Abstract
A cytogenetic FISH map of maize pachytene-stage chromosome 9 was produced with 32 maize marker-selected sorghum BACs as probes. The genetically mapped markers used are distributed along the linkage maps at an average spacing of 5 cM. Each locus was mapped by means of multicolor direct FISH with a fluorescently labeled probe mix containing a whole-chromosome paint, a single sorghum BAC clone, and the centromeric sequence, CentC. A maize-chromosome-addition line of oat was used for bright unambiguous identification of the maize 9 fiber within pachytene chromosome spreads. The locations of the sorghum BAC–FISH signals were determined, and each new cytogenetic locus was assigned a centiMcClintock position on the short (9S) or long (9L) arm. Nearly all of the markers appeared in the same order on linkage and cytogenetic maps but at different relative positions on the two. The CentC FISH signal was localized between cdo17 (at 9L.03) and tda66 (at 9S.03). Several regions of genome hyperexpansion on maize chromosome 9 were found by comparative analysis of relative marker spacing in maize and sorghum. This transgenomic cytogenetic FISH map creates anchors between various maps of maize and sorghum and creates additional tools and information for understanding the structure and evolution of the maize genome.
THE genome of maize (Zea mays L.) has been studied as a model for eukaryotic genetics, cereal crops, and monocot genome evolution (Chandler and Brendel 2002), but its size and organizational complexity complicate resolution of its structure. The presence of large gene-poor areas, segmental duplications, abundant retrotransposons, and microvariation among lines of maize all confound efforts to develop a fully assembled physical map of the entire maize genome (Kumar and Bennetzen 1999; Gaut et al. 2000; Meyers et al. 2001; Yuan et al. 2003; Messing et al. 2004; Swigonova et al. 2004; Paterson et al. 2005). Despite these complexities, several different kinds of maps have been developed to characterize its structure and function at the DNA and chromosome levels. Many linkage maps have been developed, including those based on mutant phenotypes (Emerson et al. 1935) and more recently those that include thousands of additional molecular markers such as restriction fragment length polymorphisms (RFLPs), simple sequence repeats (SSRs), single-nucleotide polymorphisms (SNPs), and insertion–deletion polymorphisms (indels) (Helentjaris et al. 1986; Coe et al. 1987; Burr et al. 1988; Causse et al. 1996; Senior and Heun 1993; Taramino and Tingey 1996; Harushima et al. 1998; Davis et al. 1999; Lee et al. 2002; Sharopova et al. 2002; Bowers et al. 2003; Fu et al. 2006). Physical maps of overlapping clones have been produced and anchored to the linkage map by means of molecular probes (De Jong et al. 1999; Bennetzen et al. 2001; Chandler and Brendel 2002; Gardiner et al. 2004; Messing et al. 2004; Bowers et al. 2005; Song et al. 2005; Hass-Jacobus et al. 2006). Another type of physical map is the cytological map produced by direct microscopic inspection of the chromosomes that make up the nuclear genome. Cytogenetic maps are valuable because they can place genetic loci directly within the entire chromosome, the ultimate contig, providing information on the location, order, and distribution of DNA sequences in relation to other genetic markers along the chromosomes (Sadder et al. 2000; Anderson et al. 2004; Kim et al. 2005a,b). In contrast to those of the well-developed linkage maps and clone- or sequence-based physical maps, the construction of high-density cytogenetic maps is nascent and relatively underdeveloped.
Cytological analysis of maize meiotic chromosomes provided fundamental insights into transmission genetics and the dynamic nature of the maize genome. The early insights included the physical basis of genetic recombination, the discovery of transposable DNA elements, the capping properties of telomeres, and evidence in support of the chromosome theory of inheritance (Creighton and McClintock 1931; Rhoades and McClintock 1935; McClintock 1941, 1978; Rhoades 1950; and reviewed by Carlson 1988). These earlier studies also provided the basis for the meiotic chromosome karyotype of maize, in which chromosome-spread preparations allow the 10 individual maize chromosomes to be recognized. Cytological chromosome stains reveal the presence of chromosomal landmarks, such as the knobs and centromeres, but the meiotic as well as the somatic karyotype of maize has lacked extensive genetic detail for many decades (Carlson 1988; Dempsey 1994; Chen et al. 2000; Adawy et al. 2004).
Advances in molecular biology and genomics offered new tools for cytological localization of DNA sequences and prospects for further cytogenetic map development in maize (De Jong et al. 1999; Harper and Cande 2000; Sadder et al. 2000). The development of cytogenetic FISH maps of maize has progressed from mapping repeat sequences such as knobs, centromeres, and telomeres to mapping RFLP markers, single-copy genes, and individual transposons on mitotic and meiotic chromosomes (Shen et al. 1987; Coe 1994; Chen et al. 2000; Sadder et al. 2000; Sadder and Weber 2001, 2002; Koumbaris and Bass 2003; Kato et al. 2004, 2005; Wang et al. 2006; Lamb et al. 2007). FISH mapping of pachytene chromosomes has proved to be useful for many other plant species such as tomato (Zhong et al. 1996a,b; Peterson et al. 1999), potato (Song et al. 2000), Arabidopsis (Fransz et al. 1996; Lysak et al. 2001), Medicago (Kulikova et al. 2001), rice (Cheng et al. 2001a,b, 2002), sorghum (Islam-Faridi et al. 2002; Kim et al. 2005a,b), Brassica (Howell et al. 2002), and soybean (Walling et al. 2006).
Mitotic and meiotic chromosomes have been successfully used to create cytogenetic maps of maize. The mitotic chromosomes are easier to prepare, but meiotic chromosomes have the advantage of longer axial fibers for improved localization within chromosome arms (Pedersen and Linde-Laursen 1994; Cheng et al. 2001a,b; Desel et al. 2001; Kulikova et al. 2001). Another advantage of meiotic chromosomes is that the pachytene-based cytogenetic maps can be compared to and integrated directly with recombination nodule-based maps and translocation breakpoint data (Anderson et al. 2004; Sheridan and Auger 2006).
Major challenges for any FISH-based mapping technique include the detection of small gene-size fragments, target chromosome identification, and probe specificity. Probe-detection limits can be overcome if large insert clones are used, such as those carried in BAC, YAC, or cosmid vectors (Woo et al. 1994; Hanson et al. 1995; Jiang et al. 1995; Ohmido et al. 1998; Zwick et al. 1998; Zhong et al. 1999; Dong et al. 2000; Kulikova et al. 2001). The advantage of increased signal strength with increasing probe size is offset, however, by the commensurate increase in the likelihood of detecting unintended targets such as repetitive sequences. This problem is acute in maize, where intergenic repetitive sequence elements abound and any given maize BAC clone may only contain a few kilobase pairs of unique, single-copy sequence (Liu et al. 2007). One approach to FISH mapping in maize is to use large maize genomic DNA fragments in conjunction with competitive in situ suppression hybridization (Sadder et al. 2000; Sadder and Weber 2002). Another approach is to find relatively large single-gene fragments (>3 kbp) for loci to be mapped (Wang et al. 2006). Yet another strategy is to use genomic BAC clones from the small-genome relative sorghum (Koumbaris and Bass 2003). The cross-hybridization of DNA probes from one species to target chromosomes of another species can help overcome detection limits if the two species have different or divergent classes of interspersed repetitive sequences, as is the case for sorghum and maize. Transgenomic mapping and large-fragment FISH have been successfully used in maize and in other plant species for comparative genomics (Hulbert et al. 1990; Fuchs et al. 1996; Gomez et al. 1997; Zwick et al. 1998; Jackson et al. 2000; Bowers et al. 2003; Koumbaris and Bass 2003).
Koumbaris and Bass (2003) developed a technique combining transgenomic and BAC–FISH mapping to overcome the probe-detection limit and establish an indirect way to define the cytogenetic location of sequences corresponding to targets such as RFLP probes, many of which are <1 kb. The method takes advantage of genomic and genetic resources in maize and sorghum. Here we report results from extension of this technique to production of the first detailed transgenomic BAC–FISH map of any maize chromosome. The locations of 32 genetically mapped markers were placed on the cytogenetic map, and their distribution revealed distinct irregularities with implications for maize genome assembly and evolution.
MATERIALS AND METHODS
Meiotic chromosome spreads:
Chromosome spreads were prepared from a disomic maize chromosome-addition line of oat (OMAd9.2b from Kynast et al. 2001), referred to as “oat–maize 9” in this study, that was grown in plant growth chambers or in a greenhouse (Mission Road Facility, Biological Science, Florida State University, Tallahassee, FL). Meiosis-stage florets were harvested and fixed in Carnoy's solution (3 parts absolute ethanol:1 part glacial acetic acid) for 1–2 days on a rotatory shaker at 4°. After fixation, the florets were rinsed with distilled water and stored in 70% ethanol at −20° until used. The meiotic stage was determined for one of the three anthers from a floret by the acetocarmine method. Anthers from florets with pachytene-stage meiocytes were collected and stored in 70% ethanol at −20° until used. Anthers were digested with enzymes as described by Zhong et al. (1996b), and during pachytene spread preparation, slides were given an extra three to five rounds of water vapor–acetic acid exposure, which allowed more spreading of pachytene chromosomes (Koumbaris and Bass 2003). The quality of spreads was evaluated by differential interference contrast microscopy. Slides with numerous well-spread chromosomes located in the middle of the slide, with good chromosome morphology and minimal cellular debris, were stored at −20° until used for FISH.
Identification and selection of sorghum BAC clones:
Sorghum BAC clones used as FISH probes were screened by hybridization with maize RFLP probes (as described below and by Koumbaris and Bass 2003) or with overgo probes (carried out by Bowers et al. 2005) designed to detect various maize marker probes. For RFLP-selected BACs, Sorghum propinquum genomic BAC grid-library filter arrays were obtained from A. H. Paterson (YRL filter pair; University of Georgia, Athens, GA). The detected S. propinquum BAC clones (typically four to seven overlapping BACs for each RFLP probe) were grown and the BAC DNA was initially isolated by the plasmid miniprep method with the QIAprep Spin Miniprep kit (no. 27104; QIAGEN, Valencia, CA). Southern blot analysis was then used on the restriction enzyme-digested miniprep DNA to verify clone identity. The criteria for BAC clone selection were (1) the BAC should belong to a group of overlapping clones detected by the probe and within one contig, (2) the BAC should be centrally located within this group of probe-detected overlapping BACs, and (3) the BAC should contain the same size restriction enzyme fragment as that observed with most or all of the other probe-detected overlapping BAC clones. A single selected sorghum BAC clone for each locus was then grown for large-scale BAC DNA preparation with the QIAGEN Large Construct kit (no. 12462), according to manufacturer's instructions. Highly purified BAC DNA was digested with EcoRI enzyme and then direct labeled for FISH by random-primed labeling. The YRL filters were reused after stripping by two to three washes for 30 min each with 15 mm sodium phosphate buffer (pH 6.9) at 80°. In addition to these BACs, we obtained others as gifts, including a S. bicolor BAC corresponding to the maize waxy1 locus (BAC 131L1, GenBank accession AF488412), from J. Ma and J. Bennetzen and several other S. bicolor BACs (sbb18256/191b4 and sb16685/174g5) that had been used as FISH probes on pachytene chromosomes (Islam-Faridi et al. 2002) from P. E. Klein (Texas A&M University, College Station, TX).
