Abstract
Cryptococcus neoformans has two varieties, var. grubii and var. neoformans, that correspond to serotypes A and D, respectively. Molecular phylogenetic analyses suggest that these two varieties have diverged from each other for ∼18 million years. The discovery of pathogenic serotype AD hybrid strains in nature indicates that intervariety mating in C. neoformans occurs in the natural environment. However, little is known about the genetic consequences of hybridization in C. neoformans. Here, we analyzed a hybrid population of 163 progeny from a cross between strains of serotypes A (CDC15) and D (JEC20), using 114 codominant nuclear PCR–RFLP markers and 1 direct PCR marker. These markers were distributed on all 14 chromosomes of the sequenced strain JEC21 that was isogenic to one of the parents (JEC20) in our cross. Our analyses identified that of the 163 progeny, 5 were heterozygous at all 115 loci, 1 was completely homozygous and identical to one of the parents (CDC15), and the remaining 157 each contained at least 1 heterozygous locus. Because all 163 progeny inherited mitochondria from the MATa parent JEC20, none of the progeny had a genotype identical to either of the two parents or to a composite of the two parents. All 115 nuclear loci showed three different genotypes in the progeny population, consistent with Mendelian segregation during meiosis. While the linkage analysis showed independent reassortment among loci on different linkage groups, there were significant differences in recombination frequencies among chromosomes and among regions within certain chromosomes. Overall, the linkage-map length from this hybrid cross was much shorter and the recombination frequency much lower than those constructed using serotype D strains, consistent with suppressed recombination in the intervariety cross between strains of serotypes A and D. We discuss the implications of our results in our understanding of the speciation and evolution of the C. neoformans species complex.
CRYPTOCOCCUS neoformans is an encapsulated basidiomycetous yeast that can infect the central nervous system to cause meningoencephalitis in immunocompromised hosts. Most of the C. neoformans strains are haploid and belong to two different serotypes, A and D, corresponding to variety grubii and variety neoformans, respectively. Molecular phylogenetic analyses have shown that var. grubii and var. neoformans have diverged from each other for ∼18.5 million years (Xu et al. 2000). Because both varieties of C. neoformans are significant opportunistic pathogens of humans and other animals, in recent years, there have been significant research activities aimed at understanding the genotypic and phenotypic differences between the varieties. However, much remains unknown. The objective of this study is to analyze the patterns of molecular-marker segregation in a hybrid cross between strains of var. grubii and var. neoformans in an effort to help improve our understanding of the genetic consequences of hybridization in this species.
Strains of C. neoformans normally grow as budding yeasts. Under certain environmental conditions, many strains can also undergo filamentous dimorphic transitions (reviewed in Alspaugh et al. 2000). C. neoformans has a defined sexual cycle with a teleomorph state called Filobasidiella neoformans. Strains of C. neoformans belong to one of two mating types (MAT) that are determined by one locus with two alternative alleles, MATa and MATα. Under suitable environmental conditions (i.e., nitrogen-limiting and low-moisture conditions), mating can occur between strains of opposite mating types. Typically, mating starts with the fusion of haploid cells of different mating types and is followed by filamentous growth of the dikaryotic cells. At the tip of the dikaryotic cells, basidia may be formed, nuclear fusion and meiosis can occur, and haploid spores are produced on the basidia (Kwon-Chung 1975, 1976).
It has been shown that strains of serotypes A and D in C. neoformans can grow and successfully mate on medium containing pigeon guano, a natural habitat for strains of these two serotypes (Staib 1981; Nielsen et al. 2007). This result suggests that mating and sexual reproduction could occur in natural environments between serotypes A and D strains. Consistent with this hypothesis, serotype AD strains have been found in natural environments and in patients (e.g., Brandt et al. 1996). These serotype AD strains are also virulent in the murine model of Cryptococcosis (Lengeler et al. 2001; Chaturvedi et al. 2002; Barchiesi et al. 2005). Indeed, gene genealogical analyses demonstrated that serotype AD strains are recent hybrids between strains of serotypes A and D and that multiple hybridization events have occurred between strains of these two serotypes (Xu et al. 2002; Xu and Mitchell 2003).
Molecular analyses have shown that most environmental and clinical strains of serotype AD are diploid or aneuploid and contain alleles typical of both serotypes A and D (Xu et al. 2000; Boekhout et al. 2001; Lengeler et al. 2001; Chaturvedi et al. 2002; Xu et al. 2002; Xu and Mitchell 2003). The abundant heterozygosity in serotype AD strains suggests that meiosis in these hybrid zygotes might be impaired, due possibly to the large genomic differences among serotypes. Indeed, significant karyotypic variations have been found in C. neoformans (Kwon-Chung et al. 1992; Wickes et al. 1994; Marra et al. 2004; Fraser et al. 2005). However, despite the medical importance of the AD hybrids and the discovery of recent natural hybridization events between serotypes A and D strains, relatively little is known about the genotypic consequences of hybridization in C. neoformans.
Here, we analyzed a hybrid progeny population generated from an intervariety cross between strains CDC15 (serotype A, MATα) and JEC20 (serotype D, MATa). These two strains differ in several phenotypic traits. For example, CDC15 is resistant to the antifungal drug Fluconazole [minimum inhibitory concentration (MIC) = 64 μg/ml] while JEC20 is not (MIC = 4 μg/ml). We obtained genotype data, 115 broadly distributed codominant molecular markers, for each of 163 progeny from this cross. The obtained hybrid linkage map was then compared to genetic linkage maps constructed from serotype D strains reported previously (Forche et al. 2000; Marra et al. 2004). The genotype and linkage map information were used to understand the patterns of marker segregation and recombination within and among chromosomes in this intervariety cross in C. neoformans.
MATERIALS AND METHODS
Mapping population:
We used a mapping population of 163 progeny, generated by a cross between C. neoformans serotype A strain CDC15 (MATα) and serotype D strain JEC20 (MATa). To construct the mapping population, parental strains were first grown on YEPD medium (1% yeast extract, 2% dextrose, 2% Bacto-peptone, 1.5% agar) at 25° for 3 days. About 108 cells from each parental strain were then thoroughly mixed on V8-juice agar medium (Xu et al. 2000). After 4 weeks of incubation at 25°, hyphal mats were visible at the edge of the mating mixture. Hyphae and basidiospores were scraped off the agar surface from the edge of the mating mixture (i.e., without any parental yeast cells), washed in sterile distilled water, diluted and spread-plated on YEPD medium. Plates were then incubated at 37° for 3 days. Well-separated single colonies without any obvious hyphal filaments from the original mating were streaked onto a new YEPD plate to obtain pure cultures. Only one colony from the second plate was picked from each original colony so as to maximize the genotypic diversity of the progeny population. DNA was extracted from each progeny as well as the parental strains according to an established procedure (Xu et al. 2000).
