Abstract
As a central component of the DNA damage checkpoint pathway, the conserved protein kinase Chk1 mediates cell cycle progression when DNA damage is generated. Msc1 was identified as a multicopy suppressor capable of facilitating survival in response to DNA damage of cells mutant for chk1. We demonstrate that loss of msc1 function results in an increased rate of chromosome loss and that an msc1 null allele exhibits genetic interactions with mutants in key kinetochore components. Multicopy expression of msc1 robustly suppresses a temperature-sensitive mutant (cnp1-1) in the centromere-specific histone H3 variant CENP-A, and localization of CENP-A to the centromere is compromised in msc1 null cells. We present several lines of evidence to suggest that Msc1 carries out its function through the histone H2A variant H2A.Z, encoded by pht1 in fission yeast. Like an msc1 mutant, a pht1 mutant also exhibits chromosome instability and genetic interactions with kinetochore mutants. Suppression of cnp1-1 by multicopy msc1 requires pht1. Likewise, suppression of the DNA damage sensitivity of a chk1 mutant by multicopy msc1 also requires pht1. We present the first genetic evidence that histone H2A.Z may participate in centromere function in fission yeast and propose that Msc1 acts through H2A.Z to promote chromosome stability and cell survival following DNA damage.
THE fission yeast Schizosaccharomyces pombe has proven to be a useful model system for studies of cell cycle events, including DNA replication, mitosis, and cytokinesis. These events must be executed with high fidelity to ensure chromosome integrity. Checkpoints monitor key events during the cell cycle and block subsequent events if earlier ones are incomplete, thereby increasing the fidelity of DNA replication and chromosome segregation (Hartwell and Weinert 1989; Murray 1992; Hartwell and Kastan 1994). The DNA damage checkpoint is activated in response to genotoxic stress such as irradiation or aberrant DNA replication (O'Connell et al. 2000) and delays cell cycle progression by inhibiting the activity of cyclin-dependent kinases (Cdk's), key regulators of cell cycle progression in all eukaryotic cells (Morgan 1997). In fission yeast, Cdc2 is the primary Cdk, which is dephosphorylated at tyrosine 15 to promote entry into mitosis (Gould and Nurse 1989). Tyrosine phosphorylation of Cdc2 is maintained by Wee1 and Mik1 (Lundgren et al. 1991), while the Cdc25 phosphatase dephosphorylates Cdc2 at the same site to initiate mitosis (Millar et al. 1991). In response to DNA damage, the protein kinase Chk1 is phosphorylated and inhibits mitotic entry by phosphorylating Wee1 and Cdc25 to prevent activation of Cdc2 (Rhind et al. 1997; Guo et al. 2000; Liu et al. 2000; Raleigh and O'Connell 2000; Lopez-Girona et al. 2001; Capasso et al. 2002). The phosphorylation of Chk1 is dependent on the protein kinase Rad3 (Walworth and Bernards 1996), which belongs to a subgroup of a phosphatidylinositol 3 kinase-like family that includes the human proteins ATM and ATR (Abraham 2001; Shiloh 2001; Melo and Toczyski 2002).
A fission yeast protein, Msc1, related to mammalian proteins RBP2 and PLU-1, was identified as a multicopy suppressor of cells defective for chk1 function (Ahmed et al. 2004). RBP2, first isolated as a protein that interacts with the tumor suppressor Rb, is postulated to act as both a transcriptional activator and a repressor, depending on the context (Chan and Hong 2001; Benevolenskaya et al. 2005). PLU-1 was first isolated as a transcript upregulated in breast cancer cells that is normally expressed mainly in the testis and during development (Lu et al. 1999; Madsen et al. 2002). PLU-1 interacts with transcription factors to repress transcription in a reporter assay system (Tan et al. 2003). Like RBP2 and PLU-1, Msc1 has multiple domains suggestive of a role in modulating chromatin structure and/or function, including three plant homeodomain (PHD) motifs and Jumonji N (JmjN) and Jumonji C (JmjC) domains, although Msc1 lacks the AT-rich interacting domain (ARID) common to RBP2 and PLU-1 (Ahmed et al. 2004). While cells lacking msc1 are relatively resistant to DNA-damaging agents, loss of msc1 function exacerbates the DNA damage sensitivity resulting from loss of chk1 function. Furthermore, msc1 null cells are sensitive to the histone deacetylase inhibitor trichostatin A (TSA), as they lose viability relative to wild-type cells. Thus, it is possible that Msc1 plays a role in maintaining proper chromatin structure and function.
The faithful segregation of chromosomes at mitosis requires the coordinated action of multiple cell cycle checkpoints that monitor replication of the genome and the attachment of sister chromatids to the mitotic spindle apparatus (Amon 1999). Attachment of spindle microtubules to chromosomes occurs through the kinetochore, a specialized protein structure that associates with the centromeric region of the chromosome (Cleveland et al. 2003; Amor et al. 2004). Nucleosomes at the centromere contain CENP-A, a centromere-specific histone H3 variant. In fission yeast, loading of CENP-A at the centromere is dependent on the Mis6 complex and is promoted by the Ams2 protein (Takahashi et al. 2000; Chen et al. 2003; Hayashi et al. 2004). Mis6, a homolog of vertebrate CENP-I (Saitoh et al. 1997; Nishihashi et al. 2002), forms a complex with Mis15, Mis17, and Sim4 (Pidoux et al. 2003; Hayashi et al. 2004). Another evolutionarily conserved kinetochore protein, Mis