Abstract
In Caenorhabditis elegans, sex is determined by the opposing actions of X-signal elements (XSEs) and autosomal signal elements (ASEs), which communicate the ratio of X chromosomes to sets of autosomes (X:A signal). This study delves more deeply into the mechanism by which XSEs transmit X chromosome dose. We determined the relative contributions of individual XSEs to the X:A signal and showed the order of XSE strength to be sex-1 > sex-2 > fox-1 > ceh-39 ≥ region 1 XSE. sex-1 exerts a more potent influence on sex determination and dosage compensation than any other XSE by functioning in two separate capacities in the pathway: sex-1 acts upstream as an XSE to repress xol-1 and downstream as an activator of hermaphrodite development and dosage compensation. Furthermore, the process of dosage compensation affects expression of the very XSEs that control it; XSEs become fully dosage compensated once sex is determined. The X:A signal is then equivalent between XO and XX animals, causing sexual differentiation to be controlled by genes downstream of xol-1 in the sex-determination pathway. Prior to the onset of dosage compensation, the difference in XSE expression between XX and XO embryos appears to be greater than twofold, making X chromosome counting a robust process.
NEARLY 60 years have passed since the initial discovery that the nematode Caenorhabditis elegans determines its sex by counting the number of X chromosomes relative to the ploidy, the number of sets of autosomes (Nigon 1951; Madl and Herman 1979). Only recently have molecular genetic approaches revealed the components of this X:A sex-determining signal. A set of X-linked genes called X signal elements (XSEs) relays X chromosome dose, and a set of autosomally linked genes called autosomal signal elements (ASEs) relays the ploidy (Akerib and Meyer 1994; Hodgkin et al. 1994; Nicoll et al. 1997; Carmi et al. 1998; Carmi and Meyer 1999; Powell et al. 2005; reviewed in Meyer 2005; Gladden and Meyer 2007, accompanying article in this issue). Both sets of elements communicate chromosome dose by controlling the sex-determining gene xol-1 (XO lethal), the direct molecular target of the X:A signal. XSEs (repressors) and ASEs (activators) conduct a molecular tug-of-war ending in xol-1 repression in XX animals and xol-1 activation in XO animals. Once active, xol-1 directs male development (Miller et al. 1988; Rhind et al. 1995). In its absence, hermaphrodite development ensues.
By regulating the activity of xol-1, signal elements control not only the choice of sexual fate, but also the rate of X-linked gene expression dictated by the process of dosage compensation (Miller et al. 1988; Akerib and Meyer 1994). This process equalizes X chromosome gene products between the sexes by reducing gene expression from both hermaphrodite X chromosomes by half (reviewed in Meyer 2005). In XO embryos, xol-1 induces the male fate by repressing the activity of the hermaphrodite-specific sdc (sex determination and dosage compensation) genes (Miller et al. 1988; Rhind et al. 1995). In XX embryos, SDC-2 acts with SDC-1 and SDC-3 to repress the male sex determination gene her-1 and to assemble the dosage compensation protein complex (DCC) onto both X chromosomes (Villeneuve and Meyer 1987, 1990; Nusbaum and Meyer 1989; Nonet and Meyer 1991; Delong et al. 1993; Klein and Meyer 1993; Davis and Meyer 1997; Dawes et al. 1999; Chu et al. 2002; McDonel et al. 2006).
To date, XSE activity has been ascribed to four genes defined by mutations and to a 2-MU interval on X (called region 1) defined by chromosomal duplications and deficiencies. SEX-1 (a nuclear hormone receptor), CEH-39 (a ONECUT homeodomain protein), SEX-2, and the region 1 XSE act synergistically to control xol-1 at the transcript level (Akerib and Meyer 1994; Nicoll et al. 1997; Carmi et al. 1998; Gladden and Meyer 2007, accompanying article; J. Powell, C. Y. Loh and B. Meyer, unpublished results). FOX-1 (an RNA-binding protein) controls xol-1 at a post-transcriptional level (Nicoll et al. 1997). Both mechanisms of repression function together to ensure the fidelity of the X chromosome counting process. The predominant form of xol-1 regulation by XSEs appears, however, to be transcriptional (Gladden and Meyer 2007). The autosomal component of the X:A signal is represented by at least two ASEs. SEA-1 (a T-box transcription factor) and SEA-2 oppose XSEs by activating xol-1 transcription (Powell et al. 2005; P. Nix and B. Meyer, unpublished results).
Recent studies (Gladden and Meyer 2007) have hinted that the earliest aspects of sex determination are controlled through a process more complex than first described (Carmi and Meyer 1999). These studies showed that individual XSEs make unequal contributions to the X:A signal but did not determine their relative contributions or assess whether the majority of XSEs had been defined. Furthermore, they showed that most XSEs appear less potent than SEX-1, but did not define the mechanism by which SEX-1 exerts its stronger influence on sex determination and dosage compensation. Finally, no prior studies have addressed whether the X signal elements used to convey X chromosome dose are themselves repressed through a feedback loop by the very dosage compensation process that they control. Our study provides answers to these questions.
MATERIALS AND METHODS
C. elegans strains:
All C. elegans strains were derived from the Bristol variant N2 and were maintained as described in Brenner (1974). Abbreviations are as follows: ceh (C. elegans homeobox), Df (deficiency), Dp (duplication), dpy (dumpy), egl (egg-laying defective), fasn (fatty acid synthase), fox (feminizing gene on X), him (high incidence of males), nhr (nuclear hormone receptor), sdc (sex determination and dosage compensation), sea (signal element on autosome), sex (signal element on X), tra (sexual transformation), unc (uncoordinated), and xol (XO lethal).
The following chromosomal aberrations and mutations were used for this study:
LG III: dpy-27(y57) (Plenefisch et al. 1989).
LG IV: him-8(e1489), mIs11, yIs58[ceh-39(+),myo-2∷gfp]. him-8(e1489) increases X chromosome nondisjunction, resulting in 37% XO, 57% XX, and 6% Dpy XXX animals (Hodgkin et al. 1979). mIs11 is a multi-construct array carrying myo-2∷gfp, pes-10∷gfp, and gut∷gfp integrated into LG IV near dpy-20. yIs58 is an integrated array carrying the wild-type ceh-39 gene and the co-injection marker myo-2∷gfp.
LG X: dpy-3(e27), unc-2(e55), ceh-39(y414), ceh-39(gk296) (Vancouver group of the C. elegans Gene Knockout Consortium), fox-1(y303) (Nicoll et al. 1997), sex-2(y324) (J. Powell and B. Meyer, unpublished results), lon-2(e678), xol-1(y9) (Miller et al. 1988), dpy-6(e14), sex-1(y263) (Carmi et al. 1998), unc-3(e151), meDf5 X (Villeneuve 1994), and yDf17 and yDf20 (Akerib and Meyer 1994).
Duplications: yDp14(X;I), yDp13(X;f) (Akerib and Meyer 1994).
Mutations not referenced are described in this study or in Riddle et al. (1997).
Western blot analysis:
For Western blot analysis of CEH-39 levels, embryos of different genotypes were prepared by washing gravid hermaphrodites with water, treating for 5 min in a 20% hypochlorite 5% sodium hydroxide solution, and then washing two times with M9. Embryonic extract was generated by boiling embryos in 3 vol of 2× SDS–PAGE loading buffer containing 7 m urea for 10 min. The supernatant was then loaded on a 10% precast polyacrylamide gel (Invitrogen, San Diego). The Western blot was performed with rabbit anti-CEH-39 peptide antibodies, mouse anti-tubulin (DM-1A) antibody (ICN Biochemicals), horseradish-peroxidase-conjugated donkey anti-rabbit antibody (Jackson ImmunoResearch Labs), and horseradish-peroxidase-conjugated donkey anti-mouse antibody (Jackson ImmunoResearch Labs). For Western blot analysis of SEX-1 levels, wild-type XX and sex-1(y424) XX embryo extracts were prepared as previously described (Chu et al. 2002), and 1 mg of protein from each extract was precipitated with trichloroacetic acid. Briefly, one-fourth volume of 4 mg/ml deoxycholic acid in trichloroacetic acid was added to an embryo lysate, vortexed, and centrifuged at 4° for 10 min at 13,000 × g. The supernatant was then removed, and 3 vol of acetone was added to the pellet, incubated at room temperature for 10 min, and centrifuged at room temperature for 10 min at 13,000 × g. After the supernatant was removed, the pellet was resuspended in 1× SDS–PAGE buffer and the equivalent of 2 μg of total protein was loaded per lane on a NuPAGE 4–12% Bis–Tris precast polyacrylamide gel (Invitrogen). After transfer to nitrocellulose, the Western blot was performed with rabbit anti-SEX-1 antibodies (Carmi et al. 1998) and horseradish-peroxidase-conjugated donkey anti-rabbit antibody (Jackson ImmunoResearch Labs). The Western blot was developed using the ECL Plus reagent (GE Healthcare), and the blot was then stripped and reprobed with anti-α-tubulin antibody, as above.
Isolation of sex-1 null mutation:
A C. elegans deletion library was constructed in the Meyer Lab and screened for a sex-1 deletion following Michael Koelle's C. elegans Gene Knockout Protocol (02/09/03 update) retrieved from his Yale University website (http://info.med.yale.edu/mbb/koelle/). sex-1 primers used were as follows:
Forward outer primer: TGCACACATGTGTAGTAGCGGC;
Reverse outer primer: CCTGGAGATTATTACGCAACCACG;
Forward inner primer: ACATGTGAAGGACTATACTAGG;
Reverse inner primer: TAGCCGCTTGCCTTCACTTTCG;
Forward poison primer: CTCCATACTACAGCCCTTCTGG;
Reverse poison primer: GTGCATACAGAAAGCGGTGTGTCAGG.
RNA interference:
RNAi was conducted as described in Gladden and Meyer (2007, accompanying article).