Slide pretreatment and FISH with pachytene chromosome spreads:
The overall FISH procedure was performed as described by Koumbaris and Bass (2003) with minor modifications. Pachytene chromosomes were denatured in 70% formamide in 2× SSC at 70° for 3–5 min and then dehydrated for 3 min in each concentration of an ice-cold ethanol series (70, 90, and 100%). The BAC clones were labeled with ChromaTide Alexa Fluor 546-16-OBEA-dCTP (Invitrogen, San Diego). The labeled FISH probes were concentrated by ethanol precipitation, redissolved in TE, and stored at −20° until used. The probe mix was denatured for 10 min at 90°, quick cooled on ice, and then combined with formamide, SSC, and dextran sulfate. The final FISH probe mixture consisted of 100 μg/ml Alexa fluor 488-labeled maize DNA (inbred KWF), centromere-specific CentC probe (10 μg/ml Alexa 647-labeled CentC repeat (Ananiev et al. 1998) DNA or 5–10 μg/ml of the oligonucleotide probe MCCY (labeled with Cy5), 200–300 μg/ml Alexa 546-labeled sorghum BAC DNA, 1 μg/ml calf thymus DNA, and 180 μg/ml S. bicolor (genotype Tx623) genomic DNA in 2× SSC, 50% formamide, and 10% dextran sulfate. The probe mix was added to the slide bearing the target pachytene chromosomes. The SSC content was decreased to as little as 0.5× SSC in cases where high background was initially observed. Hybridization was carried out at 37° on slides (20–25 μl per slide) with rubber cement-sealed coverslips with the twin tower block of the DNA engine tetrad (PTC-225; MJ Research, Watertown, MA) for 18–20 hr. After hybridization, slides were washed three times (5 min each) with 50% formamide plus 0.5–2× SSC at 37° and then washed at room temperature with 2× SSC three times (5 min each), washed with 1× PBS three times (5 min each), stained with 3–5 μg/ml DAPI, washed with 1 mm DTT in 1× PBS, and then mounted with Vectashield (Vector Labs, Burlingame, CA) for microscopy.
Data collection and image processing:
The FISH preparations were analyzed with an Olympus microscope equipped with a CCD camera (Applied Precision, Issaquah, WA). Three-dimensional (3D) images of oat–maize chromosome fibers, maize pachytene chromosome 9, sorghum BAC–FISH signal, and maize centC MCCY signal were acquired on the DAPI, FITC, RHOD, and Cy5 channels, respectively. The 3D image stacks (with Z sections spaced at 0.2–0.3 μm for a total depth of 5–8 μm) were subjected to iterative 3D deconvolution, and nuclei showing well-stained maize pachytene fibers were used to trace and computationally straighten maize 9 chromosomes for cytogenetic mapping. Pachytene fibers with little or no background staining were straightened and analyzed as described in Koumbaris and Bass (2003).
FISH locus determination and nomenclature:
The arm of interest was divided into 20–40 bins of equal length, and any FISH signal on the arm was assigned to a bin on the basis of its fractional distance from the centromere (position 0.0) to the telomere (position 1.0). Frequency histograms were inspected to identify regions with significant, above-background signals as described by Koumbaris and Bass (2003). The measured positions for all FISH signals that fell within these peak regions were averaged. The mean value, standard error (SEM), and sample size (n) are given in Table 1 for each locus in centiMcClintock (cMC) units. The nomenclature system (as described in http://www.maizegdb.org/CMMprotocols.php) for a FISH locus consists of (1) the sorghum BAC source (sbb for S. bicolor or spb for S. propinquum), (2) location on the linkage map (CBM for core bin marker or just the resident bin number for other markers), and (3) the cytogenetic map position, followed by (4) the maize RFLP marker name in parentheses.
TABLE 1.
Identification and selection of sorghum BACs
| Locus | Bin | RFLP size (bp) | Probe typea | Probe name | Sorghum FPC contigs | No. of BACs hit | Selected sorghum BACsb | FISH probe selected | cMc | Mean ± SEM (n)d | FISH locus (cMC)e | Cytolocus name |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| agrr118b | 9.00 | 500 | R | agrr118 | YN | 0.0 | ||||||
| bnl9.07a | 9.00 | 2400 | R | bnl9.07 | YN | 10.1 | ||||||
| umc109 (CBM 9.01)f | 9.01 | 800 | R | umc109 | 1220 | 7 | a0055A15, a0055B21, a0064C07, a0004M18, a0068L03, a0085L22, a0040A04 | a0004M18 | 15.5 | 9S.79 | spb-CBM9.01_S79 (umc109) | |
| umc148 | 9.01 | 520 | R | umc148 | 389 | 5 | a0030K10, a0038L23, a0038N20, a0094A07, a0013E22 | a0038L23 | 16.4 | FN | ||
| rz144a | 9.01 | 1500 | O | SOG1672 | 389 | 1 | a0030K10 | a0030K10 | 17.4 | 0.815 ± 0.004 (15) | 9S.82 | spb-9.01_S82 (rz144a) |
| rz144c | 9.01 | 1500 | O | SOG1672 | 389 | 1 | a0030K10 | a0030K10 | 21.4 | 0.745 ± 0.004 (12) | 9S.75 | spb-9.01_S75 (rz144c) |
| php10005 | 9.01 | 850 | R | php10005 | 389 | 5 | a0073F07, a0014C11, a0068G09, a0045A20, a0017D11 | a0045A20 | 24.8 | 0.730 ± 0.002 (31) | 9S.73 | spb-9.01_S73 (php10005) |
| csu95a | 9.01 | 1100 | O | SOG0620 | 130 | 1 | a0012H11 | a0012H11 | 27.7 | 0.678 ± 0.002 (10) | 9S.68 | spb-9.01_S68 (csu95a) |
| csu95a | 9.01 | 1100 | R | csu95 | 390 | 8 | a0018G19, a0094O05, a0095J06, a0095O12, a0060M11, a0096I01, a0095L23, a0042B21 | |||||
| umc248a | 9.01 | 600 | R | umc248 | YN | 27.7 | ||||||
| sh1 | 9.01 | 2500 | R | p246A | 392 | 7 | a0084C09, a0094F18, a0021O07, a0015M03, a0088L22, a0021O04, a0091L24 | a0015M03 | 36.4 | 0.661 ± 0.001 (18) | 9S.66 | spb-9.01_S66 (sh1) |
| bz1 (CBM 9.02)f | 9.02 | 1500 | R | pBZ | 1308 | 2 | a0028C12, a0020G06 | a0020G06 | 39.3 | 9S.65 | spb-CBM9.02_S65 (bz1) | |
| csu471 | 9.02 | 850 | R | csu471 | 393 | 11 | a0005O05, a0034A09, a0043E13, a0044I06, a0057D08, a0063M03, a0061M22, a0068O17, a0075C01, a0016H09, a0015L15 | a0075C01 | 42.8 | 0.636 ± 0.002 (55) | 9S.64 | spb-9.02_S64 (csu471) |
| umc256a | 9.02 | 700 | R | umc256 | 805 | 7 | SN | 43.2 | ||||
| csu486a | 9.02 | 1350 | R | csu486 | 394 | 8 | a0064N21, a0086K22, a0014H21, a0053H23, a0005O15, a0086N18, a0007E04, a0024L14 | a0086K22 | 45.5 | 0.385 ± 0.003 (34) | 9S.39 | spb-9.02_S39 (csu486) |
| prc1 | 9.02 | 1080 | R | 5C02A05 | 394 | 9 | a0064N21, a0086K22, a0014H21, a0053H23, a0005O15, a0086N18, a0007E04, a0024L14, a0076L04 | a0076L04 | 50.2 | 0.373 ± 0.002 (42) | 9S.37 | spb-9.02_S37 (prc1) |
| umc105a | 9.02 | 610 | R | umc105 | 396 | 3 | a0016B24, a0084H21, a0054A13 | a0054A13 | 54.7 | 0.276 ± 0.003 (17) | 9S.28 | spb-9.02_S28 (umc105) |
| csu228(pfk) | 9.02 | 1190 | R | csu228 | 397 | 6 | a0038I11, a0043G06, a0025D24, a0044C12, a0039P12, a0095E12 | a0043G06 | 59.7 | 0.269 ± 0.003 (24) | 9S.27 | spb-9.02_S27 [csu228(pfk)] |
| wx1 (CBM 9.03) | 9.03 | 2300 | G | umc25 | 131L1 (Bennetzen, UGA) | 131L1 | 63.7 | 0133 ± 0.005 (16) | 9S.13 | sbb-CBM9.03_S13 (wx1) | ||
| wx1 (CBM 9.03) | 9.03 | 2300 | R | umc25 | YN | |||||||
| tda66d | 9.03 | 300 | R | tda66 | 89 | 3 | a0074N08, a0054I24, a0064A03 | a0054I24 | 63.7 | 0.031 ± 0.001 (25) | 9S.03 | spb-9.03_S03 (tda66d) |
| cdo17 | 9.03 | 900 | O | SOG0101 | 874 | 5 | a0011N04, a0047J04, a0060M19, a0082G20, a0093K24 | a0047J04 | 67.7 | 0.027 ± 0.003 (15) | 9L.03 | spb-9.03_L03 (cdo17) |
| rf2 | 9.03 | 1200 | R | rf2a | 383 | 3 | a0060G13, a0070K10, a0090D12 | a0090D12 | 67.7 | FN | ||
| csu680d | 9.03 | 1550 | R | csu680 | 202 | 6 | a0047C15, a0005O02, a0060L03, a0058B07, a0064I11, a0051F17 | a0058B07 | 67.7 | FN | ||
| csu680d | 9.03 | 1550 | O | SOG1356 | 202 | 3 | a0058B07, a0064I11, a0051F17 | a0051F17 | ||||
| bnl7.24a | 9.03 | 2100 | R | bnl7.24 | 1 | 3 | SN | 68.7 | ||||
| bnl5.33c | 9.03 | 2100 | R | bnl5.33 | 53 | 5 | a0067P21, a0026E11, a0093B10, a0020L09, a0032H22 | a0020L09 | 68.7 | 0.043 ± 0.002 (64) | 9L.04 | spb-9.03_L04 (bnl5.33c) |
| csu321 | 9.03 | 750 | R | csu321 | 1361 | 5 | a0082L07, a0081N24, a0020F10, a0024F19, a0041D08 | a0020F10 | 69.1 | 0.035 ± 0.002 (40) | 9L.04 | spb-9.03_L04 (csu321) |
| rgpr3235a | 9.03 | ND | O | SOG1297 | 327 | 1 | a0040B08 | a0040B08 | 69.1 | 0.049 ± 0.003 (25) | 9L.05 | spb-9.03_L05 (rgpr3235a) |
| umc81 | 9.03 | 820 | R | umc81 | 14 | 5 | SN | 69.6 | ||||
| csu193 | 9.03 | 1270 | R | csu193 | 797 | 5 | a0014C14, a0025K08, a0060B09, a0057H24, a0095I20 | a0014C14 | 70.6 | FN | ||