Codominant molecular markers and genotyping:
PCR primers were designed on the basis of the published sequence of C. neoformans strain JEC21 (Loftus et al. 2005), which is isogenic to one of the parental strains in our cross, JEC20, except at the mating-type locus (JEC21 is MATα while JEC20 is MATa). For each chromosome of the annotated JEC21 genome, starting from one terminus, genes were selected at the frequency of about one gene every 150 kb. Primers were designed for each selected gene using an online program (http://seq.yeastgenome.org/cgi-bin/web-primer). PCR primer pairs that successfully amplified the expected sized-DNA fragments in both parental strains were selected. The PCR products from the two parental strains, as well as from an equal mixture of the genome DNA of the two parental strains (the positive control for heterozygosity), were then digested separately with each of 12 restriction enzymes. For each PCR product, the enzymatic digestion that produced band patterns easily distinguishable between the two parents was chosen to further genotype the entire mapping progeny population. For primer pairs that did not work with either parental strain or failed to produce codominant enzymatic digestion patterns, we designed new primer pairs from genes that were located close to the initially selected genes to try to find suitable PCR–RFLP markers. For some regions where suitable PCR–RFLP markers could not be found after several tries, no molecular marker was included in the analysis. Overall, we tried to have at least 2 markers from each chromosome located within 200 kb from both ends of the chromosome. The only exception was chromosome 5, for which the closest marker was ∼300 kb away from one end of this chromosome. A total of 114 PCR–RFLP markers were developed for this study (Table 1).
TABLE 1.
Summary information of individual linkage groups in the intervariety cross between strains of serotypes A and D in C. neoformans
| Linkage group | Corresponding chromosome in JEC21 | No. of markers | Length (cM) | Physical distance covered by markers (kb) | Ratio between physical distance and genetic distance (kb/cM) |
|---|---|---|---|---|---|
| 1 | 1 | 13 | 38.5 | 2135 | 55.45 |
| 2 | 2 | 9 | 12.5 | 1401 | 112.08 |
| 3 | 3 | 11 | 11.8 | 1604a | 195.61b |
| 4 | 4 | 11 | 32.3 | 1612 | 49.91 |
| 5 | 5 | 9 | 13.9 | 1144 | 82.30 |
| 6 | 6 | 7 | 12.6 | 1307 | 103.73 |
| 7 | 7 | 12 | 11.5 | 1266 | 110.09 |
| 8 | 8 and 12 | 10 | 22.2 | NCc | NCc |
| 9 | 9 | 7 | 9.1 | 1146 | 125.93 |
| 10 | 10 | 7 | 7.4 | 986 | 133.24 |
| 11 | 11 | 8 | 13.6 | 948 | 69.71 |
| 12 | 13 | 6 | 8.3 | 722 | 86.99 |
| 13 | 14 | 2 | 2.2 | 177 | 80.45 |
| 14 | 14 | 2 | 0.9 | 102 | 113.33 |
| Total | 114 | 196.8 |
Marker CNC07180 was not included, because it was identified as a translocation (see results).
Genetic distance involving marker CNC07180 was not included (see reason above). Therefore, a genetic distance of 8.2, not 11.8, was used to calculate the ratio.
Not calculated, because markers in LG 8 were located on two different chromosomes in JEC21.
Genotypes at the mating-type locus (MAT) for these progeny were determined by direct PCR using the MATa- and MATα-specific PCR primers for the ste20 gene, as described previously (Lengeler et al. 2001; Yan et al. 2002).
The 163 progeny were genotyped for each of the 115 markers. For each marker, we used “1” to represent the allele (i.e., the enzymatic digestion pattern) from parent CDC15, “2” to represent the allele from JEC20, and “3” to represent the heterozygote that contains alleles from both parental strains (i.e., a composite enzymatic digestion pattern that includes DNA fragments from both parental strains). PCR amplification, enzyme digestion, gel electrophoresis, and data scoring followed those in Xu et al. (1999) and Lan and Xu (2006).
Data analysis and linkage-map construction:
Of the 163 progeny, 162 have at least one heterozygous locus (see results below), suggesting that these progeny were either diploid or aneuploid. Therefore, in our marker-segregation and linkage analysis, these progeny were treated as diploid. For each locus, the observed homozygosity for each of the two parental alleles as well as heterozygosity in the progeny population were calculated as simple ratios of the number of progeny in each genotype over the total number of analyzed progeny. The potential bias of the two parental alleles in the progeny population was examined for each locus using the χ2 goodness-of-fit test (χ2-test). The null hypothesis in these tests was that the two parental alleles in the progeny population should be in equal frequency.
Linkage and mapping analyses were performed using the MAPMAKER software, version 3.0 (Lander and Green 1987; Lander et al. 1987). To generate the linkage map, different values of LOD thresholds and maximum mapping distances (MDs) between adjacent markers were tested to determine the optimal parameters for the MAPMAKER program, using the chromosomal organization and genome sequence of JEC21 as a guide. For maximum MD between adjacent markers, we first tried 25 cM, which was similar to those that have been used previously in linkage mapping of serotype D in C. neoformans (Forche et al. 2000; Marra et al. 2004). With this cutoff, the MAPMAKER program generated a linkage map containing 14 linkage groups (LGs) that were in overall agreement with the 14 chromosomes of the published genome sequence of JEC21 (Loftus et al. 2005). Increasing MD to 30 cM or decreasing MD to 10 cM did not significantly influence the mapping results. However, when an MD of <10 cM was used, MAPMAKER generated many small linkage groups with each including only two to four markers.
To select for an optimum LOD threshold for linkage-map construction, we first used the LOD score of 5, a value used in previous mapping studies of C. neoformans (Forche et al. 2000; Marra et al. 2004). With this LOD value, we obtained a linkage map containing 10 LGs, in which 8 LGs corresponded to 8 chromosomes of JEC21 and the remaining 2 LGs corresponding to markers from multiple chromosomes, with 1 LG containing markers from 2 chromosomes and the other containing markers from 5 chromosomes of JEC21. We then increased the LOD threshold in an effort to resolve the 2 LGs with markers from multiple chromosomes. Overall, increasing the LOD score did not influence the LGs with markers from the 8 chromosomes of JEC21. However, when the LOD score was at 20, the initial LG with markers from 5 chromosomes separated into 3 LGs, with 2 of them corresponding to 2 chromosomes of JEC21 and the other containing markers from 3 different chromosomes. When the LOD score was further increased to 25, the 2 remaining LGs containing markers from multiple chromosomes (when LOD of 20 was used) were further resolved into smaller LGs that corresponded to chromosomes of JEC21. However, 1 LG still contained markers from 2 chromosomes of JEC21 (chromosome 8 and chromosome 12). Further increasing the LOD threshold to 30, divided this LG into 2 LGs with each containing markers mostly from chromosome 8 and chromosome 12, respectively. With the incremental LOD score values, several markers became unassigned to any LG. Because with the LOD of 25 and the MD of 10 cM, all the markers (except one) were assigned to LGs and the linkage map produced by MAPMAKER was overall in good agreement with the 14 chromosomes of JEC21, these two parameters were chosen to construct the linkage map.
