Abstract
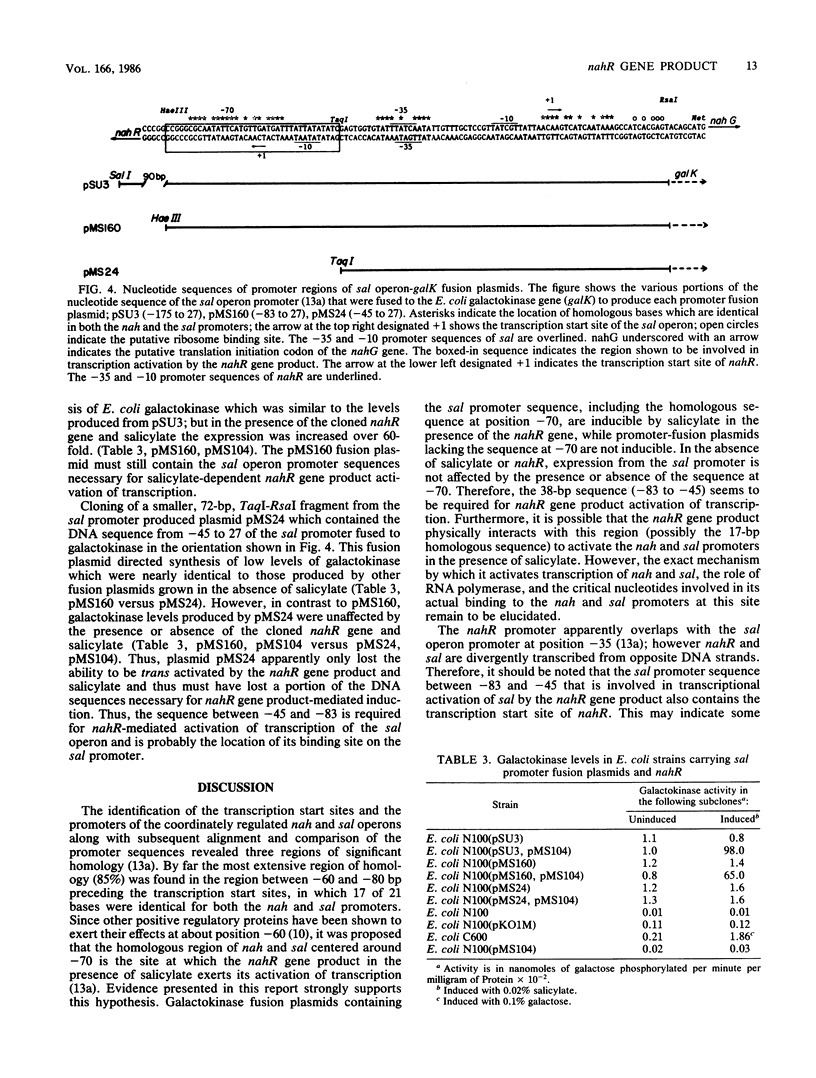
The product of the nahR gene, a salicylate-dependent activator of transcription of the nah and sal hydrocarbon degradation operons of the NAH7 plasmid, was identified and characterized after synthesis in Escherichia coli maxicells. The nahR gene product had a subunit molecular weight of 36,000, as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, whereas gel filtration analysis of the nondenatured nahR protein indicated a molecular weight in excess of 250,000. However, DNase I treatment of this high-molecular-weight complex shifted the apparent molecular weight of the nahR protein to 40,000. Various upstream portions of the sal operon promoter were transcriptionally fused to the E. coli galactokinase gene. Fusion plasmids containing the sal promoter sequence from --83 to 27 (relative to the transcription start site) showed salicylate-inducible expression of galactokinase in the presence of the cloned nahR gene, while expression of galactokinase from a fusion plasmid containing the sal promoter sequence from --45 to 27 was not induced by the nahR gene and salicylate. Results suggest that the nahR gene product is a 36-kilodalton polypeptide which exerts its salicylate-dependent activation of transcription of the sal operon by interacting with the promoter sequence in the region of --83 to --45 base pairs before the transcription start site.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barnsley E. A. The induction of the enzymes of naphthalene metabolism in pseudomonads by salicylate and 2-aminobenzoate. J Gen Microbiol. 1975 May;88(1):193–196. doi: 10.1099/00221287-88-1-193. [DOI] [PubMed] [Google Scholar]
- Dunn N. W., Gunsalus I. C. Transmissible plasmid coding early enzymes of naphthalene oxidation in Pseudomonas putida. J Bacteriol. 1973 Jun;114(3):974–979. doi: 10.1128/jb.114.3.974-979.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grund A. D., Gunsalus I. C. Cloning of genes for naphthalene metabolism in Pseudomonas putida. J Bacteriol. 1983 Oct;156(1):89–94. doi: 10.1128/jb.156.1.89-94.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Nakazawa A., Nakazawa T. Molecular cloning of regulatory gene xylR and operator-promoter regions of the xylABC and xylDEGF operons of the TOL plasmid. J Bacteriol. 1983 Sep;155(3):1192–1199. doi: 10.1128/jb.155.3.1192-1199.