Abstract
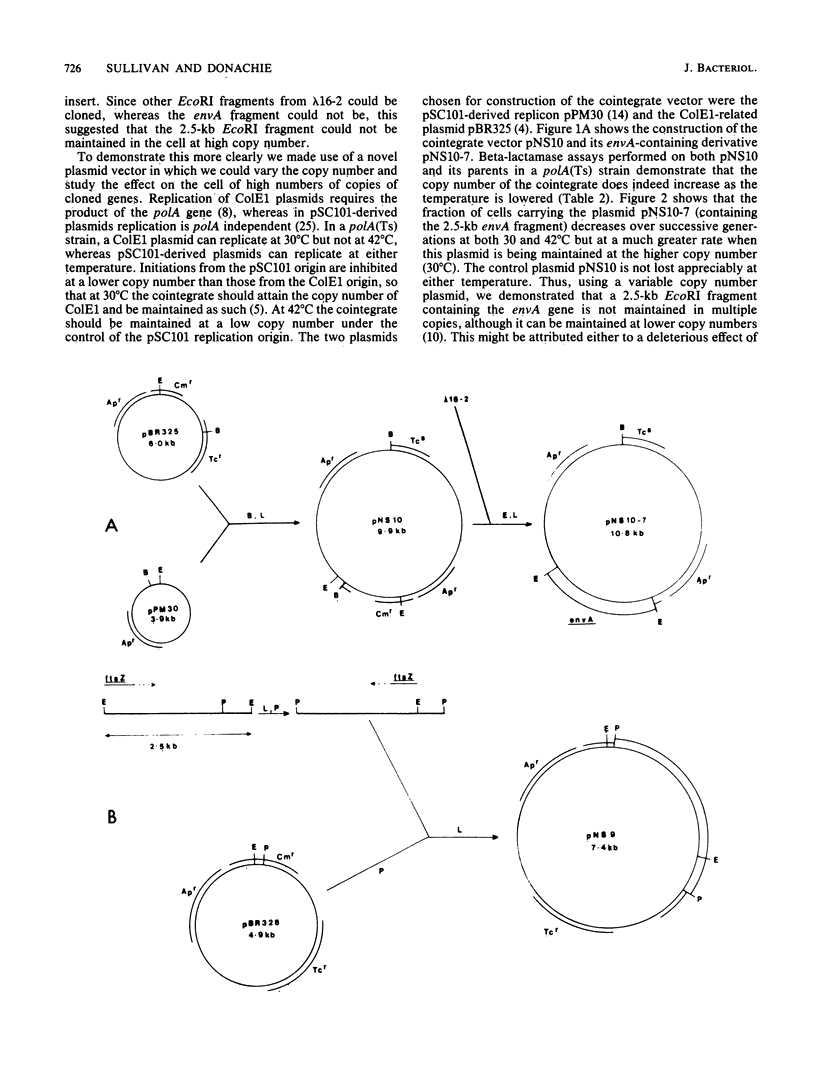
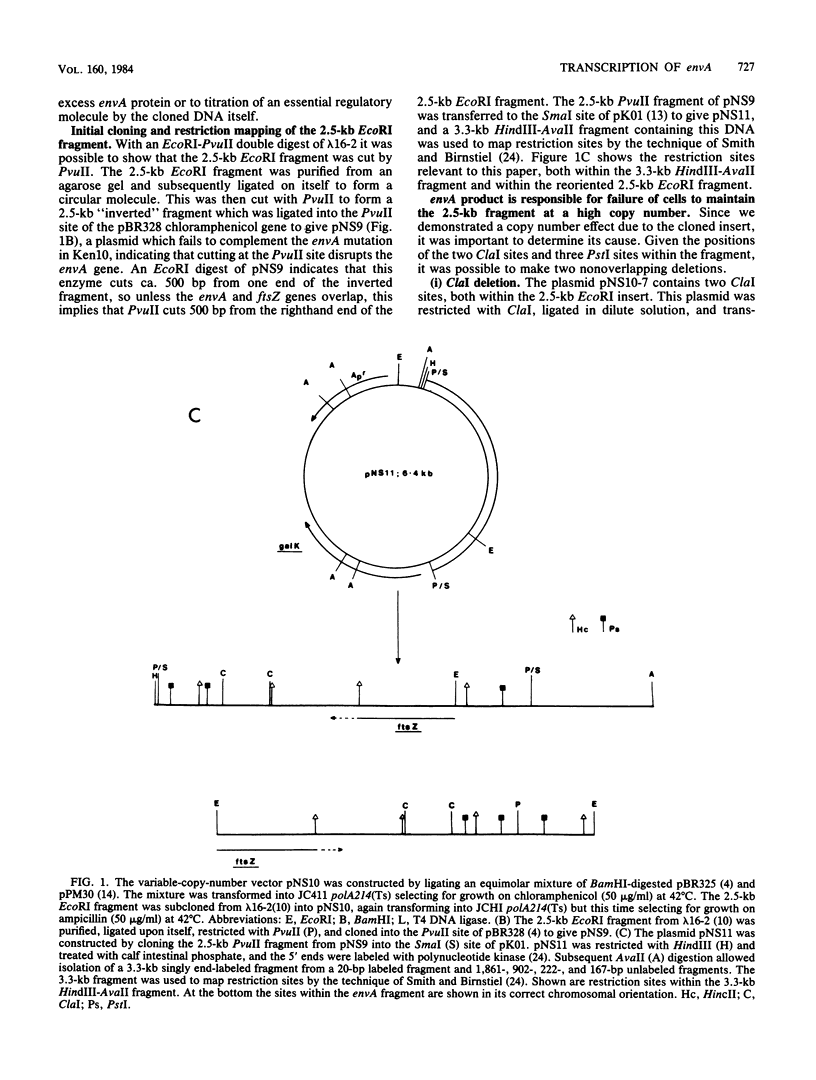
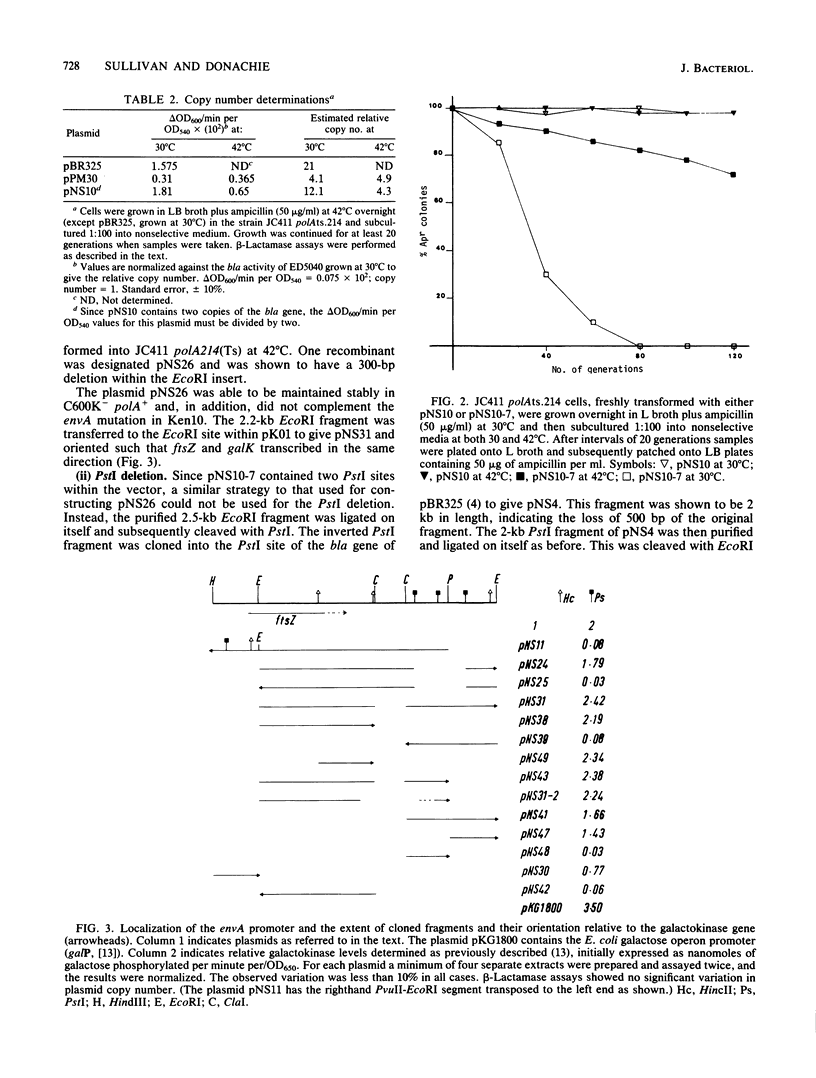
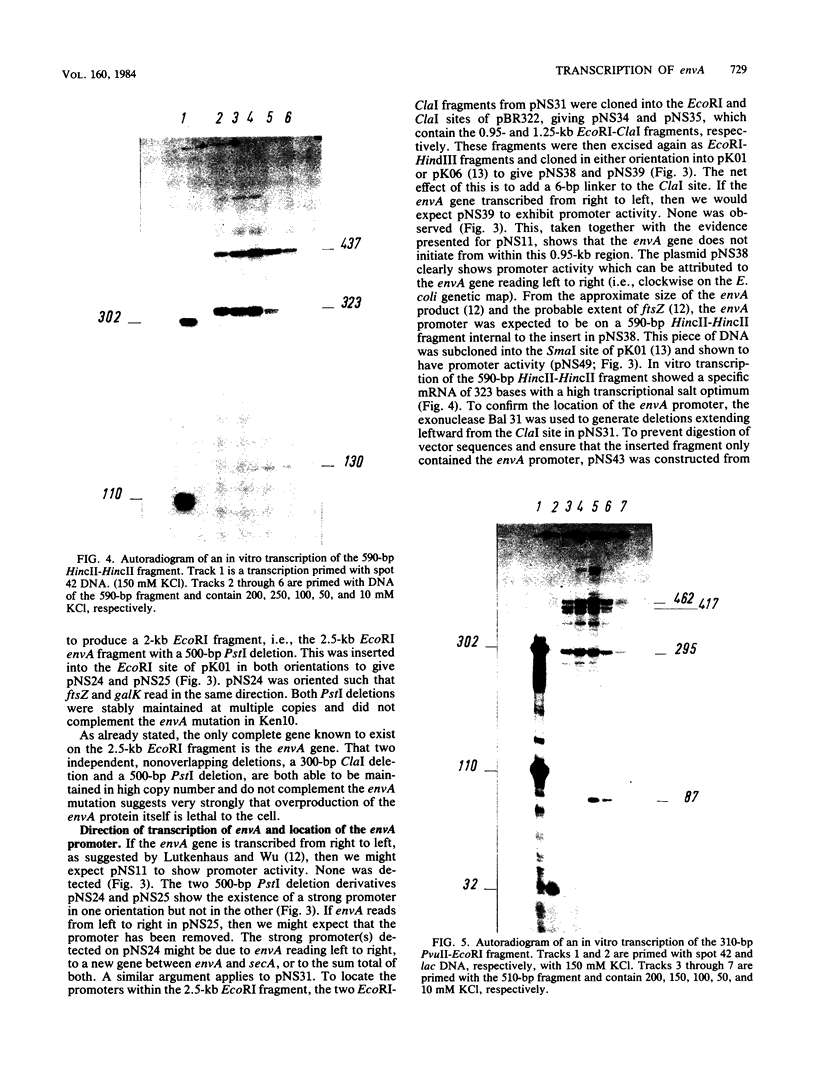
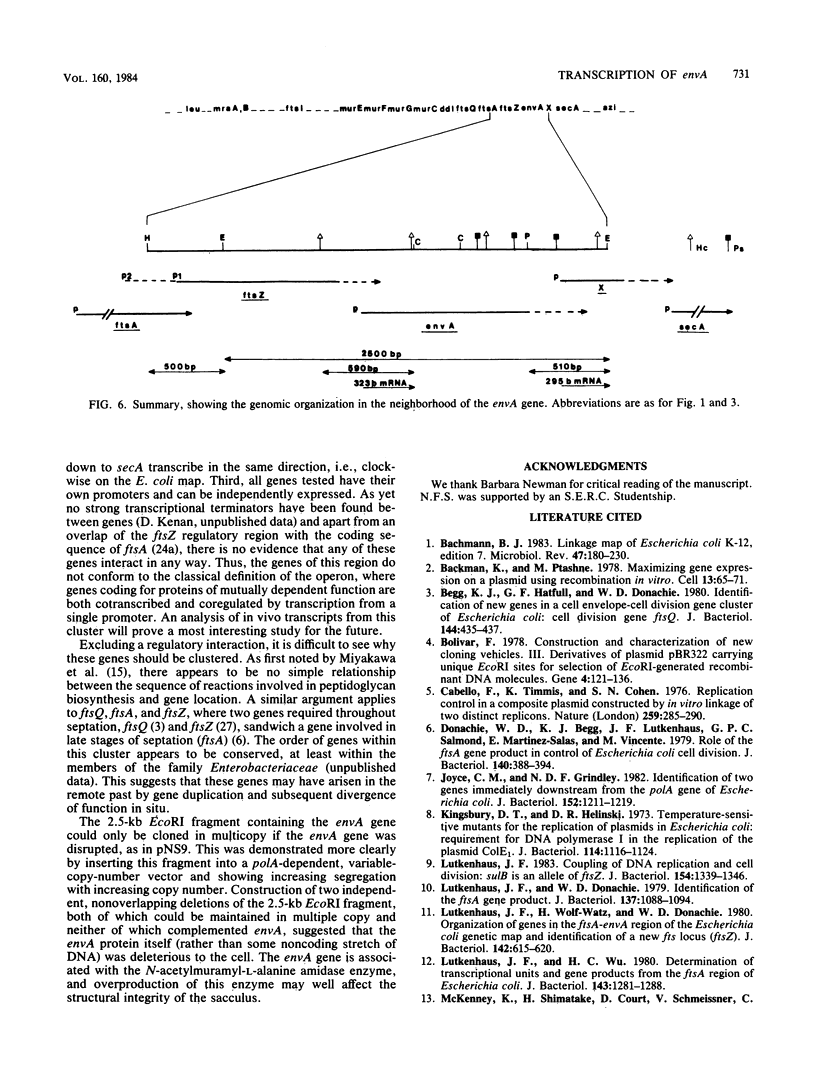
A cluster of at least 14 genes, each concerned with some aspect of cell envelope growth, morphogenesis, or function, is located at 2 min on the genetic map of Escherichia coli. We located the envA cell division gene and its promoter within the cluster and determined the direction of transcription of the gene by constructing fusions between its promoter and the galK coding sequence. In addition, we identified the promoter of a possible new gene lying between envA and the secA gene. We also present evidence from gene fusion studies which shows the direction of transcription of the ftsZ(sulB) division gene. The direction of transcription is the same for all three promoters and is the same as that of all other cluster genes for which this is known. We discuss the significance of this observation, together with the fact that every gene examined in sufficient detail within the cluster appears to have its own promoter and to be able to be expressed from isolated cloned fragments. Using a novel variable-copy plasmid vector, we demonstrate that the DNA fragment containing the envA gene is not stably maintained in multiple copies. The construction of two independent, nonoverlapping deletions allows us to conclude that the envA product itself is responsible for this effect.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backman K., Ptashne M. Maximizing gene expression on a plasmid using recombination in vitro. Cell. 1978 Jan;13(1):65–71. doi: 10.1016/0092-8674(78)90138-1. [DOI] [PubMed] [Google Scholar]