To analyze the genotype distribution among the 163 progeny and to examine their relationships among each other and with the parents, we performed a simple cluster analysis using the unweighted pair group method with arithmetic mean (UPGMA) algorithm through the PAUP* computer software (Swofford 2002).
Confirmation of chromosomal rearrangements:
To confirm the identified putative inversions and translocations (see results below), we compared the locations and orders of the markers involved in the rearrangements in the published JEC21 genome and compared them to the locations and orders of their orthologs in the H99 genome using BLAST. Because the H99 genome sequence is still not completely assembled into chromosomes (there are 210 super contigs at present in the database), an exhaustive test of potential chromosomal rearrangements identified here is not possible at present. Instead, we designed specific primers to target regions that showed clear rearrangements between the JEC21 and H99 genomes to confirm the putative chromosome rearrangement between JEC20 and CDC15 using PCR (see below).
Comparison of recombination frequency between intervariety and intravariety crosses:
Among the 115 markers analyzed in our study, 15 were used previously in a mapping study of serotype D strains of C. neoformans (Marra et al. 2004). This sharing of markers gave us an opportunity to make a direct comparison of recombination frequency between the intervariety and the intravariety crosses of C. neoformans. To make the comparison, we first obtained the genetic distance data for each marker pair in the two studies. Genetic distances from the same study were put into one group. Then the two groups of genetic distance data were compared using the Mann–Whitney U-test (Wilcoxon rank-sum test) to determine whether recombination frequency was significantly lower in the intervariety cross than that in the intravariety cross.
RESULTS AND DISCUSSION
PCR–RFLP marker development:
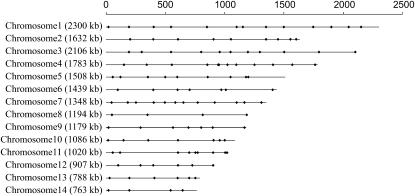
In total, we successfully obtained 114 codominant PCR–RFLP markers that easily distinguish the two parental strains as well as the heterozygote. In addition, one marker, the ste20 gene located within the mating-type region, was assayed on the basis of direct PCR using mating-type specific primers. Alleles at this locus were detected on the basis of the presence/absence of a PCR product using the MATa- and MATα-specific PCR primers. The ste20 primers were originally designed on the basis of the unique ste20 sequences at the MATa and MATα loci, respectively, by Lengler et al. (2001). Overall, these 115 markers cover all 14 chromosomes of the genome of JEC21 (Figure 1). The physical distances between 2 adjacent markers ranged from 16 to 488 kb, with an average of 169 kb (supplemental Table 1 at http://www.genetics.org/supplemental/).
Figure 1.—
Distribution of markers along the 14 chromosomes used in this study. The lengths of the chromosomes as well as the positions of the markers analyzed here on each chromosome are shown along the top bar.
Progeny genotypes:
Our progeny population of 163 was derived from single-colony isolation and not from single basidiospores dissected using a micromanipulator. Previous studies have shown that basidiospores from crosses between strains of serotypes A and D have very low viability (<10%, Lengeler et al. 2001). Therefore, to have a sufficient number of progeny for linkage-map construction and analyses, we used the alternative method of first growing single spores into colonies and then through streaking and purification to obtain pure cultures representing single spores. The size of our progeny population is similar to or larger than those used in linkage analyses in many other studies. For example, in the two studies in which genetic linkage maps of C. neoformans var. neoformans were generated, 100 and 94 progeny were analyzed, respectively (Forche et al. 2000; Marra et al. 2004). The genetic linkage map for the Oomycete plant pathogen Phytophthora infestans was constructed using 73 progeny (Van Der Lee et al. 1997). For the button mushroom Agaricus bisporus, 52 progeny were used to construct its linkage map (Kerrigan et al. 1993).
All 163 progeny inherited their mitochondrial genomes from the JEC20 parent using PCR–RFLP markers developed previously (Xu 2002; data not shown). This result is consistent with uniparental mitochondrial inheritance in C. neoformans from the MATa parent (Xu et al. 2000; Yan and Xu 2003). UPGMA cluster analysis based on genotypic data of the 115 nuclear marker loci revealed that among the 163 progeny analyzed here, there were 119 different genotypes (Figure 2). One hundred three genotypes were each represented by only 1 progeny while 2 genotypes were each represented by 10 progeny (genotypes 1 and 2). Among the remaining 14 genotypes, 1 was represented by 6 progeny (genotype 3), 1 by 5 progeny (genotype 4), 1 by 4 progeny (genotype 5), 3 by 3 progeny each, and 8 by 2 progeny each (Figure 2). Among the 163 progeny, 1 (progeny R010) had a nuclear genotype at the 115 loci identical to that of the parent strain CDC15 (but with a different mitochondrial genome—that of JEC20, see above) and 5 progeny were heterozygous (having alleles from both parents) at all 115 nuclear loci (genotype 4) screened (Figure 2). The prevalence of recombinant genotypes in our progeny population suggests widespread recombination during meiosis in this intervariety cross.
Figure 2.—
The UPGMA phenogram showing the overall genetic similarity among the 163 progeny as well as their relationships to the two parental strains CDC15 and JEC20 and their composite genotype P1P2. Genotypes represented by >3 progeny are marked to the right.
Marker segregation:
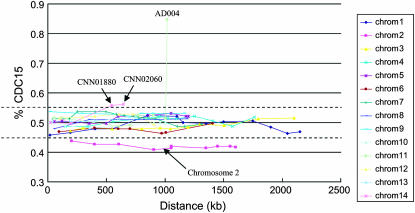
Of the 115 marker loci, 102 had allele frequencies not significantly different from 1:1 in the progeny population (χ2 < 3.84, P > 0.05) while the remaining 13 exhibited significantly skewed segregation ratios (χ2 > 3.84, P < 0.05) (Figure 3). Of these 13 loci that showed skewed allele frequencies, 10 were located on chromosome 2, with alleles from parent JEC20 significantly more prevalent in the progeny population than those from parent CDC15. Indeed, all analyzed markers located on chromosome 2 showed skewed segregations toward JEC20 (Figure 3). The remaining 3 loci were skewed toward CDC15, with 1 located on chromosome 11 and 2 on chromosome 14 (Figure 3).