Quantification of transcript levels:
Quantitative RT–PCR (qRT–PCR) was performed as described in Gladden and Meyer (2007, accompanying article). In all cases, three independent growths of the stains were used for the measurements. RNA was isolated as described in Gladden and Meyer (2007) except that sdc-2(y93, RNAi) was grown on NG–agar plates overlaid with HT115(DE3), and the following strains were grown on NG–agar plates overlaid with HB101 bacteria: dpy-27(y57), xol-1(y9), sex-1(y424), and xol-1(y9) sex-1(y424). For dpy-27(y57) and sex-1(y424) animals, plates were monitored, and young males were removed prior to reaching adulthood.
Primers for qRT–PCR are as follows:
ceh-39+, CGAGGTTCGAGGAATTGGTG;
ceh-39−, TGGAACTGGAACTGGTAGTGC;
fasn-1+, GATCCATTTGCAACTGATTCC;
fasn-1−, GCTTGGTAAGGATGGTGGC;
fox-1+, ATGGGACAAACGCAGATTGG;
fox-1−, GGGATATTCGATACGTGAAGTC;
her-1+, ACCAGCCCTTCCATCGACGC;
her-1−, GCAGTATTCTTCGAATTGGAGC;
nhr-64+, TAGAGGAAATGCGACAACGG;
nhr-64−, CCCTCATTTGGTAGCATCAG;
sex-1+, ATGACATGCCGCATTGACGG;
sex-1−, AGGCAACGGAAGTGTTGAGAG;
xol-1+, TGTAATCGCCAAGTTCGAGC;
xol-1−, TTGAAATGCTCCGTTGTCCC.
Immunofluorescence microscopy:
Embryos were fixed and stained as described in Gladden and Meyer (2007, accompanying article). Over 1000 embryos were examined for each genotype.
RESULTS
Relative contributions of individual XSEs to the X signal:
Prior studies showed that individual XSEs do not contribute equally to the X signal but did not determine their relative contributions (Carmi and Meyer 1999; Gladden and Meyer 2007, accompanying article). This disparity in XSE strength was revealed initially by the wide range in XX lethality (0–30%) caused by the loss of individual XSEs. However, the extent of XX lethality cannot be used as the sole criterion to gauge relative XSE strength, because several XSEs, including ceh-39, fox-1, and sex-2, cause insignificant lethality when disrupted. Therefore, we assessed relative strength using a more sensitive assay that measured the degree to which an XSE mutation enhanced the XX lethality caused by the partial disruption of dosage compensation (Table 1). Incomplete dosage compensation was achieved by RNAi depletion of sdc-2, a central dosage compensation gene. No XX animals survive the complete loss of sdc-2 function achieved by a null mutation, yet 81% of sdc-2(RNAi) XX animals survive, affording ample range for mutations to enhance lethality.
TABLE 1.
The contributions of individual XSEs to the X signal are of different strengths
| Genotypea | Hermaphrodite viability (%)b | nc |
|---|---|---|
| ceh-39(y414)d | 101 | 1008 |
| ceh-39(y414) + control RNAie | 100 | 900 |
| ceh-39(gk296)d | 102 | 1021 |
| ceh-39(gk296) + control RNAie | 100 | 1134 |
| fox-1(y303)d | 99 | 1054 |
| fox-1(y303) + control RNAie | 101 | 938 |
| sex-1(y263)d | 70 | 884 |
| sex-1(y263) + control RNAie | 70 | 867 |
| sex-2(y324)d | 99 | 1032 |
| sex-2(y324) + control RNAie | 99 | 794 |
| sdc-2(RNAi) | 81 | 1044 |
| ceh-39(y414) sdc-2(RNAi) | 54 | 1120 |
| ceh-39(gk296) sdc-2(RNAi) | 50 | 1079 |
| fox-1(y303) sdc-2(RNAi) | 37 | 1258 |
| sex-1(y263) sdc-2(RNAi) | 0 | 1072 |
| sex-2(y324) sdc-2(RNAi) | 0 | 1087 |
| sex-2(y324) xol-1(y9) sdc-2(RNAi)f | 93 | 1093 |
| xol-1(y9) sdc-2(RNAi) | 98 | 1211 |
Animals were fed bacteria that produced dsRNA to sdc-2.
Hermaphrodite viability was calculated by the formula: (no. of adult hermaphrodites)/(total no. of embryos) × 100.
n is the total number of embryos from at least six independent sets of progeny counts.
Data are from Gladden and Meyer (2007, accompanying article).
Mutants were grown on bacteria carrying the L4440 empty vector. Data were acquired concurrently with the data in footnote d.
This strain also has the unc-76(e911) and unc-10(e102) mutations.
Both ceh-39 alleles enhanced the XX lethality caused by sdc-2(RNAi) (Table 1). XX viability was reduced to 54 and 50% for ceh-39(y414) sdc-2(RNAi) and ceh-39(gk296) sdc-2(RNAi), respectively. fox-1 appeared to have a more potent effect on the X signal: only 37% of fox-1(y303) sdc-2(RNAi) XX animals were viable (P < 0.01 for y303 comparison to y414 or gk296) (Table 1). Both sex-1(y263) and sex-2(y324) caused complete lethality in combination with sdc-2(RNAi), indicating that both sex-1 and sex-2 contribute more strongly to the signal than either ceh-39 or fox-1 (Table 1). Since sex-2(y324) causes only 1% XX lethality, but sex-1(y263) causes 30% XX lethality, sex-1 appears to be a stronger XSE than sex-2. All five XSE mutant strains appeared unaffected by administration of control dsRNA, making it unlikely that the differences in viability reflect nonspecific RNAi effects (Table 1). This combined analysis shows that the XSEs defined by point mutations make the following relative contributions to the X signal: sex-1 > sex-2 > fox-1 > ceh-39.
A more refined distinction in XSE strength between fox-1 and ceh-39 was made by comparing the extent to which fox-1(RNAi) or ceh-39(RNAi) suppressed the complete XO-specific lethality caused by two copies of the X duplication yDp14 (Figure 1; Table 2). The XO lethality caused by yDp14 is due to the increased dose of fox-1, ceh-39, and other potential XSEs, which repress xol-1 and thereby activate dosage compensation. fox-1(RNAi) permitted 69% of him-8; yDp14/yDp14 XO animals to live, and ceh-39(RNAi) permitted only 4% of XO animals to live. In contrast, no XO Dp animals were rescued by control dsRNA made from an RNAi vector lacking a cloned gene. Thus, fox-1 appears to be a stronger XSE than ceh-39.
Figure 1.—
Genetic map of the X chromosome and structure of the sex-1 gene. (A) The X map shows XSEs and xol-1 above the line and highlights three regions (numbered boxes) previously shown by duplications (double lines) and deficiencies (single line) to contain X signal elements. The free duplication yDp13 covers region 1 (XSE not identified), region 2 (ceh-39), and region 3 (fox-1). The attached duplication yDp14 covers regions 2 and 3. The deficiency meDf5 removes region 1, yDf17 removes regions 1–3, and yDf20 removes regions 2 and 3. (B) The genomic region spanning sex-1. Exons are indicated by boxes and introns by lines. Exon sequences encoding the DNA-binding domain are solid and those encoding the ligand-binding domain are shaded. The sex-1(y424) null allele reported in this study removes 504 bp of 5′ regulatory sequences, including the ATG translation initiation site and at least part of the promoter. The partial loss-of-function mutations y263 and gm41 introduce G-to-A transitions that mutate splice acceptor sites, causing translation products to be out of frame.
TABLE 2.
fox-1 is a stronger XSE than ceh-39
| yDp14/yDp14; him-8 XO + RNAi of genea | Male viability (%)b | nc |
|---|---|---|
| No RNAi | 0 | 929 |
| dsRNA from vector with no gened | 0 | 1055 |
| fox-1(RNAi)d | 69 | 1174 |
| ceh-39(RNAi)d | 4 | 1030 |
| fox-1(RNAi)e | 72 | 1083 |
| ceh-39(RNAi)e | 4 | 1060 |
The relative strength of the XSEs fox-1 and ceh-39 was examined by treating hermaphrodites of genotype yDp14/yDp14 (X;I); him-8(e1489) IV; unc-2(e55) X with dsRNA corresponding to the gene listed, and the viability of progeny males was assessed. RNAi-mediated knockdown of an XSE should decrease the male lethality caused by the increase in XSE dose from yDp14. yDp14 is an X duplication attached to LG I that can exist in one copy (yDp14/+) or two copies (yDp14/yDp14) (Akerib and Meyer 1994). him-8 XX animals produce 37% XO males, 57% XX hermaphrodites, and 6% Dpy XXX hermaphrodites (Hodgkin et al. 1979).
Male viability was calculated by the formula: (no. of adult males)/(expected no. of males) × 100. The number of expected males was (0.37)n.
n is the total number of embryos from at least six independent sets of progeny counts.
Hermaphrodites were injected with dsRNA corresponding to the indicated gene, and the viability of progeny males was assessed.
Hermaphrodites were fed bacteria producing dsRNA corresponding to the indicated gene, and the viability of progeny males was assessed.
Using a separate assay, Carmi and Meyer (1999) compared the relative contributions of fox-1 and the region 1 XSE to the X signal. Relative XSE strengths were assessed by determining the extent to which XSE loss rescued the complete XO lethality caused by the single-copy X duplication yDp13, which increased the dose of the region 1 XSE, ceh-39 (region 2), and fox-1 (region 3) (Figure 1A). Region 1 deficiency meDf5 permitted 38% of meDf5; yDp13 XO animals to live. In contrast, 90% of fox-1(y303); yDp13 XO animals lived, indicating that fox-1 is more potent than the region 1 XSE. The combination of our data and that of Carmi and Meyer (1999) indicate that sex-1, sex-2, and fox-1 make stronger contributions to the X signal than the region 1 XSE and that the overall order of XSE strength is sex-1 > sex-2 > fox-1 > ceh-39 ≥ region 1 XSE.