- Begg K. J., Hatfull G. F., Donachie W. D. Identification of new genes in a cell envelope-cell division gene cluster of Escherichia coli: cell division gene ftsQ. J Bacteriol. 1980 Oct;144(1):435–437. doi: 10.1128/jb.144.1.435-437.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique Eco RI sites for selection of Eco RI generated recombinant DNA molecules. Gene. 1978 Oct;4(2):121–136. doi: 10.1016/0378-1119(78)90025-2. [DOI] [PubMed] [Google Scholar]
- Cabello F., Timmis K., Cohen S. N. Replication control in a composite plasmid constructed by in vitro linkage of two distinct replicons. Nature. 1976 Jan 29;259(5541):285–290. doi: 10.1038/259285a0. [DOI] [PubMed] [Google Scholar]
- Donachie W. D., Begg K. J., Lutkenhaus J. F., Salmond G. P., Martinez-Salas E., Vincente M. Role of the ftsA gene product in control of Escherichia coli cell division. J Bacteriol. 1979 Nov;140(2):388–394. doi: 10.1128/jb.140.2.388-394.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce C. M., Grindley N. D. Identification of two genes immediately downstream from the polA gene of Escherichia coli. J Bacteriol. 1982 Dec;152(3):1211–1219. doi: 10.1128/jb.152.3.1211-1219.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury D. T., Helinski D. R. Temperature-sensitive mutants for the replication of plasmids in Escherichia coli: requirement for deoxyribonucleic acid polymerase I in the replication of the plasmid ColE 1 . J Bacteriol. 1973 Jun;114(3):1116–1124. doi: 10.1128/jb.114.3.1116-1124.