Figure 3.—
Segregation ratios of the molecular markers along chromosomes. Each colored curve represents a chromosome. The x-axis shows the physical locations of the markers on the chromosome. The y-axis represents the frequency of CDC15 alleles. Dotted lines represent the boundaries for the 95% confidence interval around the null hypothesis of random segregation (1:1 ratio). Markers showing significantly skewed segregations are indicated by black arrows: all markers on chromosome 2 and two markers, CNN01880 and CNN02060, on chromosome 12.
Compared to the two genetic linkage maps constructed using serotype D strains by Forche et al. (2000) and Marra et al. (2004), the percentage of markers showing distorted segregation in our study was higher than that found in the Marra et al. study (1.7%), but lower than that found in the Forche et al. study (18.2%). In comparison to other fungal linkage mapping studies, the percentage of skewed markers found in our study was lower than most others. For example, in mapping the Agaricus bisporus genome, Kerrigan et al. (1993) found that 32.8% loci had skewed ratios. In constructing the genetic linkage map of the maize pathogen Cochliobolus heterostrophus, Tzeng et al. (1992) found segregation distortions in 15.9% of the polymorphic loci. Zhong et al. (2002) found that 19.6% of the polymorphic loci showed skewed segregation during linkage-map construction for the ascomycete plant pathogen C. sativus.
There may be two possibilities contributing to the observed segregation distortions. First, there might be different fitness properties between alleles from the two parents that could have directly influenced differential spore germination, viability, or growth rates among progeny carrying different alleles. Second, there might be tight linkage between the distorted marker loci and other loci with alleles differing in fitness. In our study, we found all the markers located on LG 2 (also chromosome 2) showed skewed allele frequencies in favor of the parent JEC20 allele, with the percentage of JEC20 alleles at each locus ranging between 56.1 and 59.2%. At present, we do not know the genetic basis for this observed chromosomewide segregation distortion. A similar phenomenon was reported by Jurgenson et al. (2002) in their linkage-mapping study of the maize fungal pathogen Gibberella zeae.
Among the 115 nuclear loci, the heterozygosity (the percentage of heterozygotes in the progeny population of 163) at individual loci ranged between 25.16 and 88.34%. The percentage of homozygotes with allele from parent CDC15 ranged between 3.68 and 72.26% among the loci. The percentage of homozygotes with the JEC20 allele ranged between 2.58 and 26.38%. When marker loci with skewed segregation ratios were excluded, the heterozygosity at these loci ranged between 49.69 and 88.34%, the percentage of homozygotes with the CDC15 allele ranged between 6.75 and 25.15%, and the percentage of homozygotes with the JEC20 allele ranged between 3.07 and 26.38% (supplemental Table 1 at http://www.genetics.org/supplemental/).
Interestingly, when heterozygosity was mapped onto the linkage map and compared to its chromosomal organizations, in 13 of the 14 chromosomes, the lowest heterozygosity was found closest to the proposed centromere regions in the published genome sequence of JEC21 (Loftus et al. 2005, supplemental Figure 1 at http://www.genetics.org/supplemental/). Lower levels of heterozygosity around these markers suggest higher levels of recombination frequency in the regions around them. The mechanism for such a low level heterozygosity surrounding the centromeres is not clear.
Linkage groups:
Linkage analysis was performed using MAPMAKER with a LOD threshold of 25 and maximum distances between adjacent markers of 10 cM (see materials and methods). Among the 115 markers, 114 were assigned to LGs. The only exception was marker AD004, which showed highly skewed segregation toward the allele from parent CDC15 (χ2 = 150.5, P < 0.0005). Marker AD004 was then excluded from further linkage analyses. These remaining 114 markers were assigned to 14 LGs, with 12 LGs having at least 6 markers each and the other 2 LGs having 2 markers each (Table 1). Eleven of the 14 LGs corresponded to 11 chromosomes of JEC21. Overall, the orders of marker loci in these 11 LGs were congruent with their physical locations in each chromosome, except a few putative inversions and translocations with each involving one to three marker loci (Figure 4 and supplemental Figure 1; see below). Our result is consistent with the physical mapping results by Schein et al. (2002) in which they compared physical maps of C. neoformans serotype A and D strains on the basis of restriction site mapping and Southern hybridizations. They found a high degree of conservation of synteny between strains JEC21 (serotype D) and H99 (serotype A) as well as some chromosomal rearrangements including both inversions and translocations.
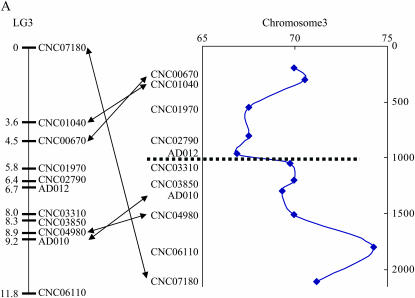
Figure 4.—
A representative linkage group and the confirmation of a translocation breakpoint in this linkage group constructed from an intervariety cross in C. neoformans. (A) LG 3 of the hybrid genetic linkage map of C. neoformans. Marker names are indicated on the right of the linkage group. Numbers on the left are genetic distances in centimorgans. The diagram on the right of the linkage group is the distributions of heterozygosity at each marker locus located on LG 3, with the marker orders following their physical locations in chromosome 3 of JEC21. The x-axis shows the level of heterozygosity and the y-axis shows the physical distance from one end of chromosome 3. Arrows indicate markers with positions in the linkage groups different from their physical locations in chromosomes 3 of JEC21 (for putative inversions and translocations, see results). Dotted lines indicate the approximate locations of the candidate centromeric region in chromosome 3 of JEC21 as identified by Loftus et al. (2005). (B) The locations of primers used to confirm the putative translocation of marker CNC07180 in LG 3. Arrows indicate the positions and directions of primers in chromosome 3 of strain JEC21 and contig 32.1 of strain H99. (C) Gel electrophoresis of PCR products amplified from different strains using different primer pairs to detect translocation. The strain names are indicated at the top of each lane. The primer pairs used for PCR are indicated at the bottom. Primer sequences are listed in Table 2.
Our LG 8 contained markers from both chromosomes 8 and 12 of the JEC21 genome. Increasing LOD threshold to 30 separated LG 8 into two LGs: one LG (LG 8-A) contained three markers from chromosome 8 only and the other LG (LG 8-B) contained all the markers from chromosome 12 except one (marker CNH03700) from chromosome 8 (supplemental Figure 1 at http://www.genetics.org/supplemental/). It has been reported by Fraser et al. (2005) that chromosome 8 and chromosome 12 in JEC21 originated through the breakage of a dicentric chromosome resulting from telomere–telomere fusion of two chromosomes from strain B3501A. During this process, a chromosomal translocation occurred and as a result, chromosome 8 and chromosome 12 of JEC21 shared a large segmental duplication spanning ∼62 kb and containing 22 predicted genes. In our study, marker CNH03700 was located in this duplicated region. When we increased the LOD threshold to 30, marker CNH03700 could be unambiguously assigned to a LG in which all the other markers were from chromosome 12 (supplemental Figure 1). In our linkage map, CNH3700 was separated by 1.6 cM from another marker, CNL06830, which was identified as a putative translocation (see below). These two markers were well separated from other markers by 5.8 and 5.4 cM, respectively, from their flanking regions (supplemental Figure 1). When these two markers were excluded in the linkage analysis, the remaining markers from chromosome 8 and chromosome 12 were assigned to two distinct LGs with each containing only markers from the same chromosome at LOD threshold 25 (data not shown). On the basis of these analyses, we can conclude that the assignment of markers from chromosomes 8 and 12 into one LG was due to misalignments of the duplicated regions between these two chromosomes during meiosis.