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Nakazawa A., Nakazawa T. Nucleotide sequence of the promoter region of the xylDEGF operon on TOL plasmid of Pseudomonas putida. Gene. 1984 Sep;29(3):323–330. doi: 10.1016/0378-1119(84)90061-1. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McKenney K., Shimatake H., Court D., Schmeissner U., Brady C., Rosenberg M. A system to study promoter and terminator signals recognized by Escherichia coli RNA polymerase. Gene Amplif Anal. 1981;2:383–415. [PubMed] [Google Scholar]
- Raibaud O., Schwartz M. Positive control of transcription initiation in bacteria. Annu Rev Genet. 1984;18:173–206. doi: 10.1146/annurev.ge.18.120184.001133. [DOI] [PubMed] [Google Scholar]
- Sancar A., Hack A. M., Rupp W. D. Simple method for identification of plasmid-coded proteins. J Bacteriol. 1979 Jan;137(1):692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell M. A. Cloning and expression in Escherichia coli of the naphthalene degradation genes from plasmid NAH7. J Bacteriol. 1983 Feb;153(2):822–829. doi: 10.1128/jb.153.2.822-829.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell M. A. Homology between nucleotide sequences of promoter regions of nah and sal operons of NAH7 plasmid of Pseudomonas putida. Proc Natl Acad Sci U S A. 1986 Jan;83(2):369–373. doi: 10.1073/pnas.83.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell M. A. Transcriptional control of the nah and sal hydrocarbon-degradation operons by the nahR gene product. Gene. 1985;36(3):301–309. doi: 10.1016/0378-1119(85)90185-4. [DOI] [PubMed] [Google Scholar]
- Schell M. A., Wilson D. B. Cloning and expression of the yeast galactokinase gene in an Escherichia coli plasmid. Gene. 1979 Apr;5(4):291–303. doi: 10.1016/0378-1119(79)90104-5. [DOI] [PubMed] [Google Scholar]
- Schell M. A., Wilson D. B. Purification and properties of galactokinase from Saccharomyces cerevisiae. J Biol Chem. 1977 Feb 25;252(4):1162–1166. [PubMed] [Google Scholar]
- Yen K. M., Gunsalus I. C. Plasmid gene organization: naphthalene/salicylate oxidation. Proc Natl Acad Sci U S A. 1982 Feb;79(3):874–878. doi: 10.1073/pnas.79.3.874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K. M., Gunsalus I. C. Regulation of naphthalene catabolic genes of plasmid NAH7. J Bacteriol. 1985 Jun;162(3):1008–1013. doi: 10.1128/jb.162.3.1008-1013.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]