| std6a(dba) | 9.03 | 1200 | R | pAS8 | 797 | 6 | SN | 70.6 | ||||
| gl15 | 9.03 | 1150 | R | HS1100.43 | 376 | 4 | a0073J07, a0075D13, a0084N16, a0009B14 | a0075D13 | 70.6 | 0.063 ± 0.002 (48) | 9L.06 | spb-9.03_L06 (gl15) |
| gl15 | 9.03 | 1150 | O | SOG0488 | 376 | 3 | a0073J07, a0075D13, a0084N16 | |||||
| csu147 (CBM 9.04) | 9.04 | 800 | O | SOG1865 | 69 | 4 | a0022A12, a0049N08, a0054F07, a0068M13 | a0049N08 | 72.8 | 0.073 ± 0.003 (21) | 9L.07 | spb-CBM9.04_L07 (csu147) |
| csu147 (CBM 9.04) | 9.04 | 800 | R | csu147 | YN | |||||||
| csu183b(cdc48) | 9.04 | 850 | R | csu183 | 401 | 3 | SN | 78.0 | ||||
| csu43 | 9.04 | 600 | R | csu43 | 550 | 4 | SN | 83.1 | ||||
| sus1 | 9.04 | 6000 | R | p21.2 | YN | 83.1 | ||||||
| csu694a(uce) | 9.04 | 950 | O | SOG0872 | 1336 | 1 | a0093O18 | a0093O18 | 84.5 | 0.355 ± 0.003 (14) | 9L.36 | spb-9.04_L36 [csu694a(uce)] |
| csu694a(uce) | 9.04 | 950 | R | csu694 | 179 | 6 | a0053I22, a0086B07, a0076H16, a0023A07, a0073A14, a0094G22 | |||||
| umc95 (CBM 9.05) | 9.05 | 660 | R | umc95 | 182 | 7 | a0015A13, a0041F23, a0043K19, a0063J06, a0069D12, a0075C14, a0094N21 | a0063J06 | 86.1 | 0.377 ± 0.002 (34) | 9L.38 | spb-CBM9.05_L38 (umc95) |
| csu392a | 9.05 | 1000 | O | SOG0184 | 539 | 11 | a0010M15, a0012B01, a0016C21, a0030A02, a0031D14, a0038B08, a0043J23, a0044E15, a0059K01, a0073M05, a0080L20 | a0010M15 | 97.0 | 0.413 ± 0.005 (20) | 9L.41 | spb-9.05_L41 (csu392a) |
| csu392a | 9.05 | 1000 | O | SOG0184 | 188 | 5 | a0004C15, a0010J13, a0026E03, a0041K24, a0079J11 | |||||
| csu392a | 9.05 | 1000 | R | csu392 | 188 | 6 | a0076K14, a0074B22, a0067J12, a0004C15, a0026E03, a0072C14 | |||||
| csu710e(apx) | 9.05 | 700 | O | SOG0484 | 211 | 5 | a0019J05, a0030K14, a0044G10, a0064D18, a0067N15 | a0019J05 | 101.1 | 0.456 ± 0.003 (25) | 9L.46 | spb-9.05_L46 [csu710e(apx)] |
| csu710e(apx) | 9.05 | 700 | R | csu710 | 188 | 5 | a0047M11, a0066O15, a0066B02, a0058C12, a0093L24 | |||||
| csu219(tgd) | 9.05 | 500 | R | csu219 | 189 | 3 | a0059O14, a0071H18, a0067I12 | a0059O14 | 102.3 | 0.506 ± 0.006 (43) | 9L.51 | spb-9.05_L51 [csu219(tgd)] |
| csu219(tgd) | 9.05 | 500 | O | SOG0074 | 189 | 2 | a0059O14, a0067I12 | |||||
| csu61a (CBM 9.06) | 9.06 | 500 | R | csu61 | 403 | 6 | a0018G11, a0019H12, a0018J02, a0041I20, a0063F18, a0090O20 | a0018G11 | 104.4 | FN | ||
| csu61a (CBM 9.06) | 9.06 | 500 | O | SOG1475 | 403 | 4 | a0018G11, a0019H12, a0018J02, a0041I20 | |||||
| csu59a | 9.06 | 500 | O | SOG0619 | 270 | 3 | a0019C22, a0074G20, a0084M22 | a0074G20 | 104.4 | 0.523 ± 0.002 (24) | 9L.52 | spb-9.06_L52 (csu59a) |
| csu59a | 9.06 | 500 | R | csu59 | 189 | 3 | a0059O14, a0071H18, a0067I12 | a0071H18 | ||||
| ibp1 | 9.06 | 1150 | R | pOD3g | YN | 104.4 | ||||||
| csu145a(pck) | 9.06 | 700 | R | csu145 | 191 | 8 | a0043G12, a0041H16, a0046O17, a0052M07, a0067L02, a0068L14, a0079N13, a0093D20 | a0093D20 | 105.4 | 0.534 ± 0.003 (26) | 9L.53 | spb-9.06_L53 [csu145a(pck)] |
| csu145a(pck) | 9.06 | 700 | O | SOG0076 | 191 | 11 | a0043G12, a0041H16, a0046O17, a0052M07, a0067L02, a0068L14, a0079N13, a0093D20, a0006K22, a0011D09, a0087G20 | |||||
| csu145a(pck) | 9.06 | 700 | O | SOG0076 | 538 | 10 | a0035K11, a0055A21, a0065B16, a0069H02, a0071B24, a0081E07, a0084G14, a0088L04, a0089F23, a0095K18 | a0055A21 | 0.527 ± 0.003 (13) | 9L.53 | spb-9.06_L53 [csu145a(pck)] | |
| dba4 | 9.06 | 900 | R | pAS14 | YN | 109.8 | ||||||
| csu28a(rpS22) | 9.06 | 700 | R | csu28 | 191 | 5 | a0010A04, a0032K15, a0076F19, a0087A24, a0093D21 | a0093D21 | 109.8 | 0.537 ± 0.003 (50) | 9L.54 | spb-9.06_L54 [csu28a(rpS22)] |
| asg44 | 9.06 | 500 | R | asg44 | 191 | 7 | a0047G04, a0081A19, a0033A22, a0091J11, a0064A12, a0038E07, a0032I20 | a0033A22 | 110.9 | 0.624 ± 0.002 (28) | 9L.62 | spb-9.06_L62 (asg44) |
| cdo1387(emp70) | 9.06 | nd | O | SOG0100 | 56 | 4 | a0007F13, a0012H21, a0036J08, a0087E20 | a0036J08 | 113.5 | 0.717 ± 0.002 (50) | 9L.72 | spb-9.06_L72 [cdo1387(emp70)] |
| cdo1387(emp70) | 9.06 | nd | O | SOG0100 | 191 | 4 | a0016I16, a0036F07, a0036L01, a0093N18 | a0036F07 | 0.722 ± 0.002 (38) | 9L.72 | spb-9.06_L72 [cdo1387(emp70)] | |
| csu1004 | 9.06 | 1800 | R | csu1004 | 1048 | 5 | a0027H23, a0046F16, a0046P22, a0054L18, a0067H10 | a0046P22 | 119.9 | 0.766 ± 0.001 (18) | 9L.77 | spb-9.06_L77 (csu1004) |
| asg12 (CBM 9.07) | 9.07 | 700 | R | asg12 | 386 | 3 | a0007D16, a0064E21, a0094P12 | a0064E21 | 121.2 | 0.779 ± 0.003 (35) | 9L.78 | spb-CBM9.07_L78 (asg12) |
| R | asg12 | 194 | 3 | a0036L10, a0074H17, a0076L16 | ||||||||
| std2a(dba) | 9.07 | 600 | R | pAS9 | YN | 122.7 | ||||||
| csu1118 | 9.07 | 700 | R | csu1118 | 197 | 1 | a0078D03 | a0078D03 | 124.3 | 0.828 ± 0.003 (25) | 9L.83 | spb-9.07_L83 (csu1118) |
| asg59b | 9.07 | 750 | R | asg59 | 890 | 5 | SN | 132.8 | ||||
| csu285(his2B) | 9.07 | 800 | R | csu285 | 9 | 8 | a0049P09, a0078L14, a0064J04, a0026P15, a0020P12, a0066J18, a0032B18, a0062J05 | a0026P15 | 147.4 | 0.939 ± 0.003 (27) | 9L.94 | spb-9.07_L94 [csu285(his2B)] |
| csu54b (CBM 9.08)g | 9.08 | 1400 | R | csu54 | 203 | 5 | a0021O07, a0061D07, a0074A03, a0080B23, a0076H02 | a0074A03 | 150.4 | 9L.95 | spb-CBM9.08_L95 (csu54b) | |
| csu804a(dnp) | 9.08 | 500 | R | csu804 | 203 | 3 | a0021O17, a0061D07, a0074A03 | a0061D07 | 150.4 | FN | ||
| Xtxa325 | G | sbb18256/191b4 (P. E. Klein, TAMU) | sbb18256 | 0.262 ± 0.009 (9) | 9L.26 | sbb_9.04-L26 (Xtxa325) | ||||||
| Xtxp32 | G | sbb16685/174g5 (P. E. Klein, TAMU) | sbb16685 | 0.530 ± 0.006 (8) | 9L.53 | sbb_9.06-L53 (Xtxp32) |
CBM, core bin marker; TAMU, Texas A&M University.
Probe type: R, RFLP; O, overgo probe; G, gift (source is indicated).
YN, YRL negative (failure to detect hybridization of RFLP probe on BAC-grid filter arrays); SN, Southern blot negative (failure to detect hybridization of RFLP probe on BAC validation Southern blot).
Centimorgan) value from the UMC 98 9 linkage map.
Numbers represent the mean of the fractional distance of the above-background peak (see materials and methods) FISH signals on the chromosome arm. SEM, standard error of measurement at 95% confidence level; (n) is the number of FISH signals used to calculate the mean.
FN, FISH negative, failed to detect FISH signals suitable for mapping on two to three slides.
Locus previously mapped by Koumbaris and Bass (2003).
IBP2 cDNA (from pOD3, H. W. Bass, unpublished data).
RESULTS
Selection of maize markers for cytogenetic mapping:
A detailed pachytene-stage cytogenetic FISH map of maize chromosome 9 was created with sorghum BACs that correspond to well-characterized maize RFLP probes. Maize RFLP probes have been used in hundreds of linkage studies but most of these probes are smaller than the lower limit of FISH detection on meiotic chromosomes as recently determined by Wang et al. (2006). We used an indirect mapping strategy that overcomes this detection limit by treating maize marker-selected syntenic sorghum BACs as surrogate FISH probes. This strategy was initially described by Koumbaris and Bass (2003) for three loci and is extended here for an additional 32 cytogenetic loci.
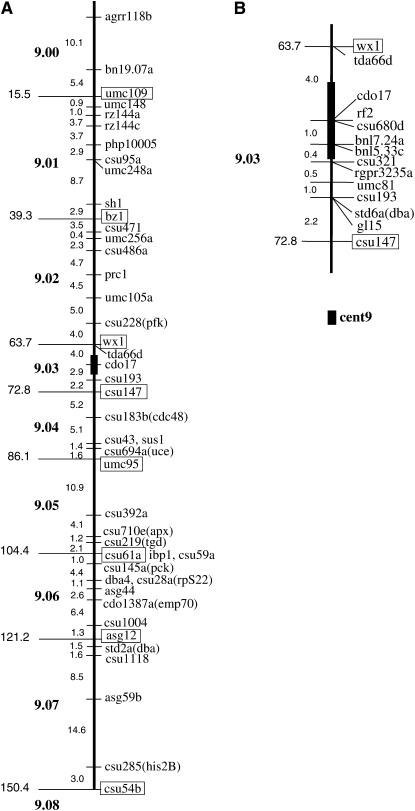
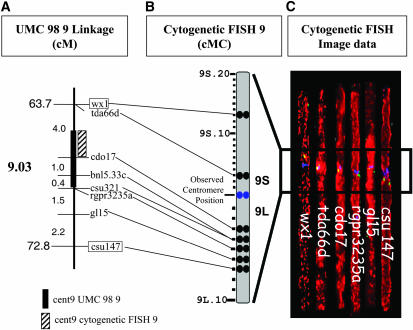
We initially selected 52 markers that are spaced ∼5 cM apart covering the entire linkage map, as shown in Figure 1, using the University of Missouri-Columbia (UMC) 98 linkage map as our base linkage map (Davis et al. 1999). This high-density map is saturated with maize RFLP marker loci for which many public probes are available (T. A. Musket and G. L. Davis, University of Missouri-Columbia RFLP Laboratory). Figure 1 shows the genetic map positions of the maize 9 markers initially chosen for FISH mapping. Very closely linked markers whose order is not resolved are listed together, separated by commas or by branching lines (e.g., sus1, csu43). The estimated position of the centromere (Figure 1, cent9, solid box) resides in bin 9.03 and its region is shown enlarged (Figure 1B). On the basis of this map, the centromere for chromosome 9 was not specifically mapped but was placed between wx1 and csu193 (Davis et al. 1999).