The dose of more than four XSEs must be reduced by half to toggle the X signal from the XX to the XO mode:
In view of the modest contribution of each XSE to the X signal, it was important to address whether the set of genetically defined XSEs constitutes the entire X signal. Since the twofold lower copy number of XSEs in XO embryos vs. XX embryos permits high xol-1 expression and the male fate, one can determine whether the known XSEs constitute the entire X signal by asking whether halving the dose of these XSEs in XX animals activates xol-1 and induces the male fate. Previous studies using large X deficiencies to reduce XSE dose in XX animals showed that halving the dose of three XSE regions caused mild-to-severe defects in sex determination and dosage compensation and that halving the dose of four regions was sufficient to switch the X:A signal to the XO mode, causing complete masculinization and lethality of XX animals (Carmi and Meyer 1999). To better estimate the minimum number of XSEs that contribute to the X signal, we assessed the effects of heterozygous XSE mutations on XX animals. In contrast to results using deficiencies, our results show that halving the dose of three or four XSEs only partially toggles the X signal toward the XO mode, suggesting that the deficiencies removed several additional, unidentified XSEs and that the X signal includes more XSEs than previously thought.
In our study, fox-1+/+ sex-1 animals were wild type, and all ceh-39 + +/+ fox-1 sex-1 animals were viable but exhibited a mild Dpy, Egl phenotype, showing that halving the dose of three XSEs reduces the overall X signal (Table 3). However, the mild Dpy Egl phenotype was much less severe than the phenotype reported previously for yDf20 +/+ sex-1 XX hermaphrodites, in which the doses of at least ceh-39, fox-1, and sex-1 were halved. Only 29% of the animals were viable, and escapers were Egl and Dpy (Table 3; Figure 1; Carmi and Meyer 1999). The phenotypic discrepancy between the two strains means either that yDf20 removes other undefined XSEs or that yDf20 removes other factors essential for the viability of hermaphrodites. Evidence presented in Gladden and Meyer (2007) indicated that ceh-39 and fox-1 are not likely to be the only XSEs in this region of X, thus making the former possibility the most probable.
TABLE 3.
Multiple XSEs must be reduced by half to perturb dosage compensation in XX animals
| Genotypea | XX phenotype | Hermaphrodite viability (%)b | nc |
|---|---|---|---|
| fox-1 +/+ sex-1d | Wild type | 100 | NA |
| yDf20 [Δ of ceh-39, fox-1]/+d | Wild type | 99 | NA |
| yDf20 [Δ of ceh-39, fox-1] +/+ sex-1d | Dpy, Egl, Tra | 29 | NA |
| yDf17 [Δ of region 1, ceh-39, fox-1] +/+ sex-1d | Dead | 0 | NA |
| ceh-39 + +/+ fox-1 sex-1e | Wild type or mild Dpy, Egl | 107f | 1401 |
| ceh-39 + +/+ sex-2 sex-1g | Wild type | 98f | 1867 |
| ceh-39 fox-1 + +/+ + sex-2 sex-1h | Dpy, Egl | 60f | 1081 |
Alleles used were sex-1(y263), fox-1(y303), sex-2(y324), and ceh-39(y414).
Hermaphrodite viability was calculated by the formula: [no. of adult hermaphrodites]/[expected no. of hermaphrodites, (0.5)n] × 100.
n is the total number of embryos from at least six independent sets of progeny counts.
Data are from Carmi and Meyer (1999).
Crosses between fox-1 sex-1 XO males and unc-32; ceh-39 hermaphrodites yielded non-Unc cross progeny that ranged from wild type to mild Dpy Egl. The phenotype of ceh-39 + +/+ fox-1 sex-1 animals (Dpy and Egl) is more severe than that of fox-1+/+ sex-1 animals (wild type), showing that changing the dose of ceh-39 by half reduces the overall X signal.
Hermaphrodite viability was calculated as the [no. of adult hermaphrodites (either non-Unc or non-Dpy-3)]/[(0.5)(n − no. of Unc or Dpy-3 adults)] × 100. Unc or Dpy-3 adults represent self-progeny.
Crosses between sex-2 sex-1 XO males and unc-32; ceh-39 hermaphrodites yielded non-Dpy non-Unc cross progeny.
Crosses between sex-2 sex-1 XO males and ceh-39 dpy-3(e27) fox-1 hermaphrodites yielded non-Dpy-3 cross progeny that ranged from Dpy to wild type. The ceh-39 allele gk296 showed results similar to y414.
fox-1, ceh-39, and sex-1 repress xol-1 using two different mechanisms, transcriptional (sex-1 and ceh-39; see below) and post-transcriptional (fox-1). While halving the dose of both transcriptional (ceh-39 and sex-1) and post-tanscriptional (fox-1) repressors of xol-1 in XX animals caused Dpy and Egl phenotypes, halving the dose of only transcriptional regulators (sex-1, ceh-39, and sex-2) caused no obvious mutant phenotype (Table 3). Since fox-1 and sex-2 are of equivalent strength, we conclude that reducing the dose of XSEs that regulate xol-1 on multiple levels disrupts xol-1 repression more severely than reducing the dose of XSEs that regulate xol-1 on the same level. This finding reinforces previous studies (Carmi and Meyer 1999).
As expected, reducing the dose of four XSEs by half using mutant alleles of ceh-39, fox-1, sex-2, and sex-1 caused more severe phenotypes than halving the dose of only three XSEs. Approximately 60% of the heterozygous quadruple mutant ceh-39 fox-1 + +/+ + sex-2 sex-1 animals were viable, and the escapers ranged in phenotype from wild type to Dpy and Egl (Table 3). However, reducing the dose of these four XSEs caused a milder phenotype than was evident for yDf17 +/+ sex-1 XX animals, all of which were dead despite bearing one wild-type copy of region 1, ceh-39, fox-1, and sex-1 (Carmi and Meyer 1999). Since sex-2 is stronger than the region 1 XSE (Table 1; Akerib and Meyer 1994; Carmi and Meyer 1999), one would logically expect the ceh-39 fox-1 + +/++ sex-2 sex-1 strain to exhibit a more severe phenotype than the yDf17 +/+ sex-1 strain. The opposite was found, suggesting that yDf17 harbors additional XSEs and that the X signal includes more XSEs than the previously estimated number.
The sex-1 null mutation causes severe disruption of sex determination and dosage compensation:
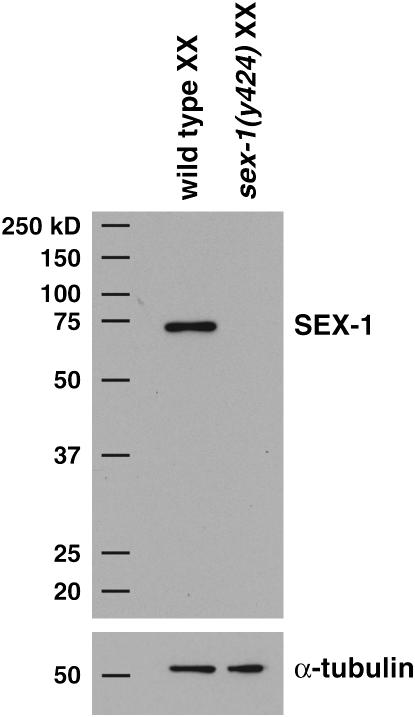
Disruption of sex-1 causes more extensive XX lethality than disruption of any other XSE, even though the sex-1(y263) allele does not appear to be a null allele. That is, sex-1(y263) reduces viability of XX animals to ∼70% (Table 4; Carmi et al. 1998), but depletion of sex-1 activity with RNAi into either wild-type animals or sex-1(y263) mutants reduces viability to 17% (Table 4). Therefore, to explore the function of sex-1 further and to determine the cause for its potent effect on sex determination and dosage compensation, we first obtained a sex-1 deletion allele by screening a C. elegans deletion library (materials and methods). sex-1(y424) reduces viability of XX animals to 20%, and the viability is not further reduced by treating the mutants with sex-1 RNAi (Table 4). Both sex-1(y424) mutants and sex-1(y424, RNAi) mutants (19% viable) are comparable in viability to sex-1(RNAi) animals (17% viability) and to sex-1(y263, RNAi) animals (17% viability), suggesting that the sex-1(y424) deletion allele removes all gene function. The y424 deletion removes 504 bp, including the ATG translational start site and part of the sex-1 promoter region (Figure 1B). Any potential translation products are predicted to be out of frame and in low abundance. Indeed, Western blot analysis detected no protein in extracts from sex-1(y424) mutants (Figure 2) even after extended exposure to film. Together, these findings indicate that sex-1(y424) is a null allele.
TABLE 4.
sex-1 null allele behaves like sex-1(RNAi)
| Genotypea | Hermaphrodite viability (%)b | nc |
|---|---|---|
| sex-1(y263) | 74 | 1107 |
| sex-1(RNAi) | 17 | 1090 |
| sex-1(y263, RNAi) | 17 | 1304 |
| sex-1(y424) | 20 | 2308 |
| sex-1(y424, RNAi) | 19 | 876 |
Animals were fed bacteria producing dsRNA generated from plasmids encoding the gene listed.
Hermaphrodite viability was calculated by the formula: (no. of adult hermaphrodites)/(total no. of embryos) × 100.
n is the total number of embryos from at least six independent sets of progeny counts.
