Abstract
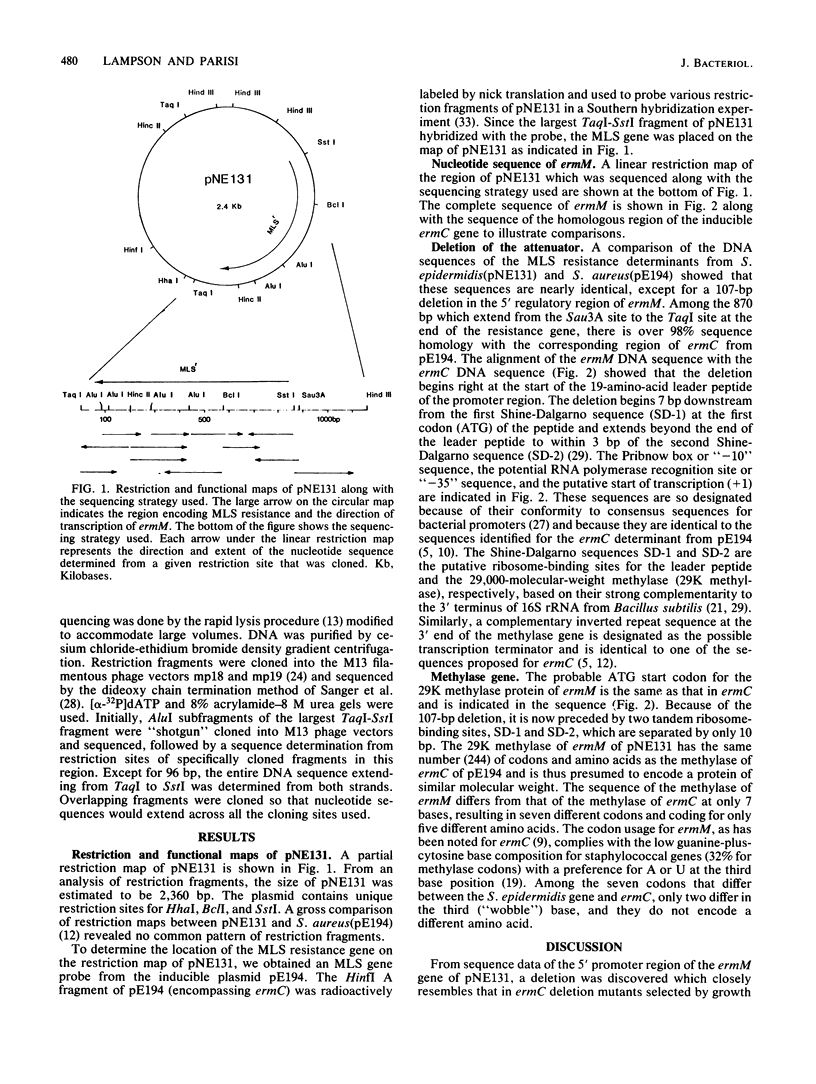
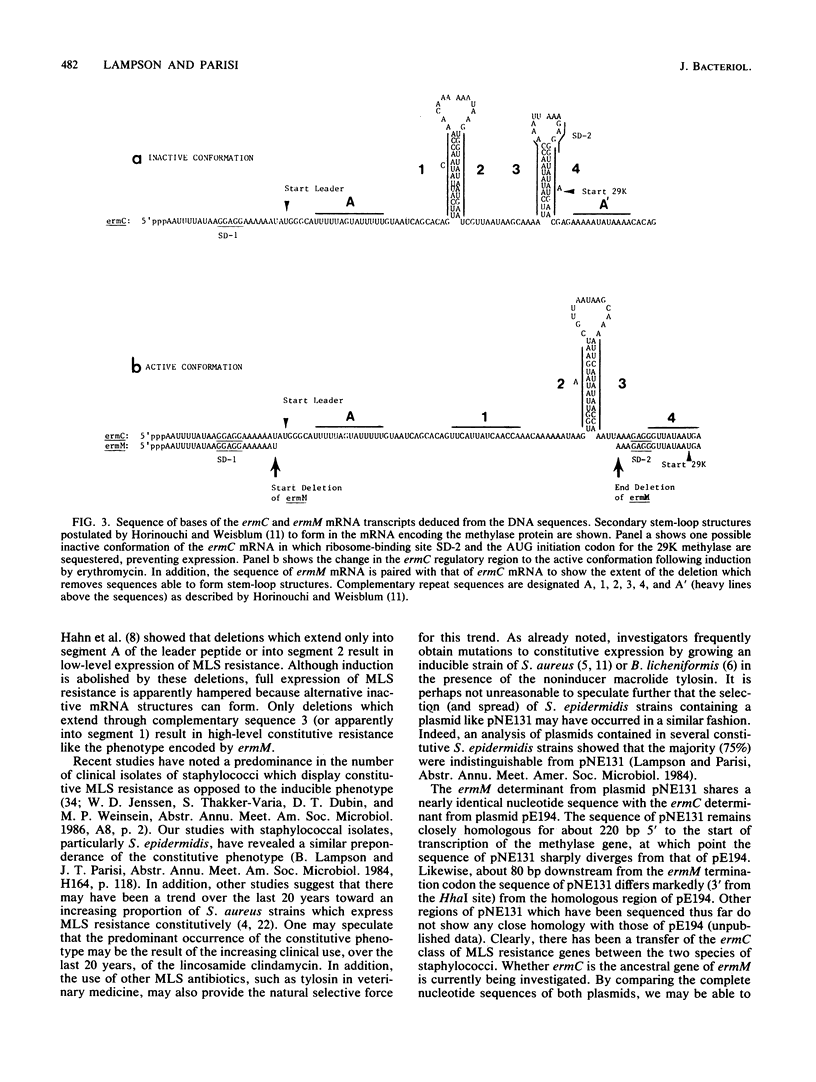
A naturally occurring constitutive macrolide-lincosamide-streptogramin B (MLS) resistance plasmid, pNE131, from Staphylococcus epidermidis was chosen to study the molecular basis of constitutive expression. Restriction and functional maps of pNE131 are presented along with the nucleotide sequence of ermM, the gene which mediates constitutive MLS resistance. Sharing 98% sequence homology within the 870-base-pair Sau3A-TaqI fragment, ermM appears to be almost identical to ermC, the inducible MLS resistance determinant from S. aureus (pE194). The two genes share nearly identical sequences, except in the 5' promoter region of ermM. Constitutive expression of ermM is due to the deletion of 107 base pairs relative to ermC; the deletion removes critical sequences for attenuation, resulting in constitutive methylase expression.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkinson B. A., Lorian V. Antimicrobial agent susceptibility patterns of bacteria in hospitals from 1971 to 1982. J Clin Microbiol. 1984 Oct;20(4):791–796. doi: 10.1128/jcm.20.4.791-796.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M. L., Wong E. S., Falkow S. Common R-plasmids in Staphylococcus aureus and Staphylococcus epidermidis during a nosocomial Staphylococcus aureus outbreak. Antimicrob Agents Chemother. 1982 Feb;21(2):210–215. doi: 10.1128/aac.21.2.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D. Induction of ermC requires translation of the leader peptide. EMBO J. 1985 Feb;4(2):533–537. doi: 10.1002/j.1460-2075.1985.tb03661.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval J. Evolution and epidemiology of MLS resistance. J Antimicrob Chemother. 1985 Jul;16 (Suppl A):137–149. doi: 10.1093/jac/16.suppl_a.137. [DOI] [PubMed] [Google Scholar]
- Gryczan T. J., Grandi G., Hahn J., Grandi R., Dubnau D. Conformational alteration of mRNA structure and the posttranscriptional regulation of erythromycin-induced drug resistance. Nucleic Acids Res. 1980 Dec 20;8(24):6081–6097. doi: 10.1093/nar/8.24.6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Israeli-Reches M., Dubnau D. Induction of macrolide-lincosamide-streptogramin B resistance requires ribosomes able to bind inducer. Mol Gen Genet. 1984;194(3):357–361. doi: 10.1007/BF00425544. [DOI] [PubMed] [Google Scholar]