Figure 1.—
Location of RFLP markers chosen for cytogenetic FISH mapping and their position on the linkage map of maize chromosome 9. (A) Partial linkage map of chromosome 9 adapted from UMC 98 9 (Davis et al. 1999). Core bin markers (CBM) are in boxes and their corresponding cumulative map position in centimorgans is indicated on the left. The genetic bins (9.00, 9.01, etc.) are indicated in boldface type and the centimorgan distance between adjacent markers is indicated (small numbers between loci). Markers with the same linkage-map positions are listed together and separated by commas or indicated by branching lines. (B) Magnified version of the linkage map between CBM 9.03 (wx1) and CBM 9.04 (csu147), showing markers around the centromere. The estimated position of the centromere is indicated (solid box, cent9).
Selection of sorghum BAC clones for use as FISH probes:
We made use of a well-developed fingerprint-contig (FPC) physical map of S. propinquum derived from restriction fragment analysis of the YRL BAC library (Lin et al. 1999). Clones from this library of partially digested HindIII restriction fragments of genomic DNA have an average insert size of 126 kb. A pair of nylon filters containing a gridded array of up to 36,864 YRL BACs are available for hybridization and these filters have been screened with genetic markers such as overgo and RFLP probes (Lin et al. 1999; Bowers et al. 2003) and used to select sorghum BACs for use as FISH probes (Koumbaris and Bass 2003). High-stringency hybridization (Tm −12°) carried out by us on the YRL sorghum BACs with maize RFLP probes resulted in the detection of an average of 5.3 BACs per probe, consistent with the sixfold genomic coverage of the two filter sets.
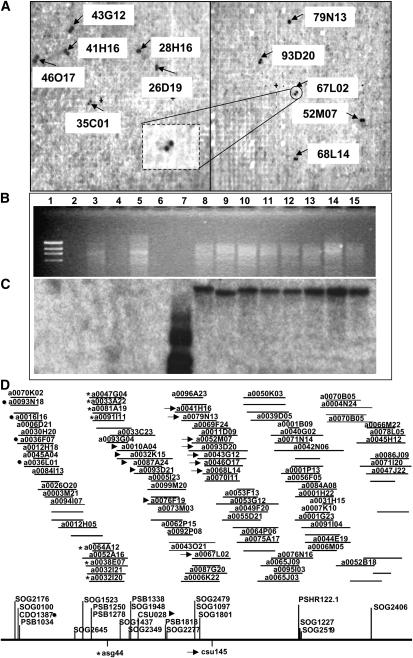
This RFLP-based BAC selection procedure is illustrated in Figure 2 for the maize marker, csu145a(pck), and the hybridization results are summarized in Table 1 for all the markers mapped in the study reported here. This marker probe hybridized to 6 BAC clones on the first filter (Figure 2A, left) and 5 on the second (Figure 2A, right). We determined the addresses of these BACs in the library and searched the online Sorghum FPC Map for them (http://www.genome.arizona.edu/fpc/sorghum/). Only 8 of the 11 BACs detected by probe csu145 (Figure 2A) were found to be overlapping in a single S. propinquum contig, no. 191 (Figure 2D, arrows). BAC a0035C01 was detected by many different maize RFLP probes used and was therefore considered a recurring false positive and excluded from the lists of loci detected. The contig to which the other two BACs (a0028H16 and a0026D19) belong could not be identified with the FPC map. We grew and isolated the remaining 8 detected BACs using a DNA miniprep procedure and subjected them to Southern blot analysis to verify that they did indeed contain a csu145-cross-hybridizing sequence (Figure 2, B and C). The same probe that was used to screen the YRL filters also hybridized to a single band in EcoRI-digested DNA minipreps (Figure 2, B and C, lanes 8–15) and to itself (lane 7) but not to other BACs on the same blot (lanes 2–5). Once confirmed by these procedures, a single BAC could be chosen for use as a FISH probe. In some cases, we used a BAC that was previously identified by overgo probe hybridization and subsequently confirmed by us using the corresponding RFLP probe.
Figure 2.—
Identification, selection, and verification of maize RFLP marker csu145-selected sorghum BAC clones. (A) Autoradiographs of Sorghum propinquum YRL filters showing six detected clones in the first filter (left) and five in the second (right). BAC a0067L02 is encircled and magnified to show one of the unique two-spot patterns from which the BAC identification is decoded. (B and C) An electrophoretic gel and autoradiograph of a blot containing the maize marker csu145, the positive control (lane 7), and the eight BACs it detected (lanes 8–15). Also included are the maize marker csu183 (lane 2), the three clones it detected (lanes 3–5), and the lambda marker (lane 1), which served as negative controls. (D) Fingerprint contig map no. 191 (http://www.genome.arizona.edu/fpc/sorghum/) showing the BACs detected by the following markers: five BACs by cdo1387 (solid circles), seven BACs by asg44 (asterisks), five BACs by csu28 (arrowheads), and eight BACs by csu145 (arrows).
We found that probes for several different but closely linked markers sometimes hybridized to BACs in the same sorghum contig or even to the same sorghum BAC clones (Table 1). The plasmid clones that identified the same contigs but selected different groups of clones are as follows: (1) csu392 and csu710 (contig 188); (2) umc148 and php10005 (contig 389); and (3) csu145, csu28, asg44, and cdo1387 (contig 191) (Figure 2D). Sorghum RFLP markers cdo1387 (Figure 2D, solid circle) and csu28 (Figure 2D, arrowhead) were already mapped on this physical contig along with other markers and overgo probes. From the positions of the BAC clones detected by maize RFLP markers asg44 (Figure 2D, asterisks) and csu145 (Figure 2D, arrows), we have determined their approximate position on the FPC map (Figure 2D, below the line). From left to right, the order of loci in the contig map for the four mentioned markers is cdo1387-asg44-csu28-csu145, the same order as that for the maize linkage map.
Overall, we set out to select BACs for 52 loci to be FISH mapped. We performed BAC filter hybridization for all of the available RFLPs (n = 47) and used the information from these or previous overgo hybridizations (Bowers et al. 2003) to select BAC probes for FISH. Nine RFLP probes resulted in a failure to detect sorghum BACs on the YRL BAC grid-array filters (n = 9, YN in Table 1) and seven RFLP probes resulted in a failure to detect sorghum BAC restriction fragments at the Southern blot verification step (n = 7, SN in Table 1). From a combination of all sources, we obtained a total of 41 BACs that were deemed suitable for FISH mapping as detailed in Table 1.
Pachytene FISH map of maize 9:
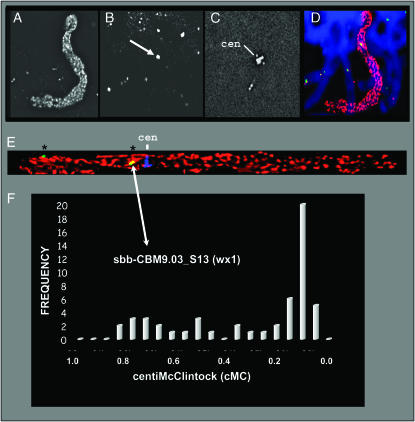
Pachytene spreads were made from oat–maize 9 anthers. To isolate maize 9 chromosome optically from among the other 21 oat pachytene fibers, we painted the entire maize 9 chromosome by genomic in situ hybridization (GISH), using labeled maize total DNA as probe. The FISH method, chromosome straightening, and cytogenetic map position value (centiMcClintock) determination are illustrated in Figure 3; the S. bicolor wx1 BAC 131L1 is used as an example.
Figure 3.—
Image collection and FISH data analysis for the Waxy1 (wx1) locus. (A) The FITC image shows GISH-painted maize pachytene chromosome 9. (B) The rhodamine image shows the signals for wx1 (arrow). (C) The Cy-5 image shows centromere FISH signals (cen). (D) RGB pseudocolor overlay of the DAPI (blue), FITC (red), and rhodamine (green) images. (E) RGB pseudocolor overlay of projections of a straightened chromosome of the FITC (red), rhodamine (green), and Cy-5 (blue) images. FISH-like signals (*) are indicated. (F) Frequency histogram graph for all FISH signals along the short arm of chromosome 9 as described in materials and methods.
All of the FISH experiments involved four-wavelength 3D image collection. Figure 3, A–C, illustrates the three differentially labeled sequences making up the FISH probe cocktail. The DAPI, FITC (maize paint), and rhodamine (BAC) images were overlaid and pseudocolored in blue, red, and green, respectively (Figure 3D). For cytogenetic FISH mapping in maize using pachytene chromosomes, each locus is assigned a centiMcClintock value on the basis of its relative position along the arm as illustrated for the wx1 locus shown in Figure 3, E and F. Two BAC–FISH signals on the short arm of chromosome 9 were observed in the example shown as green spots (Figure 3E, asterisks). We typically straightened >40 chromosomes per locus, and in this case we detected a total of 53 discrete signals on maize 9S. To distinguish true signal from background, we visualized the distribution of signals as a frequency histogram (Figure 3F). The average location of the above-background FISH signals was determined to be at 9S.13. Thus the locus is designated as sbb-CBM9.03_S13 (wx1), indicating that an S. bicolor BAC clone was used as a FISH probe for the wx1 locus that is core bin marker 9.03 and that the resulting locus is at centiMcClintock position 9S.13.
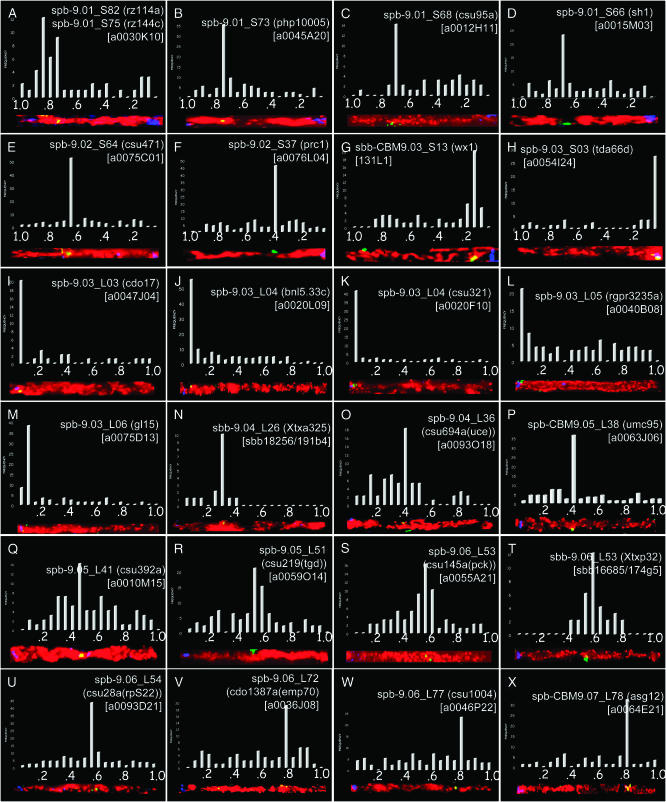
From a total of 41 BACs that were used as FISH probes, 36 of them yielded suitable FISH mapping data (Table 1). Representative images for 24 newly mapped loci are shown in Figure 4. These illustrate FISH probes distributed across the entire chromosome, including 8 from the short arm (Figure 4, A–H) and 16 from the long arm (Figure 4, I–X). Each section in Figure 4 corresponds to a single FISH probe and shows a representative straightened-arm image, the frequency histogram of FISH signals (as described for Figure 3), and the cytogenetic FISH locus name. All of the probes mapped to a single locus except for the rz144-selected BAC (a0030K10) that hybridized to two closely linked regions, 9S.75 and 9S.82 (Figure 4A).