Figure 2.—
sex-1 null mutants have undetectable levels of SEX-1. Western blots of whole-cell protein extracts from wild-type XX and sex-1(y424) XX embryos were probed with anti-SEX-1 antibody followed by anti-α-tubulin antibody for extract normalization. Positions of molecular weight markers are indicated on the left.
sex-1 controls sex determination and dosage compensation by regulating xol-1 and at least one additional target:
The first clue as to how sex-1 exerts a stronger influence on sex determination and dosage compensation than any other XSE came from correlating xol-1 transcript levels with XX lethality in mutants lacking a specific XSE. Although the XX lethality caused by sex-1 mutations is greater than that caused by any other XSE mutation, the xol-1 transcript levels in sex-1 mutants are not markedly higher than those in the other XSE mutants (Gladden and Meyer 2007, accompanying article). This finding suggested that sex-1 might regulate more targets than just xol-1 and that deregulation of the other targets might be responsible for the increased lethality of sex-1 mutants. This view was reinforced by our finding that the xol-1(y9) null mutation, which deletes the entire gene, cannot suppress all the defects caused by sex-1(y263, RNAi) or sex-1(y424) (Table 5). The xol-1 null mutation does suppress most of the XX-specific lethality caused by the sex-1(y263) partial loss-of-function mutation (from 70 to 87% viability, P < 0.01), consistent with sex-1 functioning as an XSE (Table 5 and Carmi et al. 1998), but it only partially suppresses the XX lethality caused by sex-1(y263, RNAi) (from 17 to 58% viability; Table 5) or by sex-1(y424) (from 20 to 56% viability; Table 5). The Dpy, Egl, and Tra phenotypes caused by the disruption of dosage compensation and sex determination were suppressed in all three strains to a significant degree. The incomplete suppression by xol-1(y9) of the XX lethality caused by loss of sex-1 suggests that sex-1 has a function distinct from its regulation of xol-1, possibly in controlling genes elsewhere in the sex-determination and dosage compensation regulatory pathway.
TABLE 5.
sex-1 functions as an XSE and has an XSE-independent function
| Genotypea | Hermaphrodite viability (%)b | nc |
|---|---|---|
| sex-1(y263) | 70 | 884 |
| xol-1(y9) sex-1(y263) | 87 | 690 |
| sex-1(y263, RNAi) | 17 | 1304 |
| xol-1(y9) sex-1(y263, RNAi) | 58 | 891 |
| sex-1(y424) | 20 | 2308 |
| xol-1(y9) sex-1(y424) | 56 | 1701 |
| sdc-2(RNAi)d | 84 | 1512 |
| sex-1(y263) sdc-2(RNAi)d | 0 | 1072 |
| xol-1(y9) sex-1(y263) sdc-2(RNAi)d | 6 | 2018 |
| xol-1(y9) sdc-2(RNAi)d | 98 | 1211 |
| sdc-2(RNAi)d | 86 | 1244 |
| sex-1(y424) sdc-2(RNAi)d | 0 | 912 |
| xol-1(y9) sex-1(y424) sdc-2(RNAi)d | 0 | 1256 |
| xol-1(y9) sdc-2(RNAi)d | 97 | 1283 |
| dpy-28(RNAi) | 91 | 1439 |
| dpy-28(RNAi); sex-1(y263) | 6 | 2273 |
| dpy-28(RNAi); xol-1(y9) sex-1(y263) | 4 | 1506 |
| dpy-28(RNAi); xol-1(y9) | 89 | 1315 |
| mom-2(RNAi)e | 24 | 1064 |
| mom-2(RNAi); xol-1(y9)e | 58 | 999 |
| unc-22(RNAi)e | 100% viable, 64% Unc | 550 |
| unc-22(RNAi); xol-1(y9)e | 100% viable, 32% Unc | 983 |
Animals were fed bacteria producing dsRNA generated from plasmids encoding the gene listed.
Hermaphrodite viability was calculated by the formula: (no. of adult hermaphrodites)/(total no. of embryos, n) × 100.
n is the total number of embryos from at least six independent sets of progeny counts.
All RNAi treatments and progeny counts performed simultaneously.
The xol-1(y9) mutation appears to reduce the effectiveness of RNAi against some genes. mom-2 encodes the WNT signaling molecule; loss of mom-2 function causes embryonic lethality. unc-22 encodes a muscle protein; loss of unc-22 function causes a twitching phenotype. xol-1(y9) reduces the effectiveness of mom-2(RNAi) and unc-22(RNAi). This phenomenon probably accounts for why 84% of sdc-2(RNAi) animals are viable, but 98% of xol-1; sdc-2 (RNAi) animals are viable. The xol-1(y9) mutation probably reduces the effectiveness of sdc-2(RNAi). However, xol-1(y9) does not suppress the lethality of sdc-2 mutants. The ability of xol-1(y9) to interfere with RNAi against some genes does not compromise any of our conclusions. First, xol-1(y9) suppresses the lethality caused by the sex-1(y424) null mutation (see Table 3) to the same degree that it suppresses the lethality caused by sex-1(RNAi) (this table). Second, suppression of XX lethality in sdc-2(RNAi) sex-1(y263) or dpy-28(RNAi); sex-1(y263) animals by xol-1(y9) is very poor, and the ineffectiveness of RNAi in xol-1(y9) could cause only the opposite effect: the extent of suppression would be greater than it would otherwise be.
If sex-1 acts in the pathway downstream of xol-1 or in an independent pathway, then a xol-1 mutation should fail to suppress the synergistic lethality caused by partial disruption of both sex-1 and a downstream dosage compensation gene such as sdc-2. Partial disruption of sdc-2 by RNAi reduced the viability of otherwise wild-type XX animals to 84% and the viability of sex-1(y263) XX animals to 0% (Table 5). Consistent with sex-1 playing a role in dosage compensation beyond its role in regulating xol-1, a xol-1 mutation failed to suppress the synergistic lethality between sex-1(y263) and sdc-2(RNAi). Although 87% of xol-1 sex-1(y263) XX double mutants were viable, only 6% of xol-1 sex-1(y263) sdc-2(RNAi) triple mutants were viable (Table 5). Similar results were obtained with the sex-1 null allele. sdc-2(RNAi) reduced the viability of sex-1(y424) XX mutants from 20 to 0%. Although 56% of xol-1 sex-1(y424) double mutants were viable, 0% of xol-1 sex-1(y424) sdc-2(RNAi) triple mutants were viable (Table 5).
Further results demonstrating sex-1's function in dosage compensation beyond xol-1 regulation were obtained through RNAi disruption of the dosage compensation gene dpy-28 (Table 5) in a sex-1 mutant. About 91% of dpy-28(RNAi) animals and 89% of dpy-28(RNAi); xol-1 animals were viable, but only 6% of dpy-28(RNAi); sex-1(y263) double mutants and 4% of dpy-28(RNAi); xol-1 sex-1(y263) triple mutants were viable, indicating that a xol-1 mutation cannot suppress the synergistic lethality caused by partial disruption of sex-1 and a dosage compensation gene. As an important aside, the xol-1(y9) mutation reduces the effectiveness of RNAi against some genes (Table 5). In our experiments, any reduction of RNAi by xol-1(y9) would only cause us to overestimate the suppression of lethality by xol-1(y9). Since suppression is negligible, any abrogation of RNAi by xol-1(y9) would not compromise our conclusions. Evidence presented in the next section shows that sex-1 positively regulates dosage compensation by acting downstream of xol-1.
sex-1 acts downstream of xol-1 to regulate both dosage compensation and sex determination:
xol-1 mutations induce all XO animals to activate the XX-specific sdc genes and thereby adopt the hermaphrodite mode of sex determination and dosage compensation. These feminized xol-1 XO mutants die as embryos or L1 larvae due to reduced X-linked gene expression caused by assembly of the dosage compensation complex on the single male X (Miller et al. 1988; Chuang et al. 1994; Lieb et al. 1996, 1998; Dawes et al. 1999). One can determine whether a gene functions downstream of xol-1 in the sex-determination and dosage compensation pathway and, if it does, which branch, by assessing whether mutations in the gene suppress the sex-determination defects, the dosage compensation defects, or both classes of defects in xol-1 XO mutants. For example, mutations in genes such as sdc-2, which control both sex determination and dosage compensation in XX animals, suppress both the death and feminization of xol-1 XO mutants, causing rescued XO animals to develop as males (Miller et al. 1988). In contrast, mutations in dosage compensation genes such as dpy-28 suppress the death but not the feminization of xol-1 mutants, causing the rescued XO animals to develop as hermaphrodites (Miller et al. 1988).
By themselves, neither sex-1(y263) nor sex-1(y424) rescues xol-1 XO mutants to adulthood, but both permit xol-1 XO mutants to develop beyond the L1 larval stage, indicating that a sex-1 mutation weakly disrupts dosage compensation even when its impact on the X signal has been abrogated by a xol-1 mutation (Table 6). Furthermore, a sex-1 mutation can further suppress the lethality of xol-1 XO animals whose dosage compensation machinery has been partially disrupted. RNAi of sdc-2 rescued only 6% of xol-1 XO males, but a sex-1 mutation increased the viability of sdc-2(RNAi) xol-1 XO animals to 35% (Table 6, P < 0.01). The rescued animals develop as males. These results show that sex-1 acts downstream of xol-1 as a positive regulator of dosage compensation (Table 6).
TABLE 6.
sex-1 acts downstream of xol-1 to control sex determination and dosage compensation
A complementary set of genetic experiments showed that sex-1 also controls sex determination through its action downstream of xol-1 (Table 6). dpy-28(RNAi) rescued almost all xol-1 XO animals, and all rescued dpy-28(RNAi); xol-1 XO animals developed as hermaphrodites. In contrast, 33% of the rescued dpy-28(RNAi); xol-1 sex-1(y263) XO animals developed as males, indicating that sex-1 acts downstream of xol-1 to promote the hermaphrodite sexual fate.
