuse (to RDH) and a summer research fellowship (to KMW) from the Irene and Eric Simon Brain Research Foundation.
Abbreviations
- endomorphin-1
Tyr-Pro-Trp-Phe-NH2
- endomorphin-2
Tyr-Pro-Phe-Phe-NH2
- HEK
human embryonic kidney
- MIF
melanotropin release-inhibiting hormone (Pro-Leu-Gly-NH2)
- Tyr-W-MIF
Tyr-Pro-Trp-Gly-NH2
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bailey CP, Connor M. Opioids: cellular mechanisms of tolerance and physical dependence. Current Opinions in Pharmacology. 2005;5:60–68. doi: 10.1016/j.coph.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Bradbury AF, Smyth DG, Snell CR, Birdsall NJM, Hulme EC. C fragment of lipotropin has a high affinity for brain opiate receptors. Nature. 1976;260:793–795. doi: 10.1038/260793a0. [DOI] [PubMed] [Google Scholar]
- Brantl V, Gramsch C, Lottspeich F, Mertz R, Jaeger KH, Herz A. Novel opioid peptides derived from hemoglobin: hemorphins. European Journal of Pharmacology. 1986;125:309–310. doi: 10.1016/0014-2999(86)90044-0. [DOI] [PubMed] [Google Scholar]
- Brantl V, Teschemacher H, Henschen A, Lottspeich F. Novel opioid peptides derived from casein (beta-casomorphins). I. Isolation from bovine casein peptone. Hoppe-Seyler's Zeitung fur Physiological Chemistry. 1979;360:1211–1216. doi: 10.1515/bchm2.1979.360.2.1211. [DOI] [PubMed] [Google Scholar]
- Broccardo M, Erspamer V, Falconieri-Erspamer G, Improta G, Linari G, Melchiorri P, Montecucchi PC. Pharmacological data on dermorphins, a new class of potent opioid peptides from amphibian skin. British Journal of Pharmacology. 1981;73:625–631. doi: 10.1111/j.1476-5381.1981.tb16797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi K, Christoffers KH, Singh K, Howells RD. Structure and regulation of opioid receptors. Biopolymers (Peptide Science) 2000;55:334–346. doi: 10.1002/1097-0282(2000)55:4<334::AID-BIP1006>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Chen Y, Mestek A, Liu J, Hurley JA, Yu L. Molecular cloning and functional expression of a mu-opioid receptor from rat brain. Molecular Pharmacology. 1993;44:8–12. [PubMed] [Google Scholar]
- Comb M, Seeburg PH, Adelman J, Eiden L, Herbert E. Primary structure of the human Met- and Leu-enkephalin precursor and its mRNA. Nature. 1982;295:663–666. doi: 10.1038/295663a0. [DOI] [PubMed] [Google Scholar]
- Eipper BA, Stoffers DA, Mains RE. The biosynthesis of neuropeptides: peptide α-amidation. Annual Review of Neuroscience. 1992;15:57–85. doi: 10.1146/annurev.ne.15.030192.000421. [DOI] [PubMed] [Google Scholar]
- Erchegyi J, Kastin AJ, Zadina JE. Isolation of a novel tetrapeptide with opiate, and antiopiate activity from human brain cortex. Peptides. 1992;13:623–631. doi: 10.1016/0196-9781(92)90165-y. [DOI] [PubMed] [Google Scholar]
- Espamer V, Melchiorri P, Falconieri-Erspamer G, Negri L, Corsi R, Severini C, Barra D, Simmaco M, Kreil G. Deltorphins: a family of naturally occurring peptides with high affinity and selectivity for δ opioid binding sites. Proceedings of the National Academy of Science USA. 1989;86:5188–5192. doi: 10.1073/pnas.86.13.5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans CJ, Keith DE, Jr, Morrison H, Magendzo K, Edwards RH. Cloning of a delta opioid receptor by functional expression. Science. 1992;258:1952–1955. doi: 10.1126/science.1335167. [DOI] [PubMed] [Google Scholar]