Figure 4.—
FISH signal histograms and locus names for 24 BAC–FISH probes. Each section shows the histogram, the probe name, and the representative arm projection as described in Figure 3, E and F. The sorghum BAC name is indicated in brackets for each section. The loci are located on the short (A–H) or the long (I–X) arm of maize pachytene chromosome 9.
The overgo and RFLP probes did not always detect the exact same set of BACs, as exemplified with csu145 (Table 1). In this case, we attempted FISH with two different BACs, the RFLP-selected a0093D20 and the overgo-selected a0055A21, both of which were FISH mapped to 9L.53. In a few other cases, very tightly linked maize markers even hybridized to the same BACs. For instance, the markers csu486a and prc1 independently detected eight common BACs in contig 389 (Table 1). However, one extra BAC (a0076L04) was detected only by prc1, which was then used as a FISH probe in addition to BAC a0086K22 that represented csu486a. Using these BACs, we FISH mapped prc1 to 9S.37 while csu486a mapped to 9S.39 (Table 1). Three other locus pairs (umc148 and rz144, csu219 and csu59a, and csu804a and csu54b) showed this same sort of BAC codetection result. The distances separating these locus pairs range from 0 to 5 cM, indicating that this spacing may be near the axial resolution limit for FISH mapping in some regions.
Cytological evidence for regions of genome hyperexpansion:
Three distinct noncentromeric regions of the chromosome showed evidence of hyperexpansion relative to the overall increase of 3.3-fold for maize vs. sorghum genome size. These regions are delimited by three pairs of loci, one on 9S and two on 9L. To compare them, we used the following map distance values for maize chromosome 9: 9S is 67 cM, 100 cMC, and 77 Mbp long and 9L is 84 cM, 100 cMC, and 114 Mbp long (Bennett and Laurie 1995; Davis et al. 1999; Anderson et al. 2004).
The first of these three hyperexpanded regions is in bin 9.02 on 9S between csu471 and csu486a at 9S.64–9S.39. In this segment of the chromosome, the cM/cMC ratio is 0.11, 84% lower than the whole-arm average. The second such region is in bin 9.04 on 9L between csu147 and csu694a(uce) at 9L.07–9L.36. In this segment of the chromosome, the cM/cMC ratio is 0.40, 52% lower than the whole-arm average. The third case is in bin 9.06 on 9L between csu28a(rpS22) and cdo1387a(emp70) at 9L.54–9L.72, a segment with a length of 3.7 cM and 18 cMC. The cM/cMC ratio in this region is 0.21, 76% lower than the whole-arm average. In other words, these three regions show sixfold, twofold, and fourfold higher amounts of chromosomal distance than expected from the whole-arm average ratios of 0.67 (short) and 0.84 (long) cM/cMC.
In this third region [csu28a(rpS22) to cdo1387a(emp70) in maize bin 9.06], the corresponding sorghum markers are found in a single well-defined sorghum contig (no. 191). This allows for a comparison of the genome expansion rates for this region relative to the overall expansion rate of maize relative to sorghum. These markers in sorghum are separated by ∼200 Kbp, whereas they are separated in the cytogenetic map by 18 cMC. Using the estimate of 1.14 Mbp/cMC for 9L, we calculate that this region has expanded at a rate 25-fold higher than that of the overall maize genome relative to the sorghum genome. This is one of the three major regions that were found to exhibit this unexpected pattern of marker distribution, which we refer to here as regions of genome hyperexpansion.
FISH mapping of CentC within centromeric loci:
The positions of centromeres on linkage maps are generally not well defined because they typically lack alleles or polymorphisms suitable for standard linkage mapping. For example, the location of the maize 9 centromere is an estimate that spans a large amount of bin 9.03. To map the maize 9 centromere more precisely relative to closely linked loci, we mapped the centromere-linked markers wx1, tda66d, cdo17, bnl5.33c, csu321, rgpr3235a, gl15, and csu147 as shown in Figure 5. Representative FISH images of straightened pachytene chromosomes (Figure 5C) are shown and the location of these loci on the cytogenetic map is indicated (Figure 5B). We mapped wx1 and tda66d in the short arm, whereas the other markers were in the long arm (Figure 5B). From these data, we refined the estimated location of the centromere to a smaller region (Figure 5A, hatched box), between tda66d (9S.03) and cdo17 (9L.03) (Figure 5B). This approach allows for assignment of a given locus to either the long arm or the short arm and provides an indirect method for delimiting the location of the centromere on linkage maps, assuming that the CentC repeat cluster faithfully reports the position of the functional centromere.
Figure 5.—
Refining the linkage map position of cent9, the maize 9 centromere. Integrated linkage (A) and cytogenetic (B) maps showing the new estimated map position (A, hatched box) for the maize 9 centromere are shown. Map units for A are described in the Figure 1 legend. Map units for B are centiMcClintocks, and 9S refers to the short arm and 9L refers to the long arm. (C) Straightened projections of chromosomes showing the representative of each of the centromere-linked markers, pseudocolored as described in Figure 3E.
Integrating data from sorghum and maize maps:
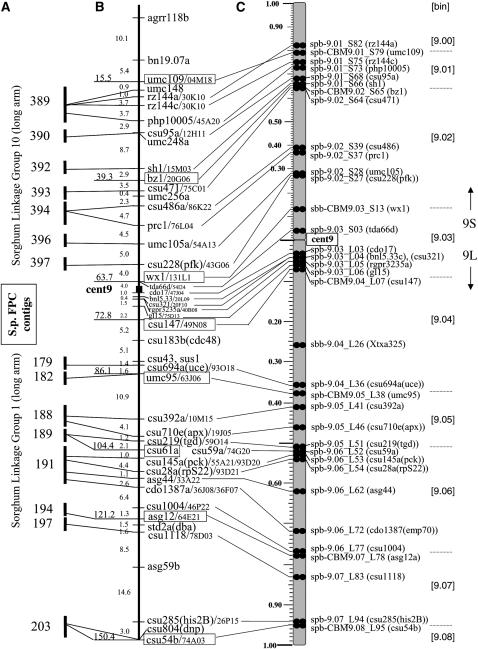
The FISH–BAC probes used in this study are unique in their applicability to multiple maps in maize and sorghum. The new cytogenetic FISH map is summarized in Figure 6 and the way in which it was created provides direct links between the physical map of sorghum (Figure 6A) and the linkage map of maize (Figure 6B). The results anchor sorghum BACs to maize pachytene chromosomes by high-stringency DNA sequence hybridization. We found nearly complete colinearity between maize linkage and cytogenetic maps, but the distributions and relative spacing between markers did not always match. One observed partial violation of agreement came from data from the sorghum BAC for rz144. In this case, rz144a and rz144c are both located proximal to umc109 on the UMC 98 9 linkage map, but the FISH signals were found on opposite sides of umc109 on the cytogenetic map. This order of loci from the telomeric end (rz144a-umc109-rz144c) is the same as that of a newer linkage map of maize, the IBM map. From these overall findings, we can conclude that this method is robust and suitable for development of a detailed cytogenetic FISH map of maize.
Figure 6.—
Integrated maps of maize chromosome 9. (A) Fingerprint contig numbers for the Sorghum FPC map and their associated linkage groups. (B) The maize 9 linkage map is shown as described in Figure 1, except that the cent9 location reflects the revised position as described in Figure 5. The names of the sorghum BACs used as a FISH probe are listed after the markers (after the slashes). (C) The new cytogenetic FISH 9 map showing the location (solid double circles) of loci along the short (9S) and the long (9L) arm. The S. bicolor BAC sbb18256 (Xtxa325) at 9L.27 is not located on the linkage map and the S. bicolor BAC sbb16685 (Xtxp32) was mapped to 9L.53, but is not shown. The new cytogenetic loci names are displayed at the right of the map in C and are detailed in Table 1.
DISCUSSION
Cytogenetic mapping provides information on the structure and evolution of genomes (Korenberg et al. 1999; Cheung et al. 2001; Trask 2002; Gonzalez et al. 2005). In addition, the BAC–FISH approach can be combined with bioinformatics to resolve inconsistencies among genome-mapping data or reveal ancestral relationships between closely related species (Ma et al. 2006; reviewed by Rocchi et al. 2006). In maize, the use of molecular cytogenetics has great potential for shedding light on the well-documented structural diversity of its chromosomes and for contributing to ongoing genome-assembly efforts (Martienssen et al. 2004; Messing and Dooner 2006). Furthermore, the development of cytogenetic resources for maize will facilitate investigations regarding the origin and evolution of the maize genome and its relationship to that of its close relatives, such as sorghum (Gaut and Doebley 1997; Gaut 2001; Kato et al. 2004, 2005; Swigonova et al. 2004; Bowers et al. 2005; Haberer et al. 2005; Bruggmann et al. 2006; Lamb and Birchler 2006; Sheridan and Auger 2006).
The development of linkage and FPC-type physical maps of maize has led to a good understanding of the distribution of genes along the chromosomes (Davis et al. 1999; Lee et al. 2002; Sharopova et al. 2002) and the physical distances between genes that reside within single FPC contigs (Nelson et al. 2005; Pampanwar et al. 2005; Yim et al. 2007). Even so, the linkage maps are based on recombination frequencies that vary widely in relation to physical distances (Anderson et al. 2003, 2004; Wang et al. 2006), and the FPC physical maps may not accurately predict the physical distance between the ends of adjacent contigs. Even as the maize genome sequence approaches completion, cytogenetic tools will remain useful for evaluating the variation between different species, subspecies, and cultivars within the genus Zea (White and Doebley 1998; Liu et al. 2003; Kato et al. 2004; Buckler et al. 2006; Lamb and Birchler 2006; Lamb et al. 2007).
The pachytene FISH map of maize 9:
Here we have described the physical organization of maize chromosome 9 by creating a new cytogenetic BAC–FISH map that integrates maize maps, provides links to sorghum maps, and uncovers several hotspots of maize genome expansion that were not anticipated from prior maps or comparative sequence analysis (Davis et al. 1999). The strategy of cross-species mapping using conserved gene sequences has proven successful in examining genome structures and relationships and predicting locations of genes and DNA markers in related species (Hulbert et al. 1990; Fuchs et al. 1996; Gomez et al. 1997; Gale and Devos 1998; Zwick et al. 1998; Jackson et al. 2000; Draye et al. 2001; Koumbaris and Bass 2003; Devos 2005). In this study, >40 sorghum BACs were used as FISH probes to create 32 new cytogenetic loci, most of which correspond to RFLP loci that are well mapped and widely used in maize linkage analysis. The success rate of this approach establishes it as a valuable and informative method for developing a detailed, integrated cytogenetic FISH map of maize. Starting with the maize 9 linkage map, 52 markers were selected and 9 of them failed at the BAC filter screening, 7 of them failed at the Southern blot stage, and 6 of them failed at the FISH stage (Table 1).