Independent molecular experiments confirmed the XSE-independent function of sex-1 and showed that this downstream effect controls sex determination by repressing transcription of the male sex-determination gene her-1, either directly or indirectly (Table 7). her-1 is one of the few genes in the sex-determination pathway that is controlled at the level of transcription and acts downstream of xol-1, making it an appropriate gene to monitor sex-1 function (Trent et al. 1991). her-1 is repressed directly through binding of SDC proteins to the promoter and second intron (Chu et al. 2002). The difference in her-1 transcript levels between XO and XX animals is estimated to be ∼20-fold (Trent et al. 1991). Using qRT–PCR, we found that normalized her-1 transcript levels were elevated 6.5-fold in sex-1 mutants (P < 0.01) and 4.6-fold in xol-1 sex-1 mutants (P < 0.01), consistent with the XSE-independent function of sex-1 acting downstream of xol-1 and upstream of her-1 (Table 7). The combined effect of the XSE-independent function of sex-1 on dosage compensation and sex determination suggests that it acts at the level of the sdc genes. Thus, sex-1 controls not only dosage compensation, but also sex determination, by acting in two separable capacities, as a repressor of xol-1 and also as a downstream inducer of the hermaphrodite fate, including the activation of dosage compensation.
TABLE 7.
sex-1 regulates her-1 transcript levels
| Transcript measured by qRT–PCRa
|
||
|---|---|---|
| Genotype | her-1b | fasn-1c |
| xol-1(y9) XX | 1.2 ± 0.2 | 1.0 ± 0.1 |
| sex-1(y424) XX | 6.5 ± 0.4 | 1.1 ± 0.1 |
| xol-1(y9) sex-1(y424) XX | 4.6 ± 0.7 | 1.0 ± 0.1 |
The levels of her-1 transcripts were measured in embryos of different genotypes (listed by genotype) by qRT–PCR and are expressed as the fold change compared to the transcript levels measured in wild-type XX embryos. For example, a value of 2.0 means that twice as many gene-specific transcripts were measured in mutant embryos than in wild-type XX embryos. All transcripts levels were normalized to the levels of the control gene, nhr-64, whose expression is not affected by dosage compensation. Fatty acid synthase fasn-1, another gene not affected by dosage compensation, was used as a control to gauge the variability and reliability of measurements made using qRT–PCR. See Van Gilst et al. (2005) for protocol. Experimental error is expressed as the standard error of the mean. Similar results were obtained for all genotypes in separate qRT–PCR experiments in which transcript levels were normalized to the levels of fasn-1.
qRT–PCR primers amplify both the short and long transcripts of her-1, which are coordinately regulated in a sex-specific manner despite being produced by two different her-1 promoters. Measurements thus reflect changes in total her-1 transcripts.
A critical control was to compare the nhr-64-normalized fasn-1 transcript levels in three independent preparations of wild-type embryos. That comparison showed the fasn-1 transcript levels to be statistically equivalent among the independent RNA preparations (fasn-1, 1.1 ± 0.2).
The XSE-independent function of sex-1 regulates dosage compensation by affecting the stability and localization of the dosage compensation complex:
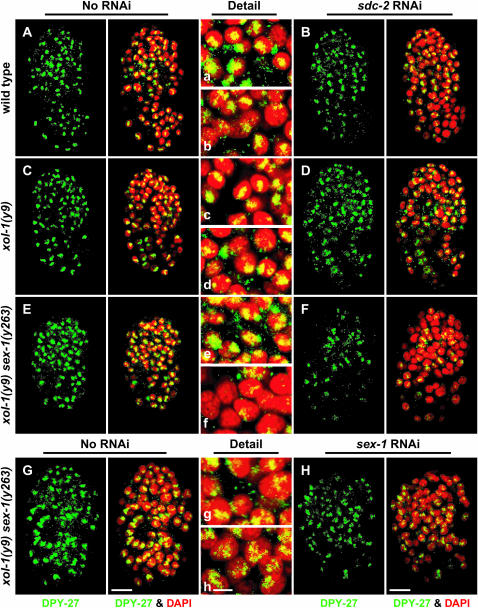
To determine the step at which the downstream function of sex-1 affects dosage compensation, the localization and abundance of the DCC component DPY-27 were examined in xol-1 XX mutants with reduced sex-1 and sdc-2 activities using DPY-27 antibodies. Disruption of sex-1 through RNAi in xol-1 sex-1(y263) mutants (58% viable, Table 5) caused only a mild decrease in DPY-27 staining (Figure 3, G and H and g and h). In contrast, the sex-1(y263) mutation reduced the DPY-27 staining and disrupted its assembly onto the X chromosomes of xol-1 sdc-2(RNAi) XX mutants (Figure 3, E and F and e and f). DPY-27 staining was undetectable in most nuclei, and the residual DPY-27 staining appeared somewhat diffuse in other nuclei. DCC localization in neither control sdc-2(RNAi) XX animals nor xol-1 sdc-2(RNAi) XX animals appeared significantly compromised (Figure 3, A–D and a–d).
Figure 3.—
X chromosome localization of the DCC is disrupted by inhibiting the XSE-independent function of sex-1. (A–H) Partial projections of false-colored confocal images of wild-type and mutant XX embryos costained with antibodies against the dosage compensation protein DPY-27 (green) and DAPI (red). (a–h) Enlargements of nuclei from A–H, respectively. (A–D) RNAi treatment of sdc-2 in wild-type and xol-1 mutant embryos mildly disrupted the X localization of DPY-27, as indicated by reduced DPY-27 levels and diffuse nuclear localization. (E and F) RNAi of sdc-2 disrupted DPY-27 more severely in xol-1 sex-1 mutants than in either wild-type or xol-1 animals, indicating that the XSE-independent function of sex-1 is important for proper stability and assembly of the DCC on X. (G and H) RNAi of sex-1 in xol-1 sex-1 XX embryos disrupts DPY-27 localization, indicating that loss of only the XSE-independent function of sex-1 impairs dosage compensation. Bars (A–H), 10 μm; (a–h), 3 μm.
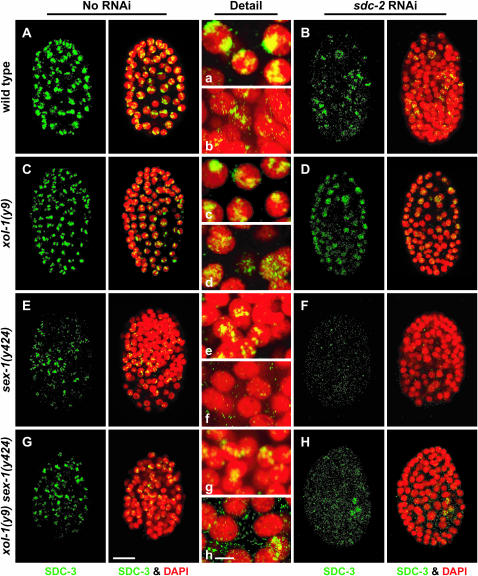
Similarly, SDC-3 staining and localization appeared only partially disrupted in sex-1(y424 null) mutants (20% viable), xol-1 sex-1(y424 null) mutants (56% viable), or sdc-2(RNAi) animals, but were severely disrupted in sex-1(y424 null) sdc-2(RNAi) mutants (0% viable) or xol-1(y9) sex-1(y424 null) sdc-2(RNAi) mutants (0% viable) Figure 4, A–H and a–h). This result further demonstrates that loss of the XSE-independent function of sex-1 disrupts the dosage compensation complex.
Figure 4.—
DCC disruption by a sex-1 null mutation and by the XSE-independent function of sex-1. (A–H) Partial projections of false-colored confocal images of wild-type and mutant XX embryos costained with antibodies against the dosage compensation protein SDC-3 (green) and DAPI (red). (a–h) Enlargements of nuclei from A–H, respectively. (A–D) RNAi disruption of sdc-2 in wild-type and xol-1 mutant embryos reduced the abundance and partially disrupted the X localization of SDC-3. (E and F) The sex-1(y424) null allele partially disrupted dosage compensation, but X-localized SDC-3 was evident in many nuclei. Some nuclei lacked SDC-3 staining. RNAi of sdc-2 into sex-1(y424) completely disrupted the DCC. Virtually no SDC-3 was detectable. (G and H) RNAi of sdc-2 in xol-1 sex-1 mutants also disrupted SDC-3 severely, similarly to xol-1(y9) sex-1(y424) sdc-2(RNAi) embryos, showing that the XSE-independent function of sex-1 is important for proper stability and assembly of the DCC on X. Bars: A–H, 10 μm; a–h, 3 μm.
The differential DCC disruption explains why partial depletion of sdc-2 activity by RNAi reduces XX viability to a greater extent in xol-1 sex-1 animals than in wild-type animals or in xol-1 mutants. These results indicate that the XSE-independent function of sex-1 promotes the stability and proper localization of the DCC. The dual functions of sex-1, as a direct repressor of xol-1 and as a downstream positive effector of hermaphrodite sexual fate and dosage compensation, account for why sex-1 mutations confer more severe phenotypes in XX animals than mutations in other XSEs such as ceh-39, which appear to relay the X signal strictly through xol-1.
After XSEs communicate X chromosome dose to determine sex, the dosage compensation process equalizes expression of XSEs between the sexes:
The twofold difference in copy number of X signal elements between XO and XX embryos forms the basis for C. elegans sex determination. Once sex is determined, the dosage compensation machinery is activated in XX animals, providing the potential for the dosage compensation process to feed back and repress expression of the very X-linked XSEs that activate the DCC. If so, the DCC can modulate expression of the X signal in a temporal manner and diminish the sex signal in XX animals. To determine whether expression of XSEs is controlled by the DCC, we first determined the expression level of each XSE in XO and XX embryos after sex had been determined using quantitative RT–PCR and then measured the XSE transcript levels in dosage-compensation-defective XX mutants.
We compared fox-1, ceh-39, and sex-1 transcript levels in XX and XO embryos of two genotypes, wild-type XX hermaphrodite embryos, and her-1; xol-1 sdc-2 XO hermaphrodite embryos (Table 8). This latter strain grows as an XO hermaphrodite strain that lacks the dosage compensation machinery. The xol-1 mutation would kill all XO animals by activating the dosage compensation machinery, but the sdc-2 mutation rescues the XO animals by preventing dosage compensation proteins from loading onto X. The her-1 mutation transforms the XO animals into hermaphrodites. The majority of animals in the population are XO, but some XX and nullo-X animals are also present. We found the XSE transcript levels to be equivalent in the XO and XX embryos, suggesting that the XSEs are dosage compensated (Table 8).