- Evans JM, Bey V, Burkey AR, Commons KG. Organization of endogenous opioids in the rostral agranular insular cortex of the rat. Journal of Comparative Neurology. 2007;500:530–541. doi: 10.1002/cne.21197. [DOI] [PubMed] [Google Scholar]
- Gates M, Tschudi G. The synthesis of morphine. Journal of the American Chemical Society. 1956;78:1380–1403. [Google Scholar]
- Girard JP, Baekkevold ES, Fellu J, Brandtzaeg P, Amalric F. Molecular cloning and functional analysis of SUT-1, a sulfate transporter from human high endothelial venules. Proceedings of the National Academy of Science USA. 1999;96:12772–12777. doi: 10.1073/pnas.96.22.12772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A, Fischli W, Lowney LT, Hunkapillar M, Hood L. Porcine pituitary dynorphin: complete amino acid sequence of the biologically active heptadecapeptide. Proceedings of the National Academy of Science USA. 1981;78:7219–7223. doi: 10.1073/pnas.78.11.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald J, Marahiel MA. Chemoenzymatic and template-directed synthesis of bioreactive macrocyclic peptides. Microbiology and Molecular Biology Reviews. 2006;70:121–146. doi: 10.1128/MMBR.70.1.121-146.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler U, Seeburg P, Hoffman BJ, Gage LP, Udenfriend S. Molecular cloning establishes proenkephalin as a precursor of enkephalin-containing peptides. Nature. 1982;295:206–208. doi: 10.1038/295206a0. [DOI] [PubMed] [Google Scholar]
- Gulland JM, Robinson R. The morphine group. Part I. A discussion of the constitutional problem. Journal of the Chemical Society. 1925;123:980–998. [Google Scholar]
- Gupta DS, von Gizycki H, Gintzler AR. Sex-/ovarian steroid-dependent release of endomorphin 2 from spinal cord. Journal of Pharmacology and Experimental Therapeutics. 2007;321:635–641. doi: 10.1124/jpet.106.118505. [DOI] [PubMed] [Google Scholar]
- Hackler L, Kastin AJ, Erchegyi J, Zadina JE. Isolation of Tyr-W-MIF-1 from bovine hypothalami. Neuropeptides. 1993;24:159–164. doi: 10.1016/0143-4179(93)90080-t. [DOI] [PubMed] [Google Scholar]
- Hackler L, Zadina JE, Ge LJ, Kastin AJ. Isolation of relatively large amounts of endomorphin-1 and endomorphin-2 from human brain cortex. Peptides. 1997;18:1635–1639. doi: 10.1016/s0196-9781(97)00259-3. [DOI] [PubMed] [Google Scholar]
- Heck SD, Faraci WS, Kelbaugh PR, Saccomano NA, Thadeio PF, Volkmann RA. Posttranslational amino acid epimerization: enzyme-catalyzed isomerization of amino acid residues in peptide chains. Proceedings of the National Academy of Science USA. 1996;93:4036–4039. doi: 10.1073/pnas.93.9.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath A, Kastin AJ. Isolation of tyrosine-melanocyte-stimulating hormone release-inhibiting factor 1 from bovine brain tissue. Journal of Biological Chemistry. 1989;264:2175–2179. [PubMed] [Google Scholar]
- Howells RD. Proenkephalin biosynthesis in the rat. In: Rapaka RS, Hawks RL, editors. Opioid peptides: Molecular Pharmacology, Biosynthesis, and Analysis, NIDA Research Monograph 70. Rockville: 1986. pp. 43–65. [PubMed] [Google Scholar]
- Hughes J, Smith TW, Kosterlitz HW, Fothergill LA, Morgan BA, Morris HR. Identification of two related pentapeptides from the brain with potent opiate agonist activity. Nature. 1975;258:577–579. doi: 10.1038/258577a0. [DOI] [PubMed] [Google Scholar]