The four markers from chromosome 14 of JEC21 were assigned to two LGs (LG 13 and LG 14) in our hybrid map (supplemental Figure 1). We experimented with different LOD thresholds to as low as 5. However, these four markers were still not assigned to the same LG. Instead, when the LOD threshold was lower than 25, two markers from LG 13 (markers CNN00060 and CNN00590) were assigned to LG 7, just like other markers from chromosome 7. These results might be due to (1) different karyotypes between the two parental strains, e.g., chromosome 14 of the JEC20 parent might correspond to two small chromosomes in parent CDC15, or (2) part of chromosome 14 in JEC20 was translocated to chromosome 7 in CDC15. Karyotypic variability within and between serotypes has been observed in C. neoformans (Kwon-Chung et al. 1992; Wickes et al. 1994; Marra et al. 2004; Fraser et al. 2005) and in other fungi, including Ustilago maydis (Kinscherf and Leong 1988), Saccharomyces cerevisiae (Ono and Ishinoarao 1988), and Candida species (Magee and Magee 1987). More markers and/or better knowledge of the chromosomal organizations for strain CDC15 might help distinguish these possibilities.
Chromosomal rearrangements:
We compared the mapping orders of marker loci on each LG with their physical locations on each chromosome in JEC21 to examine potential incongruence between the two serotypes and to infer potential chromosomal rearrangements such as inversions and translocations. If the markers on the same chromosome showed different orders between the physical map of JEC21 and their hybrid linkage map, an inversion might have occurred between the two parental genomes. For example, if the marker order on a JEC21 chromosome was A-B-C-D-E-F and we found the mapping order in this LG was A-B-E-D-C-F, we infer that there might have been one inversion including three markers C, D, and E. Similarly, translocations can be inferred within and between chromosomes when LGs and chromosomes show blocks of incongruent patterns. For example, if the physical order on the chromosome of JEC21 was A-B-C-D-E-F and we found the mapping order in the LG was A-D-E-B-C-F, there was likely one translocation between the two parents involving chromosomal regions that included markers D-E with that of B-C.
Overall, the orders of markers on each LG matched well to their physical locations on the chromosomes in JEC21. However, there were a few exceptions, likely involving putative inversions and translocations between the two parents. Specifically, eight LGs (LG 1, LG 2, LG 4, LG 5, LG 6, LG 10, LG 11, and LG 12) had the orders of marker loci identical to the physical locations of these markers in JEC21. LG 13 and LG 14 have only two markers each and their comparisons are thus meaningless. Among the other four LGs, LG 7 had one putative inversion involving two markers (AD019–CNG02290). LG 3 had two putative inversions with each involving two markers (CNC00670–CNC01040 and AD010–CNC04980). In addition, LG 3 had a putative rearrangement that involved one marker (CNC07180). LG 9 had one putative inversion that involved three markers (CNI02950–CNI03300–CNI04370). LG 8 had markers from both chromosomes 8 and 12. Comparing the marker order in LG 8 to the linear aligned order of these markers on the two chromosomes, we found one putative inversion that involved two markers (CNL04620–CNL04980) as well as one putative translocation that involved one marker (CNL06830) (Figure 4A; supplemental Figure 1 at http://www.genetics.org/supplemental/).
Confirmation of chromosomal rearrangements:
To confirm the aforementioned putative translocations and inversions, we checked the locations of the markers involved in those putative translocations and inversions in the genome databases of JEC21 and H99 to see if their orders in the two genomes are congruent. Because the H99 genome-sequence assembly is not finished, we could only compare markers that were located in the same contig in the H99 genome. Of the aforementioned seven putative inversions and translocations, two inversions (chromosome 3, CNC00670-CNC01040 and chromosome 9, CNI02950-CNI03300-CNI04370) had markers that were located on different contigs in H99; three inversions (chromosome 3, AD010-CNC04980; chromosome 7, AD019-CNG02290; chromosome 8, CNL04620-CNL04980) had the same marker orders in both the JEC21 and H99 genomes. Although we could not find definitive evidence for marker-order incongruence of the putative inversions between the JEC21 and H99 genomes, the orders of marker loci in the CDC15 genome might still be different from those of JEC20 and H99. The putative rearrangements inferred here are supported by the observation that even though the genetic distances between marker pairs involved in those putative inversions were relatively low (highest 0.9 cM and average 0.5 cM; supplemental Figure 1 at http://www.genetics.org/supplemental/), the ratio between physical distance and genetic distance was much higher than the overall average (lowest 119 kb/cM, highest 687 kb/cM, and average 367 kb/cM; see below). These results are consistent with the hypothesis that inverted regions typically experience low levels of recombination (see below).
Blast analysis of the sequence of CNL06830 against the genome sequences of JEC21 and H99 showed that this putative translocated marker had two copies in the JEC21 genome that were located on both chromosome 8 (∼170 kb away from CNH03700) and chromosome 12, respectively, but only one copy in contig 91.1 of the H99 genome. The duplicated region includes CNL06830 and spanned ∼10 kb. It has been shown in S. cerevisiae that ectopic recombination between artificial repeats of 5.5 kb in size could compete efficiently with normal allelic crossovers during meiosis (Jinks-Robertson et al. 1997). Therefore, the putative translocation of CNL06830 identified in our study could be the result of misalignment and ectopic recombination between the duplicated regions during meiosis.
Our comparison of the H99 and JEC21 genomes indicated that marker CNC07180 in H99 (chromosome 3, a putative translocation) might be located in a different chromosome region but is close to marker CNC01040 in the linkage map (Figure 4, A and B). We therefore attempted to identify the possible breakpoints of the translocation between the two genomes. To test whether such breakpoints exist among strains JEC20, H99, and CDC15, we designed PCR primers around these breakpoints on the basis of the JEC21 genome sequence. Indeed, the translocation was confirmed using PCR (Table 2; Figure 4, B and C). Specifically, primer pairs P1/P2 and P3/P4 were separated from each other on chromosome 3 of JEC21 by more than 1.4 Mb. Both primer pairs could amplify PCR products with expected sizes from JEC20 and JEC21 but not from H99 and CDC15. Using primer pair P2/P4, which comprises one primer from each of the two primer pairs, we successfully amplified PCR products with expected size from both H99 and CDC15 but not from JEC20 and JEC21, consistent with a chromosomal rearrangement between JEC20 (also JEC21) and CDC15 (also H99) (Figure 4C).
