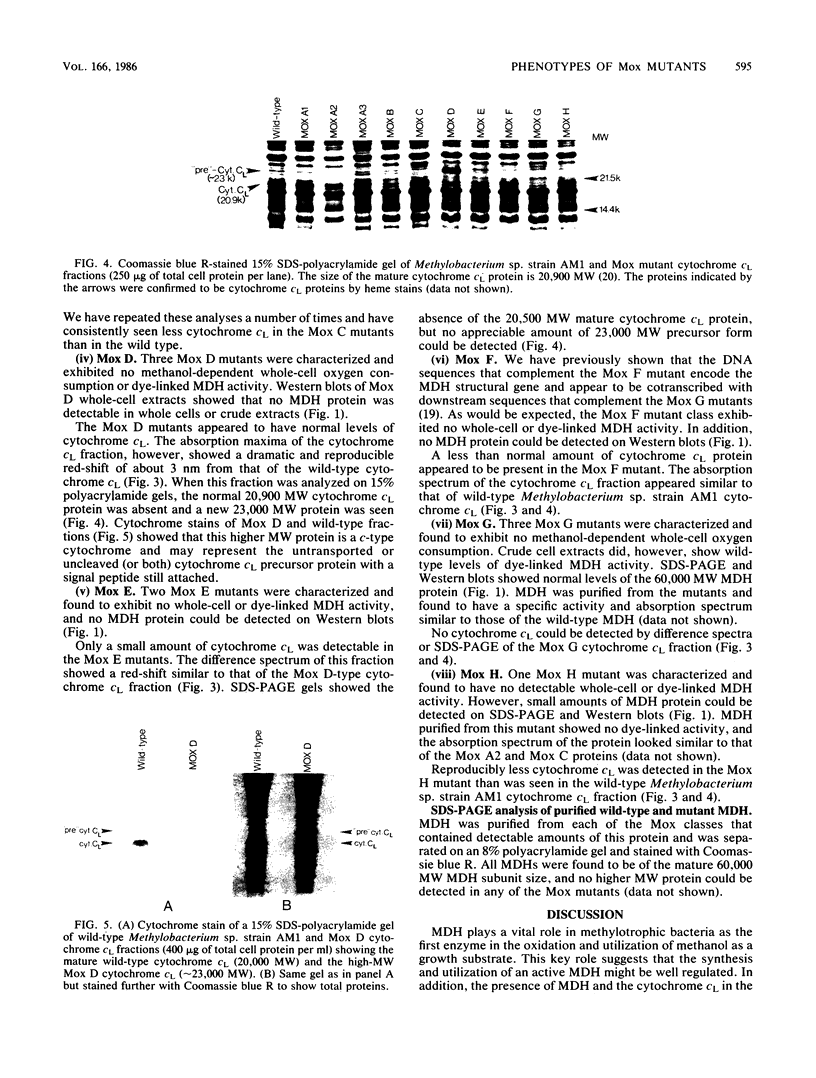
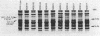Abstract
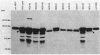
Twenty-five methanol oxidation mutants of the facultative methylotroph Methylobacterium sp. strain AM1 have been characterized by complementation analysis and assigned to 10 complementation groups, Mox A1, A2, A3, and B through H (D. N. Nunn and M. E. Lidstrom, J. Bacteriol. 166:582-591, 1986). In this study we have characterized each of the mutants belonging to the 10 Mox complementation groups for the following criteria: phenazine methosulfate-dichlorophenolindophenol dye-linked methanol dehydrogenase activity; methanol-dependent whole-cell oxygen consumption; the presence or absence of methanol dehydrogenase protein by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting; the absorption spectra of purified mutant methanol dehydrogenase proteins; and the presence or absence of the soluble cytochrome c proteins of Methylobacterium sp. strain AM1, as determined by reduced-oxidized difference spectra and sodium dodecyl sulfate-polyacrylamide gel electrophoresis. With this information, we have proposed functions for each of the genes deficient in the mutants of the 10 Mox complementation groups. These proposed gene functions include two linked genes that encode the methanol dehydrogenase structural protein and the soluble cytochrome cL, a gene encoding a secretion function essential for the synthesis and export of methanol dehydrogenase and cytochrome cL, three gene functions responsible for the proper association of the pyrrolo-quinoline quinone prosthetic group with the methanol dehydrogenase apoprotein, and four positive regulatory gene functions controlling the expression of the ability to oxidize methanol.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alefounder P. R., Ferguson S. J. A periplasmic location for methanol dehydrogenase from Paracoccus denitrificans: implications for proton pumping by cytochrome aa3. Biochem Biophys Res Commun. 1981 Feb 12;98(3):778–784. doi: 10.1016/0006-291x(81)91179-7. [DOI] [PubMed] [Google Scholar]
- Anthony C. The microbial metabolism of C1 compounds. The cytochromes of Pseudomaonas AM1. Biochem J. 1975 Feb;146(2):289–298. doi: 10.1042/bj1460289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony C., Zatman L. J. The microbial oxidation of methanol. The alcohol dehydrogenase of Pseudomonas sp. M27. Biochem J. 1965 Sep;96(3):808–812. doi: 10.1042/bj0960808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony C., Zatman L. J. The microbial oxidation of methanol. The prosthetic group of the alcohol dehydrogenase of Pseudomonas sp. M27: a new oxidoreductase prosthetic group. Biochem J. 1967 Sep;104(3):960–969. doi: 10.1042/bj1040960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson V. L., Neher J. W., Cecchini G. The biosynthesis and assembly of methanol dehydrogenase in bacterium W3A1. J Biol Chem. 1985 Aug 15;260(17):9642–9647. [PubMed] [Google Scholar]