- In Y, Minoura K, Tomoo K, Sasaki Y, Lazarus LH, Okada Y, Ishida T. Structural function of C-terminal amidation of endomorphin. FEBS Journal. 2005;272:5079–5097. doi: 10.1111/j.1742-4658.2005.04919.x. [DOI] [PubMed] [Google Scholar]
- Jaffe JH, Martin WR. Opioid analgesics and antagonists. In: Gilman AG, Rall TW, Nies AS, Taylor P, editors. Goodman and Gilman's The Pharmacological Basis of Therapeutics. 8th. Pergamon Press; New York: 2000. pp. 485–521. [Google Scholar]
- Janecka A, Fichna J, Janecki T. Opioid receptors and their ligands. Current Topics in Medicinal Chemistry. 2004;4:1–17. doi: 10.2174/1568026043451618. [DOI] [PubMed] [Google Scholar]
- Kakidani H, Furutani Y, Takanashi H, Noda M, Morimoto Y, Hirose T, Asai M, Inayama S, Nakanishi S, Numa S. Cloning and sequence analysis of cDNA for porcine β-neoendorphin/dynorphin precursor. Nature. 1982;298:245–249. doi: 10.1038/298245a0. [DOI] [PubMed] [Google Scholar]
- Kieffer BL, Befort K, Gaveriaux-Ruff C, Hirth C. The delta-opioid receptor: isolation by a cDNA by expression cloning and pharmacological characterization. Proceedings of the National Academy of Science USA. 1992;89:12048–12052. doi: 10.1073/pnas.89.24.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosterlitz HW, Paterson SJ, Robson LE. Characterization of the κ-subtype of the opiate receptor in the guinea-pig brain. British Journal of Pharmacology. 1981;73:939–949. doi: 10.1111/j.1476-5381.1981.tb08749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord JAH, Waterfield AA, Hughes J, Kosterlitz HW. Endogenous opioid peptides: multiple agonists and receptors. Nature. 1977;267:495–499. doi: 10.1038/267495a0. [DOI] [PubMed] [Google Scholar]
- Martin-Schild S, Gerall AA, Kastin AJ, Zadina JE. Endomorphin-2 is an endogenous opioid in primary sensory afferent fibers. Peptides. 1998;19:1783–1789. doi: 10.1016/s0196-9781(98)00136-3. [DOI] [PubMed] [Google Scholar]
- Martin-Schild S, Gerall AA, Kastin AJ, Zadina JE. Differential distribution of endormorphin 1- and endomorphin 2-like immunoreactivities in the CNS of the rodent. Journal of Comparative Neurology. 1999;405:450–471. [PubMed] [Google Scholar]
- Martin-Schild S, Zadina JE, Gerall AA, Vigh S, Kastin AJ. Localization of endomorphin-2-like immunoreactivity in the rat medulla and spinal cord. Peptides. 1997;18:1641–1649. doi: 10.1016/s0196-9781(97)00320-3. [DOI] [PubMed] [Google Scholar]
- McQuay H. Opioids in pain management. Lancet. 1999;353:2229–2232. doi: 10.1016/S0140-6736(99)03528-X. [DOI] [PubMed] [Google Scholar]
- Mor A, Amiche M, Nicolas P. Enter a new post-translational modification: D-amino acids in gene-encoded proteins. Trends in Biochemical Science. 1992;17:481–485. doi: 10.1016/0968-0004(92)90333-5. [DOI] [PubMed] [Google Scholar]
- Mor A, Delfour A, Amiche M, Sagan S, Nicolas P, Grassi J, Pradelles P. Dermorphin and related peptides in rat tissues. Neuropeptides. 1989;13:51–57. doi: 10.1016/0143-4179(89)90021-8. [DOI] [PubMed] [Google Scholar]
- Nair RMG, Kastin AJ, Schally AV. Isolation and structure of hypothalamic MSH release-inhibiting hormone. Biochemical and Biophysical Research Communications. 1971;43:1376–1381. doi: 10.1016/s0006-291x(71)80026-8. [DOI] [PubMed] [Google Scholar]