TABLE 2.
Primers used for the confirmation of a putative rearrangement inferred from linkage mapping analysis of the intervariety cross
| Primer | Primer sequence (5′–3′) |
|---|---|
| P1 | AACCCTCGGTCCCCCAATTA |
| P2 | TTTTATTTCCGGGCCTTTCGG |
| P3 | AAGCAAGGAGCAAGAGGCGA |
| P4 | GGCAATATTATGCAGAAGAG |
Analysis of recombination frequency:
The total length of this linkage map was 196.8 cM (Table 2). The largest LG was LG 1, with 38.5 cM in length. The smallest LG was LG 14, which was only 0.9 cM in length (Table 1). To calculate the ratio between genetic distance and physical distance as identified on the basis of the JEC21 genome sequence, we included only markers that showed clear associations between chromosomes in JEC21 and linkage groups (supplemental Figure 1 at http://www.genetics.org/supplemental/). In total, 103 of the 114 markers fit this criterion. The remaining 11 markers were excluded with 10 from LG 8 because they were from two different chromosomes and 1 (marker CNC07180) from LG 3 because it was identified as a translocation. The 103 markers covered a total physical distance of 14550 kb, ∼76% of the whole genome. The total map length covering these 103 markers was 171 cM, corresponding to 1 cM for every 85 kb.
The ratio of linkage-map distance in this hybrid cross over physical distance derived from the JEC21 genome was highly variable among chromosomes and among regions within certain chromosomes. When the map length of each individual LG was compared to the physical distance covered by its markers on chromosomes of JEC21, the ratio between physical distance and genetic distance ranged between 49.91 kb/cM (LG 4) and 195.61 kb/cM (LG 3) among the chromosomes (Table 1). When each individual marker pair was considered, the ratio between physical distance and genetic distance ranged between 13 kb/cM (LG 6, AD018–CNF03420) and 686 kb/cM (LG 6, CNF01350–CNF02070). Twelve pairs of loci showed no recombination in this cross (i.e., their pairwise genetic distance was 0). In contrast, their physical distances ranged between 20 and 149 kb in the JEC21 genome.
Comparison between the intervariety hybrid cross and a previously published intravariety cross:
Compared to the linkage maps constructed using C. neoformans serotype D strains, the total length of our hybrid linkage map (197 cM) was much shorter and the ratio between physical distance and genetic distance was much higher (85 kb/cM). For example, the linkage map constructed by Forche et al. (2000) had a total length of 1356.3 cM, corresponding to 13.6 kb/cM. The genetic map constructed by Marra et al. (2004) had a total length of ∼1500 cM, corresponding to ∼13.2 kb/cM across the genome. Fifteen PCR–RFLP markers used in the Marra et al. (2004) study were also included in our study and we compared the genetic distances between these markers in these two studies (Table 3). Only marker pairs located on the same chromosomes were compared. Of the nine marker pairs that could be compared between the two studies, one showed a smaller genetic distance in the serotype D linkage map than that in the hybrid map, a second pair showed similar genetic distances between these two maps, and the remaining seven pairs showed much greater genetic distances in the serotype D linkage map (Table 3). The differences in the seven pairs ranged from 3-fold to >30-fold. The combined analyses of all nine pairs identified that the genetic distance in the serotype D cross was significantly higher than that in the hybrid cross (Mann–Whitney U-test, P < 0.05). Because the genetic distance is a function of recombination frequency between markers, this result suggested that meiotic recombination occurred at a much higher frequency in the intraserotype D cross than in the intervariety cross.
TABLE 3.
Comparison of genetic distances for the same marker pairs between the intervariety hybridization analyzed here and the intravariety cross reported in Marra et al. (2004)
| Genetic distance
|
|||
|---|---|---|---|
| Marker pairsa | Chromosome | Intravarietyb | Intervarietyc |
| AD012 (Xba11) AD010 (Eco4) | 3 | 46.8 | 2.5 |
| AD029 (Stu4) AD030 (Hind8) | 4 | 2.2 | 2.2 |
| AD014 (Eco29) AD028 (Stu3) | 5 | 30d | 13.3 |
| AD021 (Hind21) AD020 (Hind17) | 7 | 59.4 | 3.8 |
| AD020 (Hind17) AD019 (Eco30) | 7 | 1.9 | 2.9 |
| AD019 (Eco30) AD026 (Xba21) | 7 | 70d | 4.2 |
| AD005 (Hind7) AD006 (Xho5) | 9 | 53.7 | 1.6 |
| AD001 (Eco21) AD002 (Pst28) | 11 | 49.5 | 5.8 |
| AD002 (Pst28) AD003 (Xho18) | 11 | 21.3 | 6.5 |
Marker names in parentheses were used in the serotype D mapping study (Marra et al. 2004).
Genetic distance calculated in serotype D mapping study (Marra et al. 2004).
Genetic distance calculated in this study.
This pair of markers was located on different LGs in the serotype D mapping study (Marra et al. 2004). Here the minimum possible genetic distance between this pair was derived by adding the shorter distances of each marker to the end of its LG and then adding 30 cM, the threshold swept radius used in the Marra et al. study.
The shorter genome map length and much higher physical distance to genetic distance ratios for the intervariety cross observed in our study indicated that the recombination occurred at a much lower frequency in intervariety crosses than in intravariety crosses of C. neoformans. The low level of recombination is expected during hybridization between divergent populations. Indeed, they have been considered as a driving force for speciation. Besides those aforementioned putative inversions and translocations that might have repressed recombination in those regions, several other mechanisms could also contribute to the observed low level of recombination frequency in the intervariety cross of C. neoformans. First, the DNA sequence divergence between serotype A and serotype D C. neoformans might have decreased the efficiency of homologous recombination. Recombination suppression could be achieved through the activities of mismatch repair proteins, such as Pms1 and Msh2 (Hunter et al. 1996). These genes are eukaryotic homologs of the bacterial MutS protein that have been shown to play a significant role in recombination and speciation. In S. cerevisiae, pms1 and msh6 mutants showed an increased level of mismatch binding and meiotic recombination during meiosis (Chambers et al. 1996; Hess et al. 2002). It has also been suggested that the reproductive isolation between species in Saccharomyces sensu stricto is mainly due to the mismatch repair system (Hunter et al. 1996; Greig et al. 2002; Liti et al. 2006). Genetic studies of homeologous recombination in yeast suggested that if sequences were too divergent (>10%), recombination in these regions was severely repressed, presumably due to the inability to form sufficiently stable base-paired intermediates (Selva et al. 1995; Datta et al. 1996, 1997; Chen and Jinks-Robertson 1999; reviewed in Evans and Alani 2000). Data collected in C. neoformans was consistent with this explanation. For example, sequence analyses of C. neoformans revealed that sequence divergences between serotype A and serotype D were more than 10-fold higher than those within serotype A and serotype D, up to >10% in certain regions (Xu et al. 2000). Second, theoretical studies have suggested that “recombination modifiers” that control the recombination frequency might exist. During the divergence between the two varieties, recombination modifiers from different varieties might be less or not at all compatible with each other. As a result, recombination frequencies decrease. A third possibility is the existence of strong epistasis among alleles within each of the two serotypes and these alleles can no longer segregate randomly. These three mechanisms are not mutually exclusive and it is very likely that all three could have contributed to the observed low recombination frequency in the intervariety cross of C. neoformans.