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J. F. Coupling of DNA replication and cell division: sulB is an allele of ftsZ. J Bacteriol. 1983 Jun;154(3):1339–1346. doi: 10.1128/jb.154.3.1339-1346.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J. F., Donachie W. D. Identification of the ftsA gene product. J Bacteriol. 1979 Mar;137(3):1088–1094. doi: 10.1128/jb.137.3.1088-1094.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J. F., Wolf-Watz H., Donachie W. D. Organization of genes in the ftsA-envA region of the Escherichia coli genetic map and identification of a new fts locus (ftsZ). J Bacteriol. 1980 May;142(2):615–620. doi: 10.1128/jb.142.2.615-620.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J. F., Wu H. C. Determination of transcriptional units and gene products from the ftsA region of Escherichia coli. J Bacteriol. 1980 Sep;143(3):1281–1288. doi: 10.1128/jb.143.3.1281-1288.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney K., Shimatake H., Court D., Schmeissner U., Brady C., Rosenberg M. A system to study promoter and terminator signals recognized by Escherichia coli RNA polymerase. Gene Amplif Anal. 1981;2:383–415. [PubMed] [Google Scholar]
- Meacock P. A., Cohen S. N. Partitioning of bacterial plasmids during cell division: a cis-acting locus that accomplishes stable plasmid inheritance. Cell. 1980 Jun;20(2):529–542. doi: 10.1016/0092-8674(80)90639-x. [DOI] [PubMed] [Google Scholar]