TABLE 8.
XSEs become dosage compensated after sex is determined
| Transcript measured by qRT–PCRa
|
||||
|---|---|---|---|---|
| Genotype | sex-1 | ceh-39 | fox-1 | nhr-64b |
| her-1; xol-1(y9) sdc-2(y74) unc-9 XOc | 0.8 ± 0.1 | 1.3 ± 0.1 | 0.9 ± 0.1 | 1.1 ± 0.1 |
| sdc-2(y93, RNAi) XXd | 2.9 ± 0.2 | 5.2 ± 0.6 | 2.0 ± 0.1 | 1.0 ± 0.1 |
| dpy-27(y57) XX | 1.8 ± 0.1 | 1.8 ± 0.2 | 1.3 ± 0.1 | 1.3 ± 0.1 |
| sex-1(y263) XX | 0.9 ± 0.1e | 2.8 ± 0.4 | 2.1 ± 0.5 | 1.3 ± 0.1 |
| sex-2(y324) XX | 1.4 ± 0.1 | 2.6 ± 0.3 | 2.2 ± 0.3 | 1.2 ± 0.1 |
| ceh-39(y414) XX | 1.2 ± 0.1 | 3.9 ± 0.4 | 1.8 ± 0.2 | 1.1 ± 0.1 |
| ceh-39(gk296) XX | 1.3 ± 0.1 | NA | 1.3 ± 0.1 | 1.1 ± 0.1 |
| fox-1(y303) XX | 1.2 ± 0.1 | 1.3 ± 0.1 | 1.4 ± 0.1 | 1.3 ± 0.1 |
| xol-1(y9) sex-1(y263) XX | 0.9 ± 0.1 | 1.8 ± 0.2 | 0.9 ± 0.1 | 1.1 ± 0.1 |
| yIs58[ceh-39(+)] XX | 1.0 ± 0.1 | 4.1 ± 0.5 | 1.1 ± 0.1 | 0.9 ± 0.1 |
| yIs58[ceh-39(+)]; sex-1(y263) XX | 1.0 ± 0.1 | 26.4 ± 2.8 | 3.6 ± 0.5 | 1.2 ± 0.1 |
| yIs58[ceh-39(+)]; xol-1(y9) sex-1(y263) XX | 1.0 ± 0.1 | 3.1 ± 0.3 | 2.3 ± 0.2 | 1.0 ± 0.1 |
The levels of XSE transcripts in embryos of different genotypes (listed by genotype) were measured by qRT–PCR and are expressed as the fold change compared to the transcript levels measured in wild-type XX embryos. All transcripts levels were normalized to the levels of the control gene, fatty acid synthase-1 (fasn-1), whose expression is constant throughout embryogenesis and is not affected by dosage compensation. See Van Gilst et al. (2005) for details and protocol. nhr-64, another gene not affected by dosage compensation, was used as a control to gauge the variability and reliability of measurements made using qRT–PCR. Experimental error is expressed as the standard error of the mean. Similar results were obtained for all genotypes in separate qRT–PCR experiments in which transcript levels were normalized to the levels of nhr-64.
A critical control was to compare the fasn-1-normalized nhr-64 transcript levels in three independent preparations of wild-type embryos. That comparison showed the nhr-64 transcript levels to be statistically equivalent among the independent RNA preparations (nhr-64, 1.3 ± 0.1).
The full genotype is her-1(hv1y101); xol-1(y9) sdc-2(y74) unc-9(101) XO.
XX animals carrying the weak sdc-2(y93) mutation were fed sdc-2(RNAi), and the dead, dosage-compensation-defective progeny embryos were harvested for RNA isolation.
The sex-1(y263) mutation affects sex-1 mRNA splicing and may destabilize the mutant sex-1 transcripts, causing a decrease in their overall levels, specifically in sex-1 mutants.
A further criterion for a dosage-compensated gene is that the transcript levels not only should be equivalent between the sexes, but also should be elevated in XX mutants defective in dosage compensation. The levels of XSE transcripts were first measured in the hypomorphic dpy-27(y57) XX mutant strain, which is 77% viable and therefore only partially defective in dosage compensation (Table 8). Both sex-1 and ceh-39 transcript levels were found to be elevated 1.8-fold above the levels in wild-type XX animals (P < 0.01 for sex-1, P = 0.017 for ceh-39), but the fox-1 transcript level was not significantly elevated in the dpy-27(y57) strain. The XSE transcript levels were then assessed in XX mutants more severely disrupted in dosage compensation. Fully dosage-compensation-defective XX mutants cannot be propagated as a strain; therefore we devised a means to obtain XX embryos that had died from severe disruption of dosage compensation. A large population of XX larvae carrying the very weak sdc-2(y93) mutation (100% viable, Nusbaum and Meyer 1989) were treated with sdc-2(RNAi) through feeding for one generation, and the dead, dosage-compensation-defective progeny embryos (100% inviable) were harvested for RNA isolation. This growth regime permits acquisition of reasonable quantities of severely dosage-compensation-defective animals for transcript analysis. The majority of embryos were at a developmental stage in which their sex should already have been determined. The transcript levels of all three XSEs were elevated by the severe reduction of sdc-2 activity: sex-1 by 2.9-fold (P < 0.01), ceh-39 by 5.2-fold (P < 0.01), and fox-1 by 2-fold (P < 0.01). Thus, the XSEs meet the second criterion for a dosage-compensated gene.
We also found that disruption of dosage compensation caused an elevation of xol-1 transcript levels in XX animals, suggesting that xol-1 is also repressed in response to the activation of dosage compensation. In sdc-2(y93, RNAi) XX embryos, the fasn-1-normalized xol-1 transcript level was increased 4.5 ± 0.6-fold compared to that in wild-type XX embryos, and in dpy-27(y57) XX embryos, the normalized xol-1 transcript level was increased 1.9 ± 0.2-fold.
Two important conclusions can be drawn from these experiments. First, the XSEs are dosage compensated, indicating that the very signal that communicates the difference in X chromosome dose between XO and XX animals is diminished once dosage compensation is fully activated in the embryo. Lowering xol-1 transcript levels in XX animals in response to dosage compensation could partially compensate for the reduction in expression of XSEs, the xol-1 repressors. Ultimately, however, the sex-determination decision must be maintained and transmitted to newly developing cells by sex-determination and dosage compensation genes that act downstream of xol-1. Second, XSE transcripts levels are derepressed by more than twofold when the dosage compensation complex is disrupted, implying that XSEs are repressed more than twofold by dosage compensation, and the difference in XSE expression between XO and XX embryos prior to dosage compensation is potentially greater than twofold.
Disruption of dosage compensation by mutation of one XSE feeds back to enhance expression of other XSEs:
XSE transcript levels are also affected by mutations in the XSE genes themselves (Table 8). For example, in a sex-2 mutant, ceh-39 transcript levels were elevated 2.6-fold (P < 0.01) and fox-1 transcript levels 2.2-fold (P < 0.01) above their levels in wild-type animals. Also, ceh-39 and fox-1 transcript levels were elevated 2.8-fold and 2.1-fold (P < 0.01 for both), respectively, in sex-1 mutants compared to wild-type animals. This latter elevation was suppressed in large part by a xol-1 mutation, which reduced the increase in ceh-39 transcript levels from 2.8- to 1.8-fold (P = 0.02) and the increase in fox-1 transcript levels from 2.1- to 0.9-fold (P < 0.01). The fact that wild-type xol-1 function is required to elevate the levels of XSE transcripts in XSE mutants indicates that this effect is caused by disruption of dosage compensation and not due to direct regulation among the XSEs themselves.
The most dramatic effect of an XSE mutation on the transcript level of another XSE was evident in sex-1 mutant XX embryos carrying a transgenic array expressing ceh-39 (Table 8). yIs58[ceh-39(+)] animals express ceh-39 transcripts at a level 4.1-fold higher than that in wild-type embryos, and the level increased by 6.5-fold in sex-1 mutants to a level 26.4-fold above that in wild-type animals. This increase in ceh-39 transcript levels was suppressed by a xol-1 mutation. In yIs58; xol-1 sex-1 mutants, ceh-39 transcripts drop back down to a level 3.1-fold higher than in wild-type embryos, further emphasizing that the increase in XSE transcript levels in XSE mutants is not caused by direct regulation of one XSE by another, but rather is a consequence of a dosage compensation disruption.
yIs58 contains multiple copies of the ceh-39 locus integrated into an autosome, yet it appears to be affected by the X chromosome dosage compensation process. At least two explanations might account for this phenomenon. First, the ceh-39 locus may contain a DCC recruitment site that attracts the DCC to the yIs58 integration site and permits ceh-39 to be repressed. Second, an X-linked activator of ceh-39 might be dosage compensated, and the sex-1 mutation causes this activator's expression to increase, thereby indirectly increasing ceh-39 expression in yIs58. The former possibility is unlikely because the DCC did not localize to the yIs58 locus in XX animals (data not shown). The latter case is potentially true for any X-linked gene and is a caveat that must be considered in assessing whether the process of dosage compensation represses an XSE directly or instead acts indirectly by repressing the XSE's potential regulator.