Although the overall recombination frequency observed in our study was very low, several regions showed comparable levels of recombination to the intravariety crosses of C. neoformans. For example, two regions, AD018–CNF03420 in chromosome 6 and CNK02410–CNK02590 in chromosome 11, had physical/genetic distance ratios of 13.09 and 14.17 kb/cM, respectively, similar to those found by Marra et al. (2004) and Forche et al. (2000). It has been shown that in C. neoformans, the two regions flanking the mating-type locus were recombination hotspots (Hsueh et al. 2006). In our study, one of these two regions had a physical/genetic distance ratio of 26.6 kb/cM, about three times lower than that of the genome average.
Conclusion:
In this study, we constructed and analyzed a hybrid linkage map containing 115 codominant markers using 163 progeny collected from an intervariety cross between strain CDC15 (var. grubii, serotype A, MATα) and strain JEC20 (var. neoformans, serotype D, MATa) in the important human fungal pathogen C. neoformans. Overall, the marker positions were highly similar and syntenic to those inferred from the serotype D cross and to the physical positions in the genome of strain JEC21. While our analysis identified Mendelian segregation and independent reassortment among markers during meiosis, the genetic distances between markers in the hybrid cross were much smaller than those in the serotype D cross, indicating suppressed recombination in the intervarietal cross. The low recombination frequency coupled with the presence of heterozygotes at all loci in the progeny population suggests abnormal/incomplete nuclear disjunctions during meiosis. The constructed linkage map should help the genetic analyses of divergent phenotypic traits between these two varieties.
Acknowledgments
This research is supported by the Natural Science and Engineering Research Council (NSERC) of Canada and by the Premier's Research Excellence Award. Sheng Sun has been supported by an Ontario graduate scholarship and a postgraduate scholarship from NSERC.
References
- Alspaugh, J. A., R. C. Davidson and J. Heitman, 2000. Morphogenesis of Cryptococcus neoformans, pp. 217–238 in Dimorphism in Human Pathogenic and Apathogenic Yeasts, edited by J. F. Ernst and A. Schmidt. Karger, Basel, Switzerland. [DOI] [PubMed]
- Barchiesi, F., M. Cogliati, M. C. Esposto, E. Spreghini, A. M. Schimizzi et al., 2005. Comparative analysis of pathogenicity of Cryptococcus neoformans serotypes A, D and AD in murine cryptococcosis. J. Infect. 51: 10–16. [DOI] [PubMed] [Google Scholar]
- Boekhout, T., B. Theelen, M. Diaz, J. W. Fell, W. C. J. Hop et al., 2001. Hybrid genotypes in the pathogenic yeast Cryptococcus neoformans. Microbiology 147: 891–907. [DOI] [PubMed] [Google Scholar]
- Brandt, M. E., L. C. Hutwagner, L. A. Klug, W. S. Baughman, D. Rimland et al., 1996. Molecular subtype distribution of Cryptococcus neoformans in four areas of the United States. Cryptococcal Disease Active Surveillance Group. J. Clin. Microbiol. 34: 912–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers, S. R., N. Hunter, E. J. Louis and R. H. Borts, 1996. The mismatch repair system reduces meiotic homeologous recombination and stimulates recombination-dependent chromosome loss. Mol. Cell. Biol. 16: 6110–6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi, V., J. Fan, B. Stein, M. J. Behr, W. A. Samsonoff et al., 2002. Molecular genetic analyses of mating pheromones reveal intervariety mating or hybridization in Cryptococcus neoformans. Infect. Immun. 70: 5225–5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W., and S. Jinks-Robertson, 1999. The role of the mismatch repair machinery in regulating mitotic and meiotic recombination between diverged sequences in yeast. Genetics 151: 1299–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta, A., A. Adjiri, L. New, G. F. Crouse and S. Jinks-Robertson, 1996. Crossovers between diverged sequences are regulated by mismatch repair proteins in yeast. Mol. Cell. Biol. 16: 1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta, A., M. Hendrix, M. Lipsitch and S. Jinks-Robertson, 1997. Dual roles for DNA sequence identity and the mismatch repair system in the regulation of mitotic crossing-over in yeast. Proc. Natl. Acad. Sci. USA 94: 9757–9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, E., and E. Alani, 2000. Roles for mismatch repair factors in regulating genetic recombination. Mol. Cell. Biol. 20: 7839–7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche, A., J. Xu, R. Vilgalys and T. G. Mitchell, 2000. Development and characterization of a genetic linkage map of Cryptococcus neoformans var. neoformans using amplified fragment length polymorphisms and other markers. Fungal Genet. Biol. 31: 189–203. [DOI] [PubMed] [Google Scholar]
- Fraser, J. A., J. C. Huang, R. Pukkila-Worley, J. A. Alspaugh, T. G. Mitchell et al., 2005. Chromosomal translocation and segmental duplication in Cryptococcus neoformans. Eukaryot. Cell 4: 401–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig, D., R. H. Borts, E. J. Louis and M. Travisano, 2002. Epistasis and hybrid sterility in Saccharomyces. Proc. R. Soc. Lond. Ser B 269: 1167–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess, M. T., R. D. Gupta and R. D. Kolodner, 2002. Dominant Saccharomyces cerevisiae msh6 mutations cause increased mispair binding and decreased dissociation from mispairs by Msh2-Msh6 in the presence of ATP. J. Biol. Chem. 277: 25545–25553. [DOI] [PubMed] [Google Scholar]
- Hsueh, Y., A. Idnurm and J. Heitman, 2006. Recombination hotspots flank the Cryptococcus mating-type locus: implications for the evolution of a fungal sex chromosome. PloS Genet. 2(11): e184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, N., S. R. Chambers, E. J. Louis and R. H. Borts, 1996. The mismatch repair system contributes to meiotic sterility in an interspecific yeast hybrid. EMBO J. 15: 1726–1733. [PMC free article] [PubMed] [Google Scholar]
