Abstract
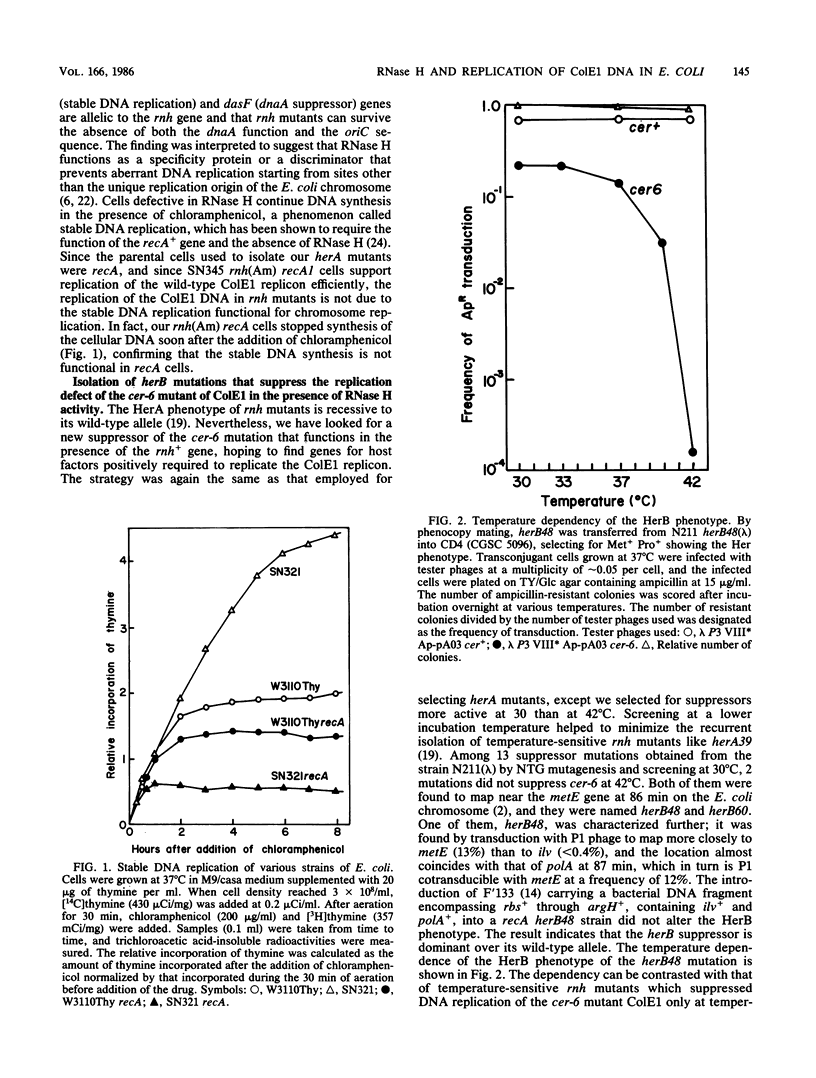
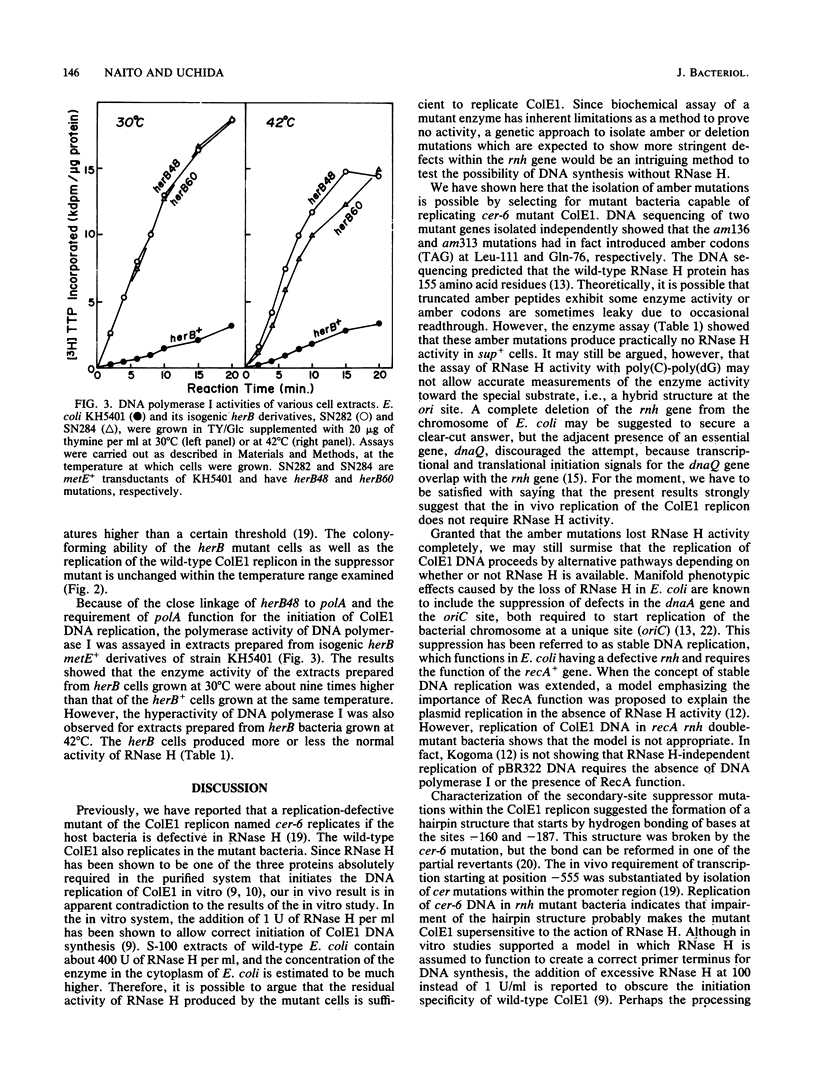
Amber mutations within the rnh (RNase H) gene of Escherichia coli K-12 were isolated by selecting for bacteria capable of replicating in a sup+ background replication-defective cer-6 mutant of the ColE1 replicon. The cer-6 mutation is an alteration of one base pair located 160 nucleotides upstream of the unique replication origin of this plasmid. Subsequently, we determined the DNA alterations present within these mutants. ColE1 DNA replicated in rnh(Am) recA cells, indicating that (i) RNase H, which has been shown to be absolutely required for in vitro initiation of ColE1 DNA replication, is dispensable in vivo, and (ii) ColE1 replication in the absence of RNase H is not dependent on "stable DNA replication," which has been reported to be an alternative mode of chromosomal DNA replication. Another class of bacterial mutations was also isolated. These mutations, named herB, suppressed cer-6 replication in rnh+ bacteria. herB mutations mapped close to the polA gene on the E. coli chromosome and increased the activity of DNA polymerase I. These findings suggest that when the DNA polymerase I has an opportunity to initiate DNA synthesis before RNase H acts, the replication-defective cer-6 mutant or the wild-type ColE1 replicates in E. coli.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csonka L. N., Clark A. J. Construction of an Hfr strain useful for transferring recA mutations between Escherichia coli strains. J Bacteriol. 1980 Jul;143(1):529–530. doi: 10.1128/jb.143.1.529-530.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lucia P., Cairns J. Isolation of an E. coli strain with a mutation affecting DNA polymerase. Nature. 1969 Dec 20;224(5225):1164–1166. doi: 10.1038/2241164a0. [DOI] [PubMed] [Google Scholar]
- Hillenbrand G., Staudenbauer W. L. Discriminatory function of ribonuclease H in the selective initiation of plasmid DNA replication. Nucleic Acids Res. 1982 Feb 11;10(3):833–853. doi: 10.1093/nar/10.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi T., Maki H., Maruyama M., Sekiguchi M. Identification of the dnaQ gene product and location of the structural gene for RNase H of Escherichia coli by cloning of the genes. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3770–3774. doi: 10.1073/pnas.78.6.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi T., Maki H., Sekiguchi M. Conditional lethality of Escherichia coli strains carrying dnaE and dnaQ mutations. Mol Gen Genet. 1981;181(1):24–28. doi: 10.1007/BF00339000. [DOI] [PubMed] [Google Scholar]
- Itoh T., Tomizawa J. Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc Natl Acad Sci U S A. 1980 May;77(5):2450–2454. doi: 10.1073/pnas.77.5.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., Tomizawa J. Initiation of replication of plasmid ColE1 DNA by RNA polymerase, ribonuclease H, and DNA polymerase I. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):409–417. doi: 10.1101/sqb.1979.043.01.047. [DOI] [PubMed] [Google Scholar]
- Kingsbury D. T., Helinski D. R. DNA polymerase as a requirement for the maintenance of the bacterial plasmid colicinogenic factor E1. Biochem Biophys Res Commun. 1970 Dec 24;41(6):1538–1544. doi: 10.1016/0006-291x(70)90562-0. [DOI] [PubMed] [Google Scholar]
- Kogoma T. Absence of RNase H allows replication of pBR322 in Escherichia coli mutants lacking DNA polymerase I. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7845–7849. doi: 10.1073/pnas.81.24.7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogoma T., von Meyenburg K. The origin of replication, oriC, and the dnaA protein are dispensable in stable DNA replication (sdrA) mutants of Escherichia coli K-12. EMBO J. 1983;2(3):463–468. doi: 10.1002/j.1460-2075.1983.tb01445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low K. B. Escherichia coli K-12 F-prime factors, old and new. Bacteriol Rev. 1972 Dec;36(4):587–607. doi: 10.1128/br.36.4.587-607.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki H., Horiuchi T., Sekiguchi M. Structure and expression of the dnaQ mutator and the RNase H genes of Escherichia coli: overlap of the promoter regions. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7137–7141. doi: 10.1073/pnas.80.23.