- Miyakawa T., Matsuzawa H., Matsuhashi M., Sugino Y. Cell wall peptidoglycan mutants of Escherichia coli K-12: existence of two clusters of genes, mra and mrb, for cell wall peptidoglycan biosynthesis. J Bacteriol. 1972 Nov;112(2):950–958. doi: 10.1128/jb.112.2.950-958.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M., Maruyama I. N., Soma M., Kato J., Suzuki H., Horota Y. On the process of cellular division in Escherichia coli: nucleotide sequence of the gene for penicillin-binding protein 3. Mol Gen Genet. 1983;191(1):1–9. doi: 10.1007/BF00330881. [DOI] [PubMed] [Google Scholar]
- Neidhardt F. C., Vaughn V., Phillips T. A., Bloch P. L. Gene-protein index of Escherichia coli K-12. Microbiol Rev. 1983 Jun;47(2):231–284. doi: 10.1128/mr.47.2.231-284.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normark S., Boman H. G., Matsson E. Mutant of Escherichia coli with anomalous cell division and ability to decrease episomally and chromosomally mediated resistance to ampicillin and several other antibiotics. J Bacteriol. 1969 Mar;97(3):1334–1342. doi: 10.1128/jb.97.3.1334-1342.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normark S. Genetics of a chain-forming mutant of Escherichia coli. Transduction and dominance of the envA gene mediating increased penetration to some antibacterial agents. Genet Res. 1970 Aug;16(1):63–78. doi: 10.1017/s0016672300002287. [DOI] [PubMed] [Google Scholar]
- Normark S., Wolf-Watz H. Cell division and permeability of unbalanced envelope mutants of Escherichia coli K12. Ann Microbiol (Paris) 1974 Sep;125 B(2):211–226. [PubMed] [Google Scholar]
- Oliver D. B., Beckwith J. Identification of a new gene (secA) and gene product involved in the secretion of envelope proteins in Escherichia coli. J Bacteriol. 1982 May;150(2):686–691. doi: 10.1128/jb.150.2.686-691.