Abstract
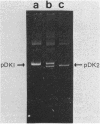
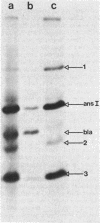
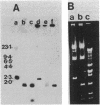
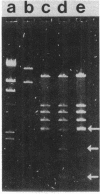
Mutants of Escherichia coli have been isolated which are resistant to beta-aspartyl hydroxamate, a lethal substrate of asparaginase II in fungi and a substrate for asparaginase II in E. coli. Among the many phenotypic classes observed, a single mutant (designated GU16) was found with multiple defects affecting asparaginases I and II and aspartase. Other asparaginase II-deficient mutants have also been derived from an asparaginase I-deficient mutant. The mutant strain, GU16, was unable to utilize asparagine and grew poorly on aspartate as the sole source of carbon; transformation of this strain with an E. coli recombinant plasmid library resulted in a large recombinant plasmid which complemented both these defects. Two subclones were isolated, designated pDK1 and pDK2; the former complemented the partial defect in the utilization of aspartate, although its exact function was not established. pDK2 encoded the asparaginase I gene (ansA), the coding region of which was further defined within a 1.7-kilobase fragment. The ansA gene specified a polypeptide, identified in maxicells, with a molecular weight of 43,000. Strains carrying recombinant plasmids encoding the ansA gene overproduced asparaginase I approximately 130-fold, suggesting that the ansA gene might normally be under negative regulation. Extracts from strains overproducing asparaginase I were electrophoresed, blotted, and probed with asparaginase II-specific antisera; no cross-reaction of the antisera with asparaginase I was observed, indicating that asparaginases I and II are not appreciably related immunologically. When a DNA fragment containing the ansA gene was used to probe Southern blots of restriction endonuclease-digested E. coli chromosomal DNA, no homologous sequences were revealed other than the expected ansA-containing fragments. Therefore, the genes encoding asparaginases I and II are highly sequence related.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams C. W., Hatfield G. W. Effects of promoter strengths and growth conditions on copy number of transcription-fusion vectors. J Biol Chem. 1984 Jun 25;259(12):7399–7403. [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns D. M., Abraham L. J., Beacham I. R. Characterization of the ush gene of Escherichia coli and its protein products. Gene. 1983 Nov;25(2-3):343–353. doi: 10.1016/0378-1119(83)90239-1. [DOI] [PubMed] [Google Scholar]
- Burns D. M., Beacham I. R. A method for the ligation of DNA following isolation from low melting temperature agarose. Anal Biochem. 1983 Nov;135(1):48–51. doi: 10.1016/0003-2697(83)90728-5. [DOI] [PubMed] [Google Scholar]
- Burns D. M., Beacham I. R. Positive selection vectors: a small plasmid vector useful for the direct selection of Sau3A-generated overlapping DNA fragments. Gene. 1984 Mar;27(3):323–325. doi: 10.1016/0378-1119(84)90077-5. [DOI] [PubMed] [Google Scholar]
- Cedar H., Schwartz J. H. Localization of the two-L-asparaginases in anaerobically grown Escherichia coli. J Biol Chem. 1967 Aug 25;242(16):3753–3755. [PubMed] [Google Scholar]
- Cedar H., Schwartz J. H. Production of L-asparaginase II by Escherichia coli. J Bacteriol. 1968 Dec;96(6):2043–2048. doi: 10.1128/jb.96.6.2043-2048.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Casale T., Sollitti P., Chesney R. H. Cytoplasmic L-asparaginase: isolation of a defective strain and mapping of ansA. J Bacteriol. 1983 Apr;154(1):513–515. doi: 10.1128/jb.154.1.513-515.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durden D. L., Distasio J. A. Comparison of the immunosuppressive effects of asparaginases from Escherichia coli and Vibrio succinogenes. Cancer Res. 1980 Apr;40(4):1125–1129. [PubMed] [Google Scholar]
- Ehrman M., Cedar H., Schwartz J. H. L-asparaginase II of Escherichia coli. Studies on the enzymatic mechanism of action. J Biol Chem. 1971 Jan 10;246(1):88–94. [PubMed] [Google Scholar]