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masukata H., Tomizawa J. Effects of point mutations on formation and structure of the RNA primer for ColE1 DNA replication. Cell. 1984 Feb;36(2):513–522. doi: 10.1016/0092-8674(84)90244-7. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miura A., Tomizawa J. I. Studies on radiation-sensitive mutants of E. coli. 3. Participation of the rec system in induction of mutation by ultraviolet irradiation. Mol Gen Genet. 1968;103(1):1–10. doi: 10.1007/BF00271151. [DOI] [PubMed] [Google Scholar]
- Naito S., Kitani T., Ogawa T., Okazaki T., Uchida H. Escherichia coli mutants suppressing replication-defective mutations of the ColE1 plasmid. Proc Natl Acad Sci U S A. 1984 Jan;81(2):550–554. doi: 10.1073/pnas.81.2.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito S., Uchida H. Initiation of DNA replication in a ColE1-type plasmid: isolation of mutations in the ori region. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6744–6748. doi: 10.1073/pnas.77.11.6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T., Okazaki T. Function of RNase H in DNA replication revealed by RNase H defective mutants of Escherichia coli. Mol Gen Genet. 1984;193(2):231–237. doi: 10.1007/BF00330673. [DOI] [PubMed] [Google Scholar]
- Ogawa T., Pickett G. G., Kogoma T., Kornberg A. RNase H confers specificity in the dnaA-dependent initiation of replication at the unique origin of the Escherichia coli chromosome in vivo and in vitro. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1040–1044. doi: 10.1073/pnas.81.4.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacon W., Sherratt D. ColE plasmid replication in DNA polymerase I-deficient strains of Escherichia coli. Mol Gen Genet. 1976 Sep 23;147(3):331–335. doi: 10.1007/BF00582885. [DOI] [PubMed] [Google Scholar]
- Torrey T. A., Kogoma T. Suppressor mutations (rin) that specifically suppress the recA+ dependence of stable DNA replication in Escherichia coliK-12. Mol Gen Genet. 1982;187(2):225–230. doi: 10.1007/BF00331121. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K., Tomizawa J. Establishment of Escherichia coli cells with an integrated high copy number plasmid. Mol Gen Genet. 1980;178(3):525–533. doi: 10.1007/BF00337857. [DOI] [PubMed] [Google Scholar]