The markers were nearly completely colinear between the genetic and cytogenetic maps, but irregularly distributed along the entire length of the chromosome. The sort of differences between linkage and physical distances along individual chromosomes that we found is well documented for maize, sorghum, and other plant species (Gill et al. 1996; Peterson et al. 1999; Kunzel et al. 2000; Sadder and Weber 2002; Kim et al. 2005a; Wang et al. 2006). For example, the centromere and other heterochromatic regions often exhibit significantly fewer linkage-map units per unit physical distance because they typically have relatively low rates of meiotic recombination (Sherman and Stack 1995; Peterson et al. 1999; Harper and Cande 2000; Anderson et al. 2003, 2004).
Local variation in marker spacing:
To compare the different maps of the maize and sorghum genomes, we calculated the overall ratio of DNA length in physical units (base pairs or centiMcClintocks) to the genetic length in map units (centimorgans). This overall ratio is calculated separately for each arm because each arm encompasses 100 cMC (Lawrence et al. 2006). The DNA content for the arms of maize chromosomes in line Seneca 60, the pollen parent for oat–maize 9, was determined by Bennett and Laurie (1995) to be 77 Mbp for 9S and 114 Mbp for 9L. The numbers of linkage-map units from the UMC 98-based “Genetic 2005 9” linkage map are 67 cM for 9S and 84 cM for 9L (http://www.maizegdb.org/cgi-bin/displaymaprecord.cgi?id=940888). From this information, we derive the values per centiMcClintock to be 0.77 Mbp/cMC for 9S, 1.14 Mbp/cMC for 9L, 0.67 cM/cMC for 9S, and 0.84 cM/cMC for 9L.
Large deviations in the cM/cMC ratios were observed when two intervals on 9S were compared: the csu95a–sh1 region (9S.68–9S.66) in bin 9.01 and the csu228(pfk)–wx1 region (9S.27–9S.13) in bin 9.03. The distance between csu95a and sh1 is 8.7 cM but corresponding to only 2.0 cMC, whereas the distance between csu228(pfk) and wx1 is 4.0 cM, corresponding to 14.0 cMC. The cM/cMC ratios for these two regions are 4.35 and 0.286, respectively, representing a 6.5-fold increase and a 57% decrease relative to the cM/cMC ratio averaged over the whole arm of 9S.
In addition to these variations in the frequency of recombination per cytological distance, additional analysis of marker distribution uncovered striking evidence of regions where the maize genome appears to exhibit accelerated expansion. One of these regions is the segment on 9L between csu28a(rpS22) and cdo1387a(emp70) in bin 9.06. These markers are only 3.7 cM apart, but separated by 18 cMC on the basis of our FISH map. This region exhibits a 25-fold higher genome expansion rate than that of the whole-genome average for maize relative to sorghum. In another region, between csu471 and csu486a on 9S in bin 9.02, we examined the physical map covered by maize FPC contigs 371, 372, and 373 (http://www.maizesequence.org). The distal marker (csu471) is anchored in maize contig 371, but the proximal marker (csu486a) is not anchored and may be in contig 372 or 373. Although this segment is only 2.7 cM in length, it spans 25 cMC on the basis of our FISH map. This physical distance is predicted to represent ∼19.25 Mbp, suggesting that the gap between maize contigs 371 and 372, or between 372 and 373, may be unusually large compared to other contig gaps. Findings such as these may be important for guiding positional cloning or other strategies that rely on chromosome walking in this area. They may also reveal hotspots for insertions or duplications of mobile or other repetitive sequence elements. It will be of interest to examine the gene content and arrangement of genes in these regions.
In considering possible mechanisms for this hyperexpansion, this region does not appear to correspond to any known heterochromatic knobs or other large blocks of gene-depleted chromatin that could account for this variation. Determining whether these regions are common to other lines of maize or unique to the pollen parent line, Seneca 60, will be of interest. Recent analysis of the maize genome has also revealed uneven chromosome contraction and expansion, but on a slightly smaller scale (Bruggmann et al. 2006). Bruggmann et al. (2006) attributed the localized chromosome expansion to the insertion of retrotransposable elements. An alternative explanation is that these expanded areas are peculiar to the oat–maize addition lines and result from chromosome rearrangements or amplifications that could have occurred after the oat–maize cross. This idea, however, is not consistent with observations from oat–maize 6 lines in which the maize 6 centromere region was found to be structurally stable in three independent addition lines (Jin et al. 2004). Comparative analysis of these regions in related species may provide insight into their origin and significance.
Concordance of cytogenetic mapping data from maize:
The linear order of markers in the cytogenetic FISH map was fully concordant with that of the linkage map except for rz144 and a very minor switch between two very tightly linked loci (bnl.5.33c and csu321). In the UMC 98/Genetic 2005 map of chromosome 9, the locus order is umc109, rz144a, rz144c, whereas their order in the IBM2 2004 neighbors 9 map is rz144a, umc109, rz144c. The sorghum BAC–FISH probe a0030K10 hybridized to two loci, one at 9S.82 and one at 9S.75. These two loci flank the position determined for the FISH signal for umc109. The only locus-order inconsistency between the cytogenetic FISH map and the UMC 98 linkage map also occurred between the two linkage maps; the FISH data matched the IBM2 map data.
Map discrepancies in maize are not uncommon because of the inherent differences in genomic structure between different lines of maize (Fu and Dooner 2002; Lee et al. 2002; Bruggmann et al. 2006). Discrepancies can also result from errors due to small sample sizes or from differences in mapping techniques. For example, the Waxy1 locus is particularly variable among different maps, having been mapped to 9S and 9L in linkage studies and to different regions on 9S in cytogenetic studies. The Waxy1 locus has been localized to ∼9S.02–9S.06 by Shen et al. (1987), 9S.06 by Anderson et al. (2004), 9S.46 by Wang et al. (2006), and 9S.13 by us. All these studies were based on pachytene chromosomes, but Anderson et al. (2004) extrapolated the position from inbred KYS recombination nodule distribution, Wang et al. (2006) used direct FISH on KYS chromosomes that were uniformly elongated by extra pepsin treatment, and the position in our study was from a sorghum BAC–FISH probe hybridized to Seneca 60 chromosomes carried in an oat-genome background. The data, thus, may not be directly comparable. The sorghum BAC probe for wx1 has been sequenced and found to be syntenic with the maize and rice waxy1 region (J. Ma and J. L. Bennetzen, personal communication). To date, the cytological maps are in good agreement with each other and the linkage maps in terms of the linear order of the loci. The map positions we detected were self-consistent, and the relative positions did not vary with absolute length of the pachytene-stage chromosomes themselves. Development of cytogenetic FISH maps for maize (Sadder and Weber 2001; Cheng et al. 2002; Koumbaris and Bass 2003; Anderson et al. 2004; Sheridan and Auger 2006; Wang et al. 2006) may eventually comprise multiple maps generated by various techniques for different genotypes.
Centromere mapping:
FISH mapping of BAC clones around the centromere provided increased resolution for extrapolating the position of the maize 9 centromere within the linkage maps. We were able to map eight markers on this region using carefully chosen sorghum BAC clones. By coincident staining of the CentC repeats and the BAC–FISH probes on individual fibers, we could make chromosome-arm assignment around the centromere. The centromere, as marked by the CentC cluster, is located between tda66d at 9S.03 and cdo17 at 9L.03. This position is close to but not the same as that determined by Luce et al. (2006), using a CenH3-based method of centromere mapping. Placing centromeres on the genetic map is an indirect form of linkage mapping and it is further complicated by the dynamic nature of centromeres and their epigenetic specification (Nagaki et al. 2004; Lee et al. 2005; Luce et al. 2006).
Mapping loci for markers <1 kbp in length:
One of the major challenges for cytogenetic mapping is sensitivity, with the goal of specifically detecting small segments of DNA. RFLP probes have been used in thousands of linkage studies in maize since they were first developed >20 years ago. Localizing these markers cytogenetically would therefore be a valuable extension of maize genetics resources. For example, the RFLP probe asg44 is 500 bp, and probe csu145 is 700 bp. In the study reported here, the advance of Koumbaris and Bass (2003) was employed to map >30 loci with an average size near 1 kbp. To date, no robust technology is available for FISH mapping of DNA sequences of this size, but use of RFLP-selected sorghum BACs as FISH probes allowed us to bypass this limitation while adding a new dimension, direct links to the sorghum genome, to the resulting cytological data.
In summary, we have produced a high-resolution FISH map of maize pachytene chromosome 9 with BAC probes from sorghum genomic DNA. We found that the loci were mostly colinear between the linkage and the cytological maps of maize and that regions of genome hyperexpansion could be detected by comparative analysis of various maps containing shared markers. These findings serve to integrate genetic data across different maize maps. They also serve to generate new links between the maps of the maize and sorghum genomes. Our results have important implications for understanding and exploring the structure and the evolution of the maize genome while developing new reagents for chromosome research in the grasses.
Acknowledgments
We thank Debbie Figueroa and Shaun Murphy for critical reading and helpful comments on the manuscript. We thank R. J. (Bobbye) Hill for her support and assistance with this project. We thank P. E. Klein (Texas A&M University) and J. L. Bennetzen (University of Georgia) for providing some of the S. bicolor BACs used in this study. This work was supported by the National Science Foundation (DBI-0321639).
References
- Adawy, S. S., R. M. Stupar and J. Jiang, 2004. Fluorescence in situ hybridization analysis reveals multiple loci of knob-associated DNA elements in one-knob and knobless maize lines. J. Histochem. Cytochem. 52: 1113–1116. [DOI] [PubMed] [Google Scholar]
- Ananiev, E. V., R. L. Phillips and H. W. Rines, 1998. Chromosome-specific molecular organization of maize (Zea mays L.) centromeric regions. Proc. Natl. Acad. Sci. USA 95: 13073–13078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, L. K., G. G. Doyle, B. Brigham, J. Carter, K. D. Hooker et al., 2003. High-resolution crossover maps for each bivalent of Zea mays using recombination nodules. Genetics 165: 849–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, L. K., N. Salameh, H. W. Bass, L. C. Harper, W. Z. Cande et al., 2004. Integrating genetic linkage maps with pachytene chromosome structure in maize. Genetics 166: 1923–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, M. D., and D. A. Laurie, 1995. Chromosome size in maize and sorghum using em serial section reconstructed nuclei. Maydica 40: 199–204. [Google Scholar]
- Bennetzen, J. L., V. L. Chandler and P. Schnable, 2001. National Science Foundation-sponsored workshop report. Maize genome sequencing project. Plant Physiol. 127: 1572–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers, J. E., C. Abbey, S. Anderson, C. Chang, X. Draye et al., 2003. A high-density genetic recombination map of sequence-tagged sites for sorghum, as a framework for comparative structural and evolutionary genomics of tropical grains and grasses. Genetics 165: 367–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers, J. E., M. A. Arias, R. Asher, J. A. Avise, R. T. Ball et al., 2005. Comparative physical mapping links conservation of microsynteny to chromosome structure and recombination in grasses. Proc. Natl. Acad. Sci. USA 102: 13206–13211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggmann, R., A. K. Bharti, H. Gundlach, J. Lai, S. Young et al., 2006. Uneven chromosome contraction and expansion in the maize genome. Genome Res. 16: 1241–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler, E. S., B. S. Gaut and M. D. Mcmullen, 2006. Molecular and functional diversity of maize. Curr. Opin. Plant Biol. 9: 172–176. [DOI] [PubMed] [Google Scholar]
- Burr, B., F. A. Burr, K. H. Thompson, M. C. Albertson and C. W. Stuber, 1988. Gene mapping with recombinant inbreds in maize. Genetics 118: 519–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, W. R., 1988. The cytogenetics of corn, pp. 259–344 in Corn and Corn Improvement, edited by G. F. Sprague and J. W. Dudley. American Society of Agronomy, Madison, WI.