- Duine J. A., Frank J., Jr The prosthetic group of methanol dehydrogenase. Purification and some of its properties. Biochem J. 1980 Apr 1;187(1):221–226. doi: 10.1042/bj1870221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunstan P. M., Anthony C., Drabble W. T. Microbial metabolism of C 1 and C 2 compounds. The role of glyoxylate, glycollate and acetate in the growth of Pseudomonas AM1 on ethanol and on C 1 compounds. Biochem J. 1972 Jun;128(1):107–115. doi: 10.1042/bj1280107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R. T., Jr, Becker R. R. Specific indication of hemoproteins in polyacrylamide gels using a double-staining process. Anal Biochem. 1984 Feb;136(2):509–514. doi: 10.1016/0003-2697(84)90253-7. [DOI] [PubMed] [Google Scholar]
- Heptinstall J., Quayle J. R. Pathways leading to and from serine during growth of Pseudomonas AM1 on C1 compounds or succinate. Biochem J. 1970 Apr;117(3):563–572. doi: 10.1042/bj1170563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. A., Quayle J. R. Microbial growth on C-1 compounds. 6. Oxidation of methanol, formaldehyde and formate by methanol-grown Pseudomonas AM-1. Biochem J. 1964 Nov;93(2):281–290. doi: 10.1042/bj0930281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasprzak A. A., Steenkamp D. J. Localization of the major dehydrogenases in two methylotrophs by radiochemical labeling. J Bacteriol. 1983 Oct;156(1):348–353. doi: 10.1128/jb.156.1.348-353.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- McNerney T., O'connor M. L. Regulation of enzymes associated with C-1 metabolism in three facultative methylotrophs. Appl Environ Microbiol. 1980 Aug;40(2):370–375. doi: 10.1128/aem.40.2.370-375.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis S., Beckwith J. Mechanism of incorporation of cell envelope proteins in Escherichia coli. Annu Rev Microbiol. 1982;36:435–465. doi: 10.1146/annurev.mi.36.100182.002251. [DOI] [PubMed] [Google Scholar]
- O'Keeffe D. T., Anthony C. The two cytochromes c in the facultative methylotroph Pseudomonas am1. Biochem J. 1980 Nov 15;192(2):411–419. doi: 10.1042/bj1920411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEEL D., QUAYLE J. R. Microbial growth on C1 compounds. I. Isolation and characterization of Pseudomonas AM 1. Biochem J. 1961 Dec;81:465–469. doi: 10.1042/bj0810465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R. N., Hou C. T., Felix A. Microbial oxidation of methane and methanol: crystallization of methanol dehydrogenase and properties of holo- and apomethanol dehydrogenase from Methylomonas methanica. J Bacteriol. 1978 Feb;133(2):641–649. doi: 10.1128/jb.133.2.641-649.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver C. A., Lidstrom M. E. Methanol dissimilation in Xanthobacter H4-14: activities, induction and comparison to Pseudomonas AM1 and Paracoccus denitrificans. J Gen Microbiol. 1985 Sep;131(9):2183–2197. doi: 10.1099/00221287-131-9-2183. [DOI] [PubMed] [Google Scholar]
- de Beer R., Duine J. A., Frank J., Large P. J. The prosthetic group of methylamine dehydrogenase from Pseudomonas AM1: evidence for a quinone structure. Biochim Biophys Acta. 1980 Apr 25;622(2):370–374. doi: 10.1016/0005-2795(80)90050-1. [DOI] [PubMed] [Google Scholar]