- Gryczan T., Israeli-Reches M., Del Bue M., Dubnau D. DNA sequence and regulation of ermD, a macrolide-lincosamide-streptogramin B resistance element from Bacillus licheniformis. Mol Gen Genet. 1984;194(3):349–356. doi: 10.1007/BF00425543. [DOI] [PubMed] [Google Scholar]
- Hahn J., Grandi G., Gryczan T. J., Dubnau D. Translational attenuation of ermC: a deletion analysis. Mol Gen Genet. 1982;186(2):204–216. doi: 10.1007/BF00331851. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Byeon W. H., Weisblum B. A complex attenuator regulates inducible resistance to macrolides, lincosamides, and streptogramin type B antibiotics in Streptococcus sanguis. J Bacteriol. 1983 Jun;154(3):1252–1262. doi: 10.1128/jb.154.3.1252-1262.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibodies. J Bacteriol. 1982 May;150(2):804–814. doi: 10.1128/jb.150.2.804-814.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Posttranscriptional modification of mRNA conformation: mechanism that regulates erythromycin-induced resistance. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7079–7083. doi: 10.1073/pnas.77.12.7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. The control region for erythromycin resistance: free energy changes related to induction and mutation to constitutive expression. Mol Gen Genet. 1981;182(2):341–348. doi: 10.1007/BF00269681. [DOI] [PubMed] [Google Scholar]
- Hutton J. P., Hamory B. H., Parisi J. T., Strausbaugh L. J. Staphylococcus epidermidis arthritis following catheter-induced bacteremia in a neutropenic patient. Diagn Microbiol Infect Dis. 1985 Mar;3(2):119–124. doi: 10.1016/0732-8893(85)90020-3. [DOI] [PubMed] [Google Scholar]
- Lai C. J., Dahlberg J. E., Weisblum B. Structure of an inducibly methylatable nucleotide sequence in 23S ribosomal ribonucleic acid from erythromycin-resistant Staphylococcus aureus. Biochemistry. 1973 Jan 30;12(3):457–460. doi: 10.1021/bi00727a015. [DOI] [PubMed] [Google Scholar]
- Lai C. J., Weisblum B. Altered methylation of ribosomal RNA in an erythromycin-resistant strain of Staphylococcus aureus. Proc Natl Acad Sci U S A. 1971 Apr;68(4):856–860. doi: 10.1073/pnas.68.4.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy F. D., Hammer S. M. Staphylococcus epidermidis infections. Ann Intern Med. 1983 Dec;99(6):834–839. doi: 10.7326/0003-4819-99-6-834. [DOI] [PubMed] [Google Scholar]