- Jinks-Robertson, S., S. Shariq and T. Murphy, 1997. Meiotic crossing over between nonhomologous chromosomes affects chromosome segregation in yeast. Genetics 146: 69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgenson, J. E., R. L. Bowden, K. A. Zeller, J. F. Leslie, N. J. Alexander et al., 2002. A genetic map of Gibberella zeae (Fusarium graminearum). Genetics 160: 1451–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrigan, R. W., J. C. Royer, L. M. Baller, Y. Kohli, P. A. Horgen et al., 1993. Meiotic behaviour and linkage relationships in the secondarily homothallic fungus Agaricus bisporus. Genetics 133: 225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinscherf, T. G., and S. A. Leong, 1988. Molecular analysis of the karyotype of Ustilago maydis. Chromosoma 96: 427–433. [DOI] [PubMed] [Google Scholar]
- Kwon-Chung, K. J., 1975. A new genus, Filobasidiella, the perfect state of Cryptococcus neoformans. Mycologia 67: 1197–1200. [PubMed] [Google Scholar]
- Kwon-Chung, K. J., 1976. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia 68: 821–823. [PubMed] [Google Scholar]
- Kwon-Chung, K. J., J. C. Edman and B. L. Wickes, 1992. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 60: 602–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, L., and J. Xu, 2006. Multiple gene genealogical analyses suggest divergence and recent clonal dispersal in the opportunistic human pathogen Candida guilliermondii. Microbiology. 152: 1539–1549. [DOI] [PubMed] [Google Scholar]
- Lander, E. S., and P. Green, 1987. Construction of multilocus genetic linkage maps in humans. Proc. Natl. Acad. Sci. USA 84: 2363–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E. S., P. Green, A. Abrahamson, A. Barlow and M. J. Daly, 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181. [DOI] [PubMed] [Google Scholar]
- Lengeler, K. B., G. M. Cox and J. Heitman, 2001. Serotype AD strains of Cryptococcus neoformans are diploid or aneuploid and are heterozygous at the mating-type locus. Infect. Immun. 69: 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti, G., D. B. H. Barton and E. J. Louis, 2006. Sequence diversity, reproductive isolation, and species concepts in Saccharomyces. Genetics 174: 839–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus, B. J., E. Fung, P. Roncaglia, D. Rowley, P. Amedeo et al., 2005. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 307: 1321–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee, B. B., and P. T. Magee, 1987. Electrophoretic karyotypes and chromosome numbers in Candida species. J. Gen. Microbiol. 133: 425–430. [DOI] [PubMed] [Google Scholar]
- Marra, R. E., J. C. Huang, E. Fung, K. Nielsen, J. Heitman et al., 2004. A genetic linkage map of Cryptococcus neoformans variety neoformans serotype D (Filobasidiella neoformans). Genetics 167: 619–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, K., A. L. De Obaldia and J. Heitman, 2007. Cryptococcus neoformans mates on pigeon guano: implications for the realized ecological niche and globalization. Eukaryot. Cell 6: 949–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono, B., and Y. Ishinoarao, 1988. Inheritance of chromosome length polymorphisms in Saccharomyces cerevisiae. Curr. Genet. 14: 413–418. [DOI] [PubMed] [Google Scholar]
- Schein, J. E., K. L. Tangen, R. Chiu, H. Shin, K. B. Lengeler et al., 2002. Physical maps for genome analysis of serotype A and D strains of the fungal pathogen Cryptococcus neoformans. Genome Res. 12: 1445–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selva, E. M., L. New, G. F. Crouse and R. S. Lahue, 1995. Mismatch correction acts as a barrier to homeologous recombination in Saccharomyces cerevisiae. Genetics 139: 1175–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staib, F., 1981. The perfect state of Cryptococcus neoformans, Filobasidiella neoformans, on pigeon manure filtrate agar. Zentralbl. Bakteriol. A 248: 575–578. [PubMed] [Google Scholar]
- Swofford, D. L., 2002. PAUP*: Phylogenetic Analysis Using Parsimony. Sinauer Associates, Sunderland, MA.
- Tzeng, T. H., L. K. Lyngholm, C. F. Ford and C. R. Bronson, 1992. A restriction fragment length polymorphism map and electrophoretic karyotype of the fungal maize pathogen Cochliobolus heterostrophus. Genetics 130: 81–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Lee, T., I. Dewitte, A. Drenth, C. Alfonso and F. Govers, 1997. AFLP linkage map of the Oomycete Phytophthora infestans. Fungal Genet. Biol. 21: 278–291. [DOI] [PubMed] [Google Scholar]
- Wickes, B. L., T. D. E. Moore and K. J. Kwon-Chung, 1994. Comparison of the electrophoretic karyotypes and chromosomal location of ten genes in the two varieties of Cryptococcus neoformans. Microbiology 140: 543–550. [DOI] [PubMed] [Google Scholar]
- Xu, J., 2002. Mitochondrial DNA polymorphisms in the human pathogenic fungus Cryptococcus neoformans. Curr. Genet. 41: 43–47. [DOI] [PubMed] [Google Scholar]
- Xu, J., T. G. Mitchell and R. Vilgalys, 1999. PCR-RFLP analyses reveal both extensive clonality and local genetic differentiation in Candida albicans. Mol. Ecol. 8: 59–74. [DOI] [PubMed] [Google Scholar]
- Xu, J., R. J. Vilgalys and T. G. Mitchell, 2000. Multiple gene genealogies reveal recent dispersion and hybridization in the human pathogenic fungus Cryptococcus neoformans. Mol. Ecol. 9: 1471–1481. [DOI] [PubMed] [Google Scholar]
- Xu, J., G. Luo, R. J. Vilgalys, M. E. Brandt and T. G. Mitchell, 2002. Multiple origins of hybrid strains of Cryptococcus neoformans with serotype AD. Microbiology 148: 203–212. [DOI] [PubMed] [Google Scholar]
- Xu, J., and T. G. Mitchell, 2003. Comparative gene genealogical analyses of strains of AD identify recombination in populations of serotypes A and D in the human pathogenic yeast Cryptococcus neoformans. Microbiology 149: 2147–2154. [DOI] [PubMed] [Google Scholar]
- Yan, Z., X. Li and J. Xu, 2002. Geographic distribution of mating type alleles of Cryptococcus neoformans in four areas of the United States. J. Clin. Microbiol. 40: 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Z., and J. Xu, 2003. Mitochondria are inherited from the MATa parent in crosses of the basidiomycete fungus Cryptococcus neoformans. Genetics 163: 1315–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, S., B. J. Steffenson, J. P. Martinez and L. M. Ciufetti, 2002. A molecular genetic map and electrophoretic karyotype of the plant pathogenic fungus Cochliobolus sativus. Mol. Plant-Microbe Interact. 15: 481–492. [DOI] [PubMed] [Google Scholar]