- Nakanishi S, Inoue A, Kita T, Nakamura M, Chang ACY, Cohen SN, Numa S. Nucleotide sequence of clonal cDNA for bovine corticotropin-β-lipotropin precursor. Nature. 1979;278:423–427. doi: 10.1038/278423a0. [DOI] [PubMed] [Google Scholar]
- Negri L, Melchiorri P, Lattanzi R. Pharmacology of amphibian opiate peptides. Peptides. 2000;21:1639–1647. doi: 10.1016/s0196-9781(00)00295-3. [DOI] [PubMed] [Google Scholar]
- Noda M, Furutani Y, Takahashi H, Toyosato M, Hirose T, Inayama S, Nakanishi S, Numa S. Cloning and sequence analysis of cDNA for bovine adrenal preproenkephalin. Nature. 1982;295:202–206. doi: 10.1038/295202a0. [DOI] [PubMed] [Google Scholar]
- Nyberg F, Sanderson K, Glamsa EI. The hemorphins: a new class of opioid peptides derived from the blood protein hemoglobin. Biopolymers. 1997;43:147–156. doi: 10.1002/(SICI)1097-0282(1997)43:2<147::AID-BIP8>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Paterson SJ, Robson LE, Kosterlitz HW. Opioid receptors. In: Udenfriend S, Meienhofer J, editors. The Peptides. Vol. 4. Academic Press; Orlando: 1984. pp. 147–189. [Google Scholar]
- Pert CA, Snyder SH. Opiate receptor: demonstration in nervous tissue. Science. 1973;179:1011–1014. doi: 10.1126/science.179.4077.1011. [DOI] [PubMed] [Google Scholar]
- Pierce TL, Grahek MD, Wessendorf MW. Immunoreactivity for endomorphin-2 occurs in primary afferents in rats and monkey. NeuroReport. 1998;9:385–389. doi: 10.1097/00001756-199802160-00005. [DOI] [PubMed] [Google Scholar]
- Pierce TL, Wessendorf MW. Immunocytochemical mapping of endomorphin-2-immunoreactivity in rat brain. Journal of Chemical Neuroanatomy. 2000;18:181–207. doi: 10.1016/s0891-0618(00)00042-9. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Research. 2007;3:D61–65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardt A, Kemme T, Wagner S, Schwarzer D, Marahiel MA, Hovemann BT. Ebony, a nonribosomal peptide synthetase for beta-alanine conjugation of biogenic amines in Drosophila. Journal of Biological Chemistry. 2003;278:41160–41166. doi: 10.1074/jbc.M304303200. [DOI] [PubMed] [Google Scholar]
- Ronai AZ, Szemenyei E, Kato E, Kocsis L, Orosz G, Al-Khrasani M, Toth G. Endomorphin synthesis in rat brain from intracerebroventricularly injected [3H]-Tyr-Pro: a possible biosynthetic route for endomorphins. Regulatory Peptides. 2006;134:54–60. doi: 10.1016/j.regpep.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Sanderson Nydahl K, Skinner K, Julius D, Basbaum AI. Co-localization of endomorphin-2 and substance P in primary afferent nociceptors and effects of injury: a light and electron microscopic study in the rat. Journal of Comparative Neurology. 2004;19:1789–1799. doi: 10.1111/j.1460-9568.2004.03284.x. [DOI] [PubMed] [Google Scholar]
- Schreff M, Schulz S, Wiborny D, Höllt V. Immunofluorescent identification of endomorphin-2-containing nerve fibers and terminals in the rat brain and spinal cord. NeuroReport. 1998;9:1031–1034. doi: 10.1097/00001756-199804200-00014. [DOI] [PubMed] [Google Scholar]
- Sertürner FW. Journal der Pharmacie fuer Aerzte und Apotheker. 1805;13:229–243. No title. [Google Scholar]
- Sertürner FW. Analyse de l'opium. De la morphine et l'acide meconique, consideres comme parties essentielles de l'opium. Annales de Chimie et de Physique. 1817;5:21–42. [Google Scholar]