- Causse, M., S. Santoni, C. Damerval, A. Maurice, A. Charcosset et al., 1996. A composite map of expressed sequences in maize. Genome 39: 418–432. [DOI] [PubMed] [Google Scholar]
- Chandler, V. L., and V. Brendel, 2002. The Maize Genome Sequencing Project. Plant Physiol. 130: 1594–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. C., C. M. Chen, F. C. Hsu, C. J. Wang, J. T. Yang et al., 2000. The pachytene chromosomes of maize as revealed by fluorescence in situ hybridization with repetitive DNA sequences. Theor. Appl. Genet. 101: 30–36. [Google Scholar]
- Cheng, Z., C. R. Buell, R. A. Wing, M. Gu and J. Jiang, 2001. a Toward a cytological characterization of the rice genome. Genome Res. 11: 2133–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Z., G. G. Presting, C. R. Buell, R. A. Wing and J. Jiang, 2001. b High-resolution pachytene chromosome mapping of bacterial artificial chromosomes anchored by genetic markers reveals the centromere location and the distribution of genetic recombination along chromosome 10 of rice. Genetics 157: 1749–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Z., C. R. Buell, R. A. Wing and J. Jiang, 2002. Resolution of fluorescence in-situ hybridization mapping on rice mitotic prometaphase chromosomes, meiotic pachytene chromosomes and extended DNA fibers. Chromosome Res. 10: 379–387. [DOI] [PubMed] [Google Scholar]
- Cheung, V. G., N. Nowak, W. Jang, I. R. Kirsch, S. Zhao et al., 2001. Integration of cytogenetic landmarks into the draft sequence of the human genome. Nature 409: 953–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe, E. H., 1994. A-a translocations: breakpoints and stocks., pp. 364–376 in The Maize Handbook, edited by M. Freeling and V. Walbot. Springer-Verlag, New York.
- Coe, E., D. Hoisington and M. Neuffer, 1987. Linkage map of corn (maize) (Zea mays L.). Maize Genet. Coop. Newslett. 61: 116–147. [Google Scholar]
- Creighton, H. B., and B. McClintock, 1931. A correlation of cytological and genetical crossing over in Zea mays. Proc. Natl. Acad. Sci. USA 17: 492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, G. L., M. D. Mcmullen, C. Baysdorfer, T. Musket, D. Grant et al., 1999. A maize map standard with sequenced core markers, grass genome reference points and 932 expressed sequence tagged sites (ESTs) in a 1736-locus map. Genetics 152: 1137–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong, J. H., P. Fransz and P. Zabel, 1999. High resolution FISH in plants - techniques and applications. Trends Plant Sci. 4: 258–263. [DOI] [PubMed] [Google Scholar]
- Dempsey, E., 1994. Traditional analysis of maize pachytene chromosomes., pp. 432–441 in The Maize Handbook, edited by M. Freeling and V. Walbot. Springer-Verlag, New York.
- Desel, C., C. Jung, D. Cai, M. Kleine and T. Schmidt, 2001. High-resolution mapping of YACs and the single-copy gene Hs1(pro-1) on Beta vulgaris chromosomes by multi-colour fluorescence in situ hybridization. Plant Mol. Biol. 45: 113–122. [DOI] [PubMed] [Google Scholar]
- Devos, K. M., 2005. Updating the ‘crop circle’. Curr. Opin. Plant Biol. 8: 155–162. [DOI] [PubMed] [Google Scholar]
- Dong, F., J. Song, S. Naess, J. Helgeson, C. Gebhardt et al., 2000. Development and applications of a set of chromosome-specific cytogenetic DNA markers in potato. Theor. Appl. Genet. 101: 1001–1007. [Google Scholar]
- Draye, X., Y. R. Lin, X. Y. Qian, J. E. Bowers, G. B. Burow et al., 2001. Toward integration of comparative genetic, physical, diversity, and cytomolecular maps for grasses and grains, using the sorghum genome as a foundation. Plant Physiol. 125: 1325–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson, R., G. Beadle and A. Fraser, 1935. A summary of linkage studies in maize. Cornell Univ. Agric. Exp. Stn. Memoir 180: 1–83. [Google Scholar]
- Fransz, P. F., C. Alonso-Blanco, T. B. Liharska, A. J. Peeters, P. Zabel et al., 1996. High-resolution physical mapping in Arabidopsis thaliana and tomato by fluorescence in situ hybridization to extended DNA fibres. Plant J. 9: 421–430. [DOI] [PubMed] [Google Scholar]
- Fu, H., and H. K. Dooner, 2002. Intraspecific violation of genetic colinearity and its implications in maize. Proc. Natl. Acad. Sci. USA 99: 9573–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y., T. J. Wen, Y. I. Ronin, H. D. Chen, L. Guo et al., 2006. Genetic dissection of intermated recombinant inbred lines using a new genetic map of maize. Genetics 174: 1671–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs, J., A. Houben, A. Brandes and I. Schubert, 1996. Chromosome ‘painting’ in plants—A feasible technique? Chromosoma 104: 315–320. [DOI] [PubMed] [Google Scholar]
- Gale, M. D., and K. M. Devos, 1998. Comparative genetics in the grasses. Proc. Natl. Acad. Sci. USA 95: 1971–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner, J., S. Schroeder, M. L. Polacco, H. Sanchez-Villeda, Z. Fang et al., 2004. Anchoring 9,371 maize expressed sequence tagged unigenes to the bacterial artificial chromosome contig map by two-dimensional overgo hybridization. Plant Physiol. 134: 1317–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B. S., 2001. Patterns of chromosomal duplication in maize and their implications for comparative maps of the grasses. Genome Res. 11: 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B. S., and J. F. Doebley, 1997. DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl. Acad. Sci. USA 94: 6809–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B. S., M. Le Thierry D'Ennequin, A. S. Peek and M. C. Sawkins, 2000. Maize as a model for the evolution of plant nuclear genomes. Proc. Natl. Acad. Sci. USA 97: 7008–7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill, K. S., B. S. Gill, T. R. Endo and E. V. Boyko, 1996. Identification and high-density mapping of gene-rich regions in chromosome group 5 of wheat. Genetics 143: 1001–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez, M., M. Nurul Islam-Faridi, S.-S. Woo, K. F. Schertz, D. Czeschin et al., 1997. FISH of a maize sh2-selected sorghum BAC to chromosomes of Sorghum bicolor. Genome 40: 475–478. [DOI] [PubMed] [Google Scholar]
- Gonzalez, J., M. Nefedov, I. Bosdet, F. Casals, O. Calvete et al., 2005. A BAC-based physical map of the Drosophila buzzatii genome. Genome Res. 15: 885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberer, G., S. Young, A. K. Bharti, H. Gundlach, C. Raymond et al., 2005. Structure and architecture of the maize genome. Plant Physiol. 139: 1612–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson, R. E., M. S. Zwick, S. Choi, M. N. Islam-Faridi, T. D. Mcknight et al., 1995. Fluorescent in situ hybridization of a bacterial artificial chromosome. Genome 38: 646–651. [DOI] [PubMed] [Google Scholar]
- Harper, L. C., and W. Z. Cande, 2000. Mapping a new frontier; development of integrated cytogenetic maps in plants. Funct. Integr. Genomics 1: 89–98. [DOI] [PubMed] [Google Scholar]
- Harushima, Y., M. Yano, A. Shomura, M. Sato, T. Shimano et al., 1998. A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics 148: 479–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass-Jacobus, B. L., M. Futrell-Griggs, B. Abernathy, R. Westerman, J. L. Goicoechea et al., 2006. Integration of hybridization-based markers (overgos) into physical maps for comparative and evolutionary explorations in the genus Oryza and in Sorghum. BMC Genomics 7: 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helentjaris, T., D. F. Weber and S. Wright, 1986. Use of monosomics to map cloned DNA fragments in maize. Proc. Natl. Acad. Sci. USA 83: 6035–6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell, E. C., G. C. Barker, G. H. Jones, M. J. Kearsey, G. J. King et al., 2002. Integration of the cytogenetic and genetic linkage maps of Brassica oleracea. Genetics 161: 1225–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulbert, S. H., T. E. Richter, J. D. Axtell and J. L. Bennetzen, 1990. Genetic mapping and characterization of sorghum and related crops by means of maize DNA probes. Proc. Natl. Acad. Sci. USA 87: 4251–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam-Faridi, M. N., K. L. Childs, P. E. Klein, G. Hodnett, M. A. Menz et al., 2002. A molecular cytogenetic map of sorghum chromosome 1: fluorescence in situ hybridization analysis with mapped bacterial artificial chromosomes. Genetics 161: 345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, S. A., Z. Cheng, M. L. Wang, H. M. Goodman and J. Jiang, 2000. Comparative fluorescence in situ hybridization mapping of a 431-kb Arabidopsis thaliana bacterial artificial chromosome contig reveals the role of chromosomal duplications in the expansion of the Brassica rapa genome. Genetics 156: 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, J., B. S. Gill, G. L. Wang, P. C. Ronald and D. C. Ward, 1995. Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc. Natl. Acad. Sci. USA 92: 4487–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, W., J. R. Melo, K. Nagaki, P. B. Talbert, S. Henikoff et al., 2004. Maize centromeres: organization and functional adaptation in the genetic background of oat. Plant Cell 16: 571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, A., J. C. Lamb and J. A. Birchler, 2004. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. USA 101: 13554–13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, A., J. M. Vega, F. Han, J. C. Lamb and J. A. Birchler, 2005. Advances in plant chromosome identification and cytogenetic techniques. Curr. Opin. Plant Biol. 8: 148–154. [DOI] [PubMed] [Google Scholar]
- Kim, J. S., M. N. Islam-Faridi, P. E. Klein, D. M. Stelly, H. J. Price et al., 2005. a Comprehensive molecular cytogenetic analysis of sorghum genome architecture: distribution of euchromatin, heterochromatin, genes and recombination in comparison to rice. Genetics 171: 1963–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. S., P. E. Klein, R. R. Klein, H. J. Price, J. E. Mullet et al., 2005. b Molecular cytogenetic maps of sorghum linkage groups 2 and 8. Genetics 169: 955–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenberg, J. R., X. N. Chen, Z. Sun, Z. Y. Shi, S. Ma et al., 1999. Human genome anatomy: BACs integrating the genetic and cytogenetic maps for bridging genome and biomedicine. Genome Res. 9: 994–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumbaris, G. L., and H. W. Bass, 2003. A new single-locus cytogenetic mapping system for maize (Zea mays L.): overcoming FISH detection limits with marker-selected sorghum (S. propinquum L.) BAC clones. Plant J. 35: 647–659. [DOI] [PubMed] [Google Scholar]
- Kulikova, O., G. Gualtieri, R. Geurts, D. J. Kim, D. Cook et al., 2001. Integration of the FISH pachytene and genetic maps of Medicago truncatula. Plant J. 27: 49–58. [DOI] [PubMed] [Google Scholar]
- Kumar, A., and J. L. Bennetzen, 1999. Plant retrotransposons. Annu. Rev. Genet. 33: 479–532. [DOI] [PubMed] [Google Scholar]