- Ferrara P., Duchange N., Zakin M. M., Cohen G. N. Internal homologies in the two aspartokinase-homoserine dehydrogenases of Escherichia coli K-12. Proc Natl Acad Sci U S A. 1984 May;81(10):3019–3023. doi: 10.1073/pnas.81.10.3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furano A. V. Direct demonstration of duplicate tuf genes in enteric bacteria. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3104–3108. doi: 10.1073/pnas.75.7.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershoni J. M., Palade G. E. Protein blotting: principles and applications. Anal Biochem. 1983 May;131(1):1–15. doi: 10.1016/0003-2697(83)90128-8. [DOI] [PubMed] [Google Scholar]
- Julian G. R., Reithel F. J. Glucose-6-phosphate dehydrogenase from bovine mammary gland. Methods Enzymol. 1975;41:183–188. doi: 10.1016/s0076-6879(75)41044-8. [DOI] [PubMed] [Google Scholar]
- Kaplan J. B., Nichols B. P. Nucleotide sequence of Escherichia coli pabA and its evolutionary relationship to trp(G)D. J Mol Biol. 1983 Aug 15;168(3):451–468. doi: 10.1016/s0022-2836(83)80295-2. [DOI] [PubMed] [Google Scholar]
- Kim K. W., Roon R. J. Asparaginase II of Saccharomyces cerevisiae: positive selection of two mutations that prevent enzyme synthesis. J Bacteriol. 1984 Mar;157(3):958–961. doi: 10.1128/jb.157.3.958-961.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maita T., Matsuda G. The primary structure of L-asparaginase from Escherichia coli. Hoppe Seylers Z Physiol Chem. 1980;361(2):105–117. doi: 10.1515/bchm2.1980.361.1.105. [DOI] [PubMed] [Google Scholar]
- Murthy N. S., Knox J. R. Small-angle x-ray scattering studies of Escherichia coli L-asparaginase. J Mol Biol. 1976 Aug 25;105(4):567–575. doi: 10.1016/0022-2836(76)90235-7. [DOI] [PubMed] [Google Scholar]
- Neu H. C. The 5'-nucleotidase of Escherichia coli. I. Purification and properties. J Biol Chem. 1967 Sep 10;242(17):3896–3904. [PubMed] [Google Scholar]
- Peterson R. G., Richards F. F., Handschumacher R. E. Structure of peptide from active site region of Escherichia coli L-asparaginase. J Biol Chem. 1977 Mar 25;252(6):2072–2076. [PubMed] [Google Scholar]
- Prentki P., Krisch H. M. A modified pBR322 vector with improved properties for the cloning, recovery, and sequencing of blunt-ended DNA fragments. Gene. 1982 Feb;17(2):189–196. doi: 10.1016/0378-1119(82)90072-5. [DOI] [PubMed] [Google Scholar]
- Sancar A., Hack A. M., Rupp W. D. Simple method for identification of plasmid-coded proteins. J Bacteriol. 1979 Jan;137(1):692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger M. J. Secretion of alkaline phosphatase subunits by spheroplasts of Escherichia coli. J Bacteriol. 1968 Sep;96(3):727–733. doi: 10.1128/jb.96.3.727-733.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz J. H., Reeves J. Y., Broome J. D. Two L-asparaginases from E. coli and their action against tumors. Proc Natl Acad Sci U S A. 1966 Nov;56(5):1516–1519. doi: 10.1073/pnas.56.5.1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer M. E., Lebeter V. M., Guest J. R. Location of the Aspartase Gene (aspA) on the linkage map of Escherichia coli K12. J Gen Microbiol. 1976 Nov;97(1):73–82. doi: 10.1099/00221287-97-1-73. [DOI] [PubMed] [Google Scholar]
- Strauch K. L., Lenk J. B., Gamble B. L., Miller C. G. Oxygen regulation in Salmonella typhimurium. J Bacteriol. 1985 Feb;161(2):673–680. doi: 10.1128/jb.161.2.673-680.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talmadge K., Gilberg W. Construction of plasmid vectors with unique PstI cloning sites in a signal sequence coding region. Gene. 1980 Dec;12(3-4):235–241. doi: 10.1016/0378-1119(80)90105-5. [DOI] [PubMed] [Google Scholar]
- Van Vliet F., Cunin R., Jacobs A., Piette J., Gigot D., Lauwereys M., Piérard A., Glansdorff N. Evolutionary divergence of genes for ornithine and aspartate carbamoyl-transferases--complete sequence and mode of regulation of the Escherichia coli argF gene; comparison of argF with argI and pyrB. Nucleic Acids Res. 1984 Aug 10;12(15):6277–6289. doi: 10.1093/nar/12.15.6277. [DOI] [PMC free article] [PubMed] [Google Scholar]