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmond G. P., Lutkenhaus J. F., Donachie W. D. Identification of new genes in a cell envelope-cell division gene cluster of Escherichia coli: cell envelope gene murG. J Bacteriol. 1980 Oct;144(1):438–440. doi: 10.1128/jb.144.1.438-440.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan N. F., Donachie W. D. Overlapping functional units in a cell division gene cluster in Escherichia coli. J Bacteriol. 1984 Jun;158(3):1198–1201. doi: 10.1128/jb.158.3.1198-1201.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis K., Cabello F., Cohen S. N. Utilization of two distinct modes of replication by a hybrid plasmid constructed in vitro from separate replicons. Proc Natl Acad Sci U S A. 1974 Nov;71(11):4556–4560. doi: 10.1073/pnas.71.11.4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. R., Kovarik A., Allen J. S., Gustafson R. A. Regulation of bacterial cell division: temperature-sensitive mutants of Escherichia coli that are defective in septum formation. J Bacteriol. 1975 Aug;123(2):693–703. doi: 10.1128/jb.123.2.693-703.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf-Watz H., Normark S. Evidence for a role of N-acetylmuramyl-L-alanine amidase in septum separation in Escherichia coli. J Bacteriol. 1976 Nov;128(2):580–586. doi: 10.1128/jb.128.2.580-586.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heijenoort J., Parquet C., Flouret B., van Heijenoort Y. Envelope-bound N-acetylmuramyl-L-alanine amidase of Escherichia coli K 12. Purification and properties of the enzyme. Eur J Biochem. 1975 Oct 15;58(2):611–619. doi: 10.1111/j.1432-1033.1975.tb02412.x. [DOI] [PubMed] [Google Scholar]