- Kunzel, G., L. Korzun and A. Meister, 2000. Cytologically integrated physical restriction fragment length polymorphism maps for the barley genome based on translocation breakpoints. Genetics 154: 397–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kynast, R. G., O. Riera-Lizarazu, M. I. Vales, R. J. Okagaki, S. B. Maquieira et al., 2001. A complete set of maize individual chromosome additions to the oat genome. Plant Physiol. 125: 1216–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, J. C., and J. A. Birchler, 2006. Retroelement genome painting: cytological visualization of retroelement expansions in the genera Zea and Tripsacum. Genetics 173: 1007–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, J. C., T. Danilova, M. J. Bauer, J. M. Meyer, J. J. Holland et al., 2007. Single-gene detection and karyotyping using small-target fluorescence in situ hybridization on maize somatic chromosomes. Genetics 175: 1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, C. J., T. E. Seigfried, H. W. Bass and L. K. Anderson, 2006. Predicting chromosomal locations of genetically mapped loci in maize using the Morgan2McClintock Translator. Genetics 172: 2007–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H. R., W. Zhang, T. Langdon, W. Jin, H. Yan et al., 2005. Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc. Natl. Acad. Sci. USA 102: 11793–11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M., N. Sharopova, W. D. Beavis, D. Grant, M. Katt et al., 2002. Expanding the genetic map of maize with the intermated B73 x Mo17 (IBM) population. Plant Mol. Biol. 48: 453–461. [DOI] [PubMed] [Google Scholar]
- Lin, Y.-R., L. Zhu, S. Ren, J. Yang, K. F. Schertz et al., 1999. A Sorghum propinquum BAC library, suitable for cloning genes associated with loss-of-function mutations during crop domestication. Mol. Breed. 5: 511–520. [Google Scholar]
- Liu, K., M. Goodman, S. Muse, J. S. Smith, E. Buckler et al., 2003. Genetic structure and diversity among maize inbred lines as inferred from DNA microsatellites. Genetics 165: 2117–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, R., C. Vitte, J. Ma, A. A. Mahama, T. Dhliwayo et al., 2007. A GeneTrek analysis of the maize genome. Proc. Natl. Acad. Sci. USA 104: 11844–11849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luce, A. C., A. Sharma, O. S. Mollere, T. K. Wolfgruber, K. Nagaki et al., 2006. Precise centromere mapping using a combination of repeat junction markers and chromatin immunoprecipitation-polymerase chain reaction. Genetics 174: 1057–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak, M. A., P. F. Fransz, H. B. Ali and I. Schubert, 2001. Chromosome painting in Arabidopsis thaliana. Plant J. 28: 689–697. [DOI] [PubMed] [Google Scholar]
- Ma, J., L. Zhang, B. B. Suh, B. J. Raney, R. C. Burhans et al., 2006. Reconstructing contiguous regions of an ancestral genome. Genome Res. 16: 1557–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen, R. A., P. D. Rabinowicz, A. O'Shaughnessy and W. R. Mccombie, 2004. Sequencing the maize genome. Curr. Opin. Plant Biol. 7: 102–107. [DOI] [PubMed] [Google Scholar]
- McClintock, B., 1941. The stability of broken ends of chromosomes in Zea mays. Genetics 26: 234–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock, B., 1978. Mechanisms that rapidly reorganize the genome. Stadler Genet. Symp. 10: 25–48. [Google Scholar]
- Messing, J., and H. K. Dooner, 2006. Organization and variability of the maize genome. Curr. Opin. Plant Biol. 9: 157–163. [DOI] [PubMed] [Google Scholar]
- Messing, J., A. K. Bharti, W. M. Karlowski, H. Gundlach, H. R. Kim et al., 2004. Sequence composition and genome organization of maize. Proc. Natl. Acad. Sci. USA 101: 14349–14354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B. C., S. V. Tingey and M. Morgante, 2001. Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res. 11: 1660–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki, K., Z. Cheng, S. Ouyang, P. B. Talbert, M. Kim et al., 2004. Sequencing of a rice centromere uncovers active genes. Nat. Genet. 36: 138–145. [DOI] [PubMed] [Google Scholar]
- Nelson, W. M., A. K. Bharti, E. Butler, F. Wei, G. Fuks et al., 2005. Whole-genome validation of high-information-content fingerprinting. Plant Physiol. 139: 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmido, N., Y. Akiyama and K. Fukui, 1998. Physical mapping of unique nucleotide sequences on identified rice chromosomes. Plant Mol. Biol. 38: 1043–1052. [DOI] [PubMed] [Google Scholar]
- Pampanwar, V., F. Engler, J. Hatfield, S. Blundy, G. Gupta et al., 2005. FPC Web tools for rice, maize, and distribution. Plant Physiol. 138: 116–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson, A. H., M. Freeling and T. Sasaki, 2005. Grains of knowledge: genomics of model cereals. Genome Res. 15: 1643–1650. [DOI] [PubMed] [Google Scholar]
- Pedersen, C., and I. Linde-Laursen, 1994. Chromosomal locations of four minor rDNA loci and a marker microsatellite sequence in barley. Chromosome Res. 2: 65–71. [DOI] [PubMed] [Google Scholar]
- Peterson, D. G., N. L. Lapitan and S. M. Stack, 1999. Localization of single- and low-copy sequences on tomato synaptonemal complex spreads using fluorescence in situ hybridization (FISH). Genetics 152: 427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M. M., 1950. Meiosis in maize. J. Hered. 61: 59–67. [DOI] [PubMed] [Google Scholar]
- Rhoades, M. M., and B. McClintock, 1935. The cytogenetics of maize. Bot. Rev. 1: 292–325. [Google Scholar]
- Rocchi, M., N. Archidiacono and R. Stanyon, 2006. Ancestral genomes reconstruction: an integrated, multi-disciplinary approach is needed. Genome Res. 16: 1441–1444. [DOI] [PubMed] [Google Scholar]
- Sadder, M., and G. Weber, 2001. Karyotype of maize (Zea mays L) mitotic metaphase chromosomes as revealed by flourescence in situ hybridization (FISH) with cytogenetic DNA markers. Plant Mol. Biol. 19: 117–123. [Google Scholar]
- Sadder, M. T., and G. Weber, 2002. Comparison between genetic and physical maps in Zea mays L. of molecular markers linked to resistance against Diatraea spp. Theor. Appl. Genet. 104: 908–915. [DOI] [PubMed] [Google Scholar]
- Sadder, M. T., N. Ponelies, U. Born and G. Weber, 2000. Physical localization of single-copy sequences on pachytene chromosomes in maize (Zea mays L.) by chromosome in situ suppression hybridization. Genome 43: 1081–1083. [PubMed] [Google Scholar]
- Senior, M., and M. Heun, 1993. Mapping maize microsatellites and polymerase chain reaction confirmation of the targeted repeats using a CT primer. Genome 36: 884–889. [DOI] [PubMed] [Google Scholar]
- Sharopova, N., M. D. Mcmullen, L. Schultz, S. Schroeder, H. Sanchez-Villeda et al., 2002. Development and mapping of SSR markers for maize. Plant Mol. Biol. 48: 463–481. [DOI] [PubMed] [Google Scholar]
- Shen, D.-L., Z.-F. Wang and M. Wu, 1987. Gene mapping on maize pachytene chromosomes by in situ hybridization. Chromosoma 95: 311–314. [Google Scholar]
- Sheridan, W. F., and D. L. Auger, 2006. Construction and uses of new compound B-A-A maize chromosome translocations. Genetics 174: 1755–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, J. D., and S. M. Stack, 1995. Two-dimensional spreads of synaptonemal complexes from solanaceous plants. VI. High-resolution recombination nodule map for tomato (Lycopersicon esculentum). Genetics 141: 683–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, B. K., K. Nadarajah, M. N. Romanov and W. Ratnam, 2005. Cross-species bacterial artificial chromosome (BAC) library screening via overgo-based hybridization and BAC-contig mapping of a yield enhancement quantitative trait locus (QTL) yld1.1 in the Malaysian wild rice Oryza rufipogon. Cell Mol. Biol. Lett. 10: 425–437. [PubMed] [Google Scholar]
- Song, J., F. Dong and J. Jiang, 2000. Construction of a bacterial artificial chromosome (BAC) library for potato molecular cytogenetics research. Genome 43: 199–204. [PubMed] [Google Scholar]
- Swigonova, Z., J. Lai, J. Ma, W. Ramakrishna, V. Llaca et al., 2004. Close split of sorghum and maize genome progenitors. Genome Res. 14: 1916–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taramino, G., and S. Tingey, 1996. Simple sequence repeats for germplasm analysis and mapping in maize. Genome 39: 277–287. [DOI] [PubMed] [Google Scholar]
- Trask, B. J., 2002. Human cytogenetics: 46 chromosomes, 46 years and counting. Nat. Rev. Genet. 3: 769–778. [DOI] [PubMed] [Google Scholar]
- Walling, J. G., R. Shoemaker, N. Young, J. Mudge and S. Jackson, 2006. Chromosome-level homeology in paleopolyploid soybean (Glycine max) revealed through integration of genetic and chromosome maps. Genetics 172: 1893–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. J., L. Harper and W. Z. Cande, 2006. High-resolution single-copy gene fluorescence in situ hybridization and its use in the construction of a cytogenetic map of maize chromosome 9. Plant Cell 18: 529–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, S., and J. Doebley, 1998. Of genes and genomes and the origin of maize. Trends Genet. 14: 327–332. [DOI] [PubMed] [Google Scholar]
- Woo, S. S., J. Jiang, B. S. Gill, A. H. Paterson and R. A. Wing, 1994. Construction and characterization of a bacterial artificial chromosome library of Sorghum bicolor. Nucleic Acids Res. 22: 4922–4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim, Y. S., P. Moak, H. Sanchez-Villeda, T. A. Musket, P. Close et al., 2007. A BAC pooling strategy combined with PCR-based screenings in a large, highly repetitive genome enables integration of the maize genetic and physical maps. BMC Genomics 8: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, Y., P. J. Sanmiguel and J. L. Bennetzen, 2003. High-Cot sequence analysis of the maize genome. Plant J. 34: 249–255. [DOI] [PubMed] [Google Scholar]
- Zhong, X. B., H. J. De Jong and P. Zabel, 1996. a Preparation of tomato meiotic pachytene and mitotic metaphase chromosomes suitable for fluorescence in situ hybridization (FISH). Chromosome Res. 4: 24–28. [DOI] [PubMed] [Google Scholar]
- Zhong, X. B., P. F. Fransz, J. Wennekes-Van Eden, P. Zabel, A. Van Kammen et al., 1996. b High-resolution mapping on pachytene chromosomes and extended DNA fibres by fluorescence in-situ hybridisation. Plant Mol. Biol. Rep. 14: 232–242. [Google Scholar]
- Zhong, X. B., J. Bodeau, P. F. Fransz, V. M. Williamson, A. Van Kammen et al., 1999. FISH to meiotic pachytene chromosomes of tomato locates the root-knot nematode resistance gene Mi-1 and the acid phosphatase gene Aps-1 near the junction of euchromatin and pericentromeric heterochromatin of chromosome arms 6S and 6L, respectively. Theor. Appl. Genet. 98: 365–370. [Google Scholar]
- Zwick, M. S., M. N. Islam-Faridi, D. G. J. Czeschin, R. A. Wing, G. E. Hart et al., 1998. Physical mapping of the liguleless linkage group in Sorghum bicolor using rice RFLP-selected sorghum BACs. Genetics 148: 1983–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]