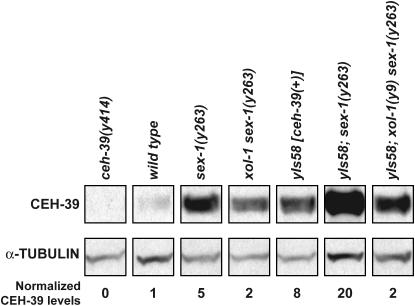
The changes in ceh-39 transcript levels measured by qRT–PCR correlated with changes in CEH-39 protein levels assessed by Western blots (Figure 5). CEH-39 levels were elevated 5-fold in sex-1 mutants compared to levels in wild-type animals and the increase in protein level was suppressed by a xol-1 mutation, as was the increase in ceh-39 transcript levels. CEH-39 levels were only 2-fold higher in xol-1 sex-1 animals. CEH-39 levels made from yIs58[ceh-39(+)] were also subjected to repression through the dosage compensation process. CEH-39 levels were 8- and 20-fold higher in yIs58[ceh-39(+)] animals and in yIs58[ceh-39(+)]; sex-1 animals, respectively, compared to levels in wild-type animals and were suppressed 2-fold higher in yIs58[ceh-39(+)]; xol-1 sex-1 animals compared to levels in wild-type animals (Figure 5). Thus, the dosage compensation disruption caused by loss of sex-1's XSE activity created a feedback loop that elevated ceh-39 expression.
Figure 5.—
CEH-39 levels are affected by the process of dosage compensation. A Western blot of whole-cell extracts from wild-type and mutant XX embryos shows that CEH-39 levels increase when sex-1 is mutant. The levels of CEH-39 in all protein extracts were normalized to the levels of α-tubulin. The normalized CEH-39 levels were then compared in mutant and wild-type (N2) extracts. CEH-39 was detected by CEH-39 antibodies in all extracts except those from ceh-39(y414) mutant embryos, which lacked the epitope for the antibody. CEH-39 level was increased by 5-fold in sex-1 mutants compared to wild-type embryos and by 2-fold in xol-1 sex-1 mutants, indicating that the increase in CEH-39 level was caused by a dosage compensation disruption. The level of CEH-39 in yIs58[ceh-39(+)] embryos was increased by 8-fold compared to that in wild-type embryos, showing that CEH-39 is indeed overexpressed in this strain. A sex-1 mutation caused CEH-39 to be expressed at an even higher level from yIs58 (20-fold); mutation of xol-1 greatly reduced the CEH-39 level (2-fold).
DISCUSSION
We have shown that the earliest aspects of sex determination are regulated through a more elaborate mechanism than previously understood. The X component of the X:A sex-determination signal includes more elements than first predicted, and each has a more modest effect on sex determination than the nuclear receptor SEX-1. The more potent influence of SEX-1 on sex determination and dosage compensation derives from its dual roles in the pathway: as an XSE to repress xol-1 and as a downstream activator of hermaphrodite fate. Furthermore, the dosage compensation process reduces expression of the XSEs once the X:A signal has determined sex. After the activation of dosage compensation, expression levels of XSEs are equivalent between XX and XO embryos, indicating that the X:A signal cannot guide sexual fate decisions later in development.
Composition of the X signal:
Prior studies using heterozygous X chromosome deletions to remove one copy of XSEs in XX animals suggested that reducing the dose of only four XSEs by half was sufficient to switch the X:A signal from its XX mode of xol-1 repression to its XO mode of xol-1 activation (Akerib and Meyer 1994; Carmi and Meyer 1999). In contrast, the severity of phenotypes that we observed by reducing the dose of XSEs using mutations rather than chromosomal deficiencies was less extreme, suggesting that the deletions also reduced the dose of additional, undefined XSEs and caused an overestimate of the contribution made by an individual XSE to the X:A signal. In our analysis, reducing the dose of four individual XSEs (sex-1, sex-2, ceh-39, and fox-1) by half was not sufficient to mimic the XO state. Moreover, the XO state is unlikely to be induced in XX animals only by halving the dose of these four XSEs plus the XSE in region 1, which is comparable in strength to ceh-39. Thus, the X component of the X:A signal likely utilizes more than five XSEs, more than previously predicted, to communicate the distinction between one X chromosome and two. This interpretation was reinforced by our reciprocal analysis of XSE mutations in XO animals. XSE mutations only partially suppressed the male lethality caused by increasing the XSE dose through X duplications, while mutations in downstream dosage compensation genes such as sdc-2 suppressed the lethality fully (Akerib and Meyer 1994), further indicating that the residual male lethality was likely caused by the activity of undefined XSEs.
In their study analyzing the effects of X duplications on the sex of polyploid animals, Madl and Herman (1979) concluded that the right side of X likely harbors sex-determining genes that participate in X:A assessment. While it remains a possibility that XSEs are encoded on that part of X and can account in part for our undefined XSE activity, the feminizing effect described by Madl and Herman (1979) is equally likely due to the partial disruption of dosage compensation caused by the duplications. The right-end duplications bind the DCC, which is limiting in supply, and titrate the DCC from X (Csankovszki et al. 2004). Those duplications, like mutations in dosage compensation genes, can suppress the extensive masculinization of sdc-3(Tra) XX mutants (Delong et al. 1993). Furthermore, those duplications abrogate the dosage compensation of genes not covered by them (Meneely and Nordstrom 1988). New genetic screens without bias for X chromosome location are needed to identify additional XSEs.
Different combinations of heterozygous XSE mutations revealed that the effects on sex determination and dosage compensation were more extreme if the XSEs with reduced dose included those that controlled xol-1 at both transcriptional and post-transcriptional levels. These experiments were carefully controlled to ensure that only combinations of XSEs with equivalent strengths were compared. Our results confirm the view that both transcriptional and post-transcriptional mechanisms of repression are important for xol-1 regulation, but make it clear that more of the XSEs control xol-1 at the transcript level.
Relative strength of XSEs:
The involvement in the X:A signal of more XSEs than previously projected is coupled to the current understanding that each XSE makes a smaller contribution to the signal than anticipated by mere extrapolation from the strength of the second-discovered XSE, sex-1. sex-1 null mutations cause 80% of XX animals to die, but mutations in other XSEs (ceh-39, fox-1, and sex-2) cause insignificant XX lethality. The relative strengths of the XSEs were determined by more sensitive assays that revealed their relative contributions to be sex-1 > sex-2 > fox-1 > ceh-39 ≥ region 1 XSE. The picture has thus emerged that multiple different X signal elements, most with a modest repressive effect on xol-1, act cumulatively to communicate X dose using two different mechanisms of xol-1 repression. The modest individual contribution of each XSE to the X:A signal accounts in part for the large number of XSEs (at least five) needed to regulate xol-1 and thereby communicate X chromosome dose.
sex-1 regulates the sex-determination pathway at multiple steps:
sex-1 has an unusually strong effect on sex determination and dosage compensation compared to other XSEs. Our work revealed the underlying cause of this phenomenon: sex-1 functions in two separate capacities at different steps in the pathway. sex-1 acts upstream in the pathway as an XSE to repress xol-1 and downstream in the pathway at the level of sdc genes to activate the hermaphrodite mode of sex determination and dosage compensation.
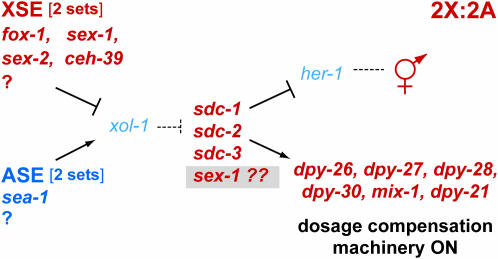
Hints of this complexity first came from the paradox that, although sex-1 mutations cause stronger dosage compensation phenotypes than mutations in other XSEs, they do not cause substantially greater derepression of xol-1 transcript levels. Isolation of a sex-1 null allele helped us to demonstrate the multi-faceted roles of sex-1. First, the XX lethality caused by the sex-1 null allele is only partially suppressed by a xol-1 null mutation, showing that while sex-1 acts as an XSE to regulate xol-1, it also functions in a separate capacity. Furthermore, mutations in all XSEs enhance the XX lethality caused by partial disruption of the downstream gene sdc-2, but xol-1 mutations do not suppress the enhanced lethality caused by sex-1 mutations, although they do suppress the enhanced lethality caused by ceh-39 and sex-2 mutations. Thus, sex-1, but apparently not other XSEs, acts independently of the X signal to regulate both sex determination and dosage compensation. Finally, the XSE-independent role of sex-1 in the sex-determination pathway acts downstream of xol-1, as revealed by two experiments. A sex-1 mutation can further suppress the lethality of xol-1 XO animals whose dosage compensation machinery has already been partially disrupted. In addition, in a xol-1 XX mutant, a sex-1 mutation causes derepression of her-1 transcript levels. her-1 is a male sex determination gene that acts downstream of xol-1 and is repressed by sdc genes in XX animals (Figure 6). The downstream function of sex-1 appears to be strong, since 44% of xol-1(null) sex-1(null) XX mutants are dead.
Figure 6.—
Current model of the sex-determination and dosage compensation pathway in C. elegans. Regulation of the sex-determination pathway is more complex than previously thought. The XSE sex-1 functions in two separate capacities at different steps in the pathway, accounting for its more potent effects on sex determination and dosage compensation than any other XSE. sex-1 acts upstream in the pathway as an XSE to repress xol-1. It also acts downstream of xol-1 in the pathway to activate the hermaphrodite mode of sex determination and dosage compensation. Although the nature of the XSE-independent function is unknown, it appears to act at the level of sdc genes. sex-1 could, for example, control an sdc gene or its protein activity, or it could function with an undefined gene at the same step as the sdc genes. Alternatively, it could control the sex-determination and dosage compensation branches of the pathway independently.
Although the nature of the XSE-independent sex-1 activity is unknown, it is functionally similar to the activities of sdc genes (Figure 6). sex-1 might, for example, control the activity of an sdc gene or the stability of the SDC protein complex, or it could function with undefined genes in the sex-determination pathway at the same step as the sdc genes. Alternatively, sex-1 could control the sex-determination and dosage compensation branches of the pathway independently. Regardless of the mechanism, the dual functions of sex-1 likely account for its more potent impact on sex determination and dosage compensation than any other XSE. These functions act synergistically. The synergistic lethality caused by the loss of both the XSE function and the XSE-independent function of sex-1 is analogous to the synergistic lethality caused by the loss of a weaker XSE (ceh-39 or fox-1) and a weak dosage compensation disruption due to sdc-2(RNAi).