- MARMUR J., FALKOW S., MANDEL M. NEW APPROACHES TO BACTERIAL TAXONOMY. Annu Rev Microbiol. 1963;17:329–372. doi: 10.1146/annurev.mi.17.100163.001553. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford M., Weisblum B. Messenger RNA from Staphylococcus aureus that specifies macrolide-lincosamide-streptogramin resistance. Demonstration of its conformations and of the leader peptide it encodes. J Mol Biol. 1985 Oct 20;185(4):769–780. doi: 10.1016/0022-2836(85)90061-0. [DOI] [PubMed] [Google Scholar]
- McLaughlin J. R., Murray C. L., Rabinowitz J. C. Unique features in the ribosome binding site sequence of the gram-positive Staphylococcus aureus beta-lactamase gene. J Biol Chem. 1981 Nov 10;256(21):11283–11291. [PubMed] [Google Scholar]
- Murphy E. Nucleotide sequence of ermA, a macrolide-lincosamide-streptogramin B determinant in Staphylococcus aureus. J Bacteriol. 1985 May;162(2):633–640. doi: 10.1128/jb.162.2.633-640.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Parisi J. T. Coagulase-negative staphylococci and the epidemiological typing of Staphylococcus epidermidis. Microbiol Rev. 1985 Jun;49(2):126–139. doi: 10.1128/mr.49.2.126-139.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi J. T., Robbins J., Lampson B. C., Hecht D. W. Characterization of a macrolide, lincosamide, and streptogramin resistance plasmid in Staphylococcus epidermidis. J Bacteriol. 1981 Nov;148(2):559–564. doi: 10.1128/jb.148.2.559-564.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivakumar A. G., Dubnau D. Characterization of a plasmid-specified ribosome methylase associated with macrolide resistance. Nucleic Acids Res. 1981 Jun 11;9(11):2549–2562. doi: 10.1093/nar/9.11.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner R., Cundliffe E., Schmidt F. J. Site of action of a ribosomal RNA methylase responsible for resistance to erythromycin and other antibiotics. J Biol Chem. 1983 Oct 25;258(20):12702–12706. [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Thabaut A., Meyran M., Huerre M. Evolution and present situation of Staphylococcus aureus sensitivity to MLS in hospital (1975-83). J Antimicrob Chemother. 1985 Jul;16 (Suppl A):205–207. doi: 10.1093/jac/16.suppl_a.205. [DOI] [PubMed] [Google Scholar]
- Weisblum B., Graham M. Y., Gryczan T., Dubnau D. Plasmid copy number control: isolation and characterization of high-copy-number mutants of plasmid pE194. J Bacteriol. 1979 Jan;137(1):635–643. doi: 10.1128/jb.137.1.635-643.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]