- Simon EJ, Hiller JM, Edelman I. Stereospecific binding of the potent narcotic analgesic [3H]etorphine to rat brain homogenate. Proceedings of the National Academy of Science USA. 1973;70:1947–1949. doi: 10.1073/pnas.70.7.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenius L. Stereospecific interaction between narcotic analgesics and a synaptic plasma membrane fraction of rat cerebral cortex. Acta Pharmacology Toxicology. 1973;32:317–320. doi: 10.1111/j.1600-0773.1973.tb01477.x. [DOI] [PubMed] [Google Scholar]
- Tooyama I, Abe H, Renda T, Erspamer V, Kimura H. [D-Ala2]Deltorphin 1-like immunoreactivity in adult rat brain: immunohistochemical localization. Proceedings of the National Academy of Science USA. 1993;90:9635–9639. doi: 10.1073/pnas.90.20.9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QP, Zadina JE, Guan JL, Kastin AJ, Funahashi H, Shioda S. Endomorphin-2 immunoreactivity in the cervical dorsal horn of the rat spinal cord at the electron microscopic level. Neuroscience. 2002;113:593–605. doi: 10.1016/s0306-4522(02)00153-7. [DOI] [PubMed] [Google Scholar]
- Weber E, Esch FS, Bohlen P, Paterson S, Corbett AD, McKnight AT, Kosterlitz HW, Barchas JD, Evans CJ. Metorphamide: isolation, structure, and biologic activity of an amidated opioid octapeptide from bovine brain. Proceedings of the National Academy of Sciences USA. 1983;80:7362–7366. doi: 10.1073/pnas.80.23.7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CA, Wu SY, Dun SL, Kwok EH, Dun NJ. Release of endomorphin-2 like substances from the rat spinal cord. Neuroscience Letters. 1999;273:25–28. doi: 10.1016/s0304-3940(99)00613-8. [DOI] [PubMed] [Google Scholar]
- Wolfe D, Hao S, Hu J, Srinivasan R, Goss J, Mata M, Fink DJ, Glorioso JC. Engineering an endomorphin-2 gene for use in neuropathic pain therapy. Pain. 2007 doi: 10.1016/j.pain.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Research. 2006;34:D187–191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav PN, Chaturvedi K, Howells RD. Inhibition of agonist-induced down-regulation of the δ-opioid receptor with a proteasome inhibitor attenuates opioid tolerance in human embryonic kidney 293 cells. Journal of Pharmacology and Experimental Therapeutics. 2007;320:1186–1194. doi: 10.1124/jpet.106.113621. [DOI] [PubMed] [Google Scholar]
- Yasuda K, Raynor K, Kong H, Breder CD, Takeda J, Reisine T, Bell GI. Cloning and functional comparison of kappa and delta opioid receptors from mouse brain. Proceedings of the National Academy of Sciences USA. 1993;90:6736–6740. doi: 10.1073/pnas.90.14.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Zhao T, Fan M, Tooyama I, Kimura H, Renda T. Production of a monoclonal antibody to deltorphin-1 and its immunohistochemical application to adult rat brain and cultured rat brain neurons. Peptides. 2000;21:1657–1662. doi: 10.1016/s0196-9781(00)00314-4. [DOI] [PubMed] [Google Scholar]
- Zadina JE, Hackler L, Ge LJ, Kastin AJ. A potent and selective endogenous agonist for the μ-opiate receptor. Nature. 1997;386:499–5502. doi: 10.1038/386499a0. [DOI] [PubMed] [Google Scholar]
- Zadina JE, Kastin AJ, Ge LJ, Hackler L. Mu, delta, and kappa opiate receptor binding of Tyr-MIF-1 and of Tyr-W-MIF-1, its active fragments, and two potent analogs. Life Sciences. 1994;55:PL 461–466. doi: 10.1016/0024-3205(94)00533-8. [DOI] [PubMed] [Google Scholar]