Dosage compensation and the regulation of XSEs:
The use of an X-chromosome-counting mechanism to determine sex creates the potential paradox that the very genes, the XSEs, that communicate X chromosome dose, could themselves be subjected to the process that they regulate, X chromosome dosage compensation. If XSEs become dosage compensated, the primary signal that communicates the difference in X chromosome dose between the sexes by repressing xol-1 in XX animals would be reduced and would likely cease to exist during embryogenesis. Either the XSEs must therefore escape dosage compensation in XX embryos or the commitment to sexual fate must be firmly established by the onset of dosage compensation.
We found the XSEs to be dosage compensated as judged by two criteria: XSE expression was equivalent in XX and XO embryos that had already determined their sex, and XSE transcript levels were elevated in dosage-compensation-defective XX mutants. Thus, the process of dosage compensation influences the X signal in a temporal manner, and after the onset of dosage compensation, the XSEs cannot be used as a reference to guide sexual differentiation. Downstream genes in the sex-determination pathway that are turned on or off in response to xol-1's activity state must maintain the initial choice of sexual fate.
The discovery that XSEs are dosage compensated fits well with the previous observation that, toward the end of gastrulation, synthesis of xol-1 becomes dispensable in XO embryos and inconsequential in XX embryos (Rhind et al. 1995). These results suggested that an irreversible commitment to sexual fate had occurred by then and that assessment of the X:A signal was no longer necessary. Our results substantiate this view and further establish that the X:A signal cannot function after dosage compensation has been implemented, a time that precedes the end of gastrulation. The partial repression of xol-1 expression in XX embryos by the dosage compensation process would help keep xol-1 properly regulated until its level of activity was no longer of consequence to the embryo. Thus far, all lines of experiments have indicated that the X:A signal is assessed during a brief time window in embryogenesis and is not assessed continuously throughout the rest of development.
Finally, we found that the expression of XSEs is derepressed by more than twofold in embryos deficient in dosage compensation, implying that, in XX embryos, the XSEs are downregulated more than twofold by the DCC. The simplest interpretation of these data is that expression of XSEs differs by greater than twofold between XX and XO animals prior to dosage compensation. The probable mechanisms underlying such surprising signal amplification are not known and might range from gene-specific forms of control such as autoregulation to more general forms of regulation, perhaps related to the strategy for dosage compensation. Whatever the mechanism, such amplification would make assessment of X chromosome dose between XX and XO embryos a more robust process.
Acknowledgments
We thank M. Van Gilst for guidance with quantitative RT–PCR, T. Cline for discussions, and M. Jow and A. Severson for critical comments on the manuscript. This work was supported by National Institutes of Heath grant GM30702 to B.J.M., a National Science Foundation Predoctoral Fellowship to J.M.G., and the American Cancer Society Postdoctoral Fellowship PF-06-027-01 to B.F. B.J.M. is an investigator of the Howard Hughes Medical Institute.
References
- Akerib, C. C., and B. J. Meyer, 1994. Identification of X chromosome regions in Caenorhabditis elegans that contain sex-determination signal elements. Genetics 138: 1105–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmi, I., and B. J. Meyer, 1999. The primary sex determination signal of Caenorhabditis elegans. Genetics 152: 999–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmi, I., J. B. Kopczynski and B. J. Meyer, 1998. The nuclear hormone receptor SEX-1 is an X-chromosome signal that determines nematode sex. Nature 396: 168–173. [DOI] [PubMed] [Google Scholar]
- Chu, D. S., H. E. Dawes, J. D. Lieb, R. C. Chan, A. F. Kuo et al., 2002. A molecular link between gene-specific and chromosome-wide transcriptional repression. Genes Dev. 16: 796–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang, P. T., D. G. Albertson and B. J. Meyer, 1994. DPY-27: a chromosome condensation protein homolog that regulates C. elegans dosage compensation through association with the X chromosome. Cell 79: 459–474. [DOI] [PubMed] [Google Scholar]
- Csankovszki, G., P. McDonel and B. J. Meyer, 2004. Recruitment and spreading of the C. elegans dosage compensation complex along X chromosomes. Science 303: 1182–1185. [DOI] [PubMed] [Google Scholar]
- Davis, T. L., and B. J. Meyer, 1997. SDC-3 coordinates the assembly of a dosage compensation complex on the nematode X chromosome. Development 124: 1019–1031. [DOI] [PubMed] [Google Scholar]
- Dawes, H. E., D. S. Berlin, D. M. Lapidus, C. Nusbaum, T. L. Davis et al., 1999. Dosage compensation proteins targeted to X chromosomes by a determinant of hermaphrodite fate. Science 284: 1800–1804. [DOI] [PubMed] [Google Scholar]
- Delong, L., J. D. Plenefisch, R. D. Klein and B. J. Meyer, 1993. Feedback control of sex determination by dosage compensation revealed through Caenorhabditis elegans sdc-3 mutations. Genetics 133: 875–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladden, J. M., and B. J. Meyer, 2007. A ONECUT homeodomain protein communicates X chromosome dose to specify Caenorhabditis elegans sexual fate by repressing a sex switch gene. Genetics 177: 1621–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin, J., H. R. Horvitz and S. Brenner, 1979. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics 91: 67–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin, J., J. D. Zellan and D. G. Albertson, 1994. Identification of a candidate primary sex determination locus, fox-1, on the X chromosome of Caenorhabditis elegans. Development 120: 3681–3689. [DOI] [PubMed] [Google Scholar]
- Klein, R. D., and B. J. Meyer, 1993. Independent domains of the SDC-3 protein control sex determination and dosage compensation in C. elegans. Cell 72: 349–364. [DOI] [PubMed] [Google Scholar]
- Lieb, J. D., E. E. Capowski, P. Meneeley and B. J. Meyer, 1996. DPY-26, a link between dosage compensation and meiotic chromosome segregation in the nematode. Science 274: 1732–1736. [DOI] [PubMed] [Google Scholar]
- Lieb, J. D., M. R. Albrecht, P. T. Chuang and B. J. Meyer, 1998. MIX-1: an essential component of the C. elegans mitotic machinery executes X chromosome dosage compensation. Cell 92: 265–277. [DOI] [PubMed] [Google Scholar]
- Madl, J. E., and R. K. Herman, 1979. Polyploids and sex determination in Caenorhabditis elegans. Genetics 93: 393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonel, P. E., J. Jans, B. K. Peterson and B. J. Meyer, 2006. Clustered DNA motifs mark X chromosomes for repression by a dosage compensation complex. Nature 444: 614–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneely, P. M., and K. D. Nordstrom, 1988. X chromosome duplications affect a region of the chromosome they do not duplicate in Caenorhabditis elegans. Genetics 119: 365–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, B. J., 2005. X–chromosome dosage compensation, in WormBook, edited by The C. elegans Research Community. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Miller, L. M., J. D. Plenefisch, L. P. Casson and B. J. Meyer, 1988. xol-1: a gene that controls the male modes of both sex determination and X chromosome dosage compensation in C. elegans. Cell 55: 167–183. [DOI] [PubMed] [Google Scholar]
- Nicoll, M., C. C. Akerib and B. J. Meyer, 1997. X-chromosome-counting mechanisms that determine nematode sex. Nature 388: 200–204. [DOI] [PubMed] [Google Scholar]
- Nigon, V., 1951. Polyploidie experimentale chez un nematode libre, Rabditis elegans Maupas. Bull. Biol. Fr. Belg. 85: 187–225. [Google Scholar]
- Nonet, M., and B. J. Meyer, 1991. Early aspects of Caenorhabditis elegans sex determination and dosage compensation are regulated by a zinc-finger protein. Nature 351: 65–68. [DOI] [PubMed] [Google Scholar]
- Nusbaum, C., and B. J. Meyer, 1989. The Caenorhabditis elegans gene sdc-2 controls sex determination and dosage compensation in XX animals. Genetics 122: 579–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plenefisch, J. D., L. Delong and B. J. Meyer, 1989. Genes that implement the hermaphrodite mode of dosage compensation in Caenorhabditis elegans. Genetics 121: 57–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell, J., M. Jow and B. Meyer, 2005. The T-box transcription factor SEA-1 is an autosomal element of the X:A signal that determines C. elegans sex. Dev. Cell 9: 339–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind, N. R., L. M. Miller, J. B. Kopczynski and B. J. Meyer, 1995. xol-1 acts as an early switch in the C. elegans male/hermaphrodite decision. Cell 80: 71–82. [DOI] [PubMed] [Google Scholar]
- Riddle, D. L., T. Blumenthal, B. J. Meyer and J. R. Priess, 1997. C. elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- Trent, C., B. Purnell, S. Gavinski, J. Hageman and W. B. Wood, 1991. Sex-specific transcriptional regulation of the C. elegans sex-determination gene her-1. Mech. Dev. 34: 43–56. [DOI] [PubMed] [Google Scholar]
- Van Gilst, M. R., H. Hadjivassiliou, A. Jolly and K. R. Yamamoto, 2005. Nuclear hormone receptor NHR-49 controls fat consumption and fatty acid composition in C. elegans. PLoS Biol. 3: e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve, A. M., 1994. A cis-acting locus that promotes crossing over between X chromosomes in Caenorhabditis elegans. Genetics 136: 887–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve, A. M., and B. J. Meyer, 1987. sdc-1: a link between sex determination and dosage compensation in C. elegans. Cell 48: 25–37. [DOI] [PubMed] [Google Scholar]
- Villeneuve, A. M., and B. J. Meyer, 1990. The role of sdc-1 in the sex determination and dosage compensation decisions in Caenorhabditis elegans. Genetics 124: 91–114. [DOI] [PMC free article] [PubMed] [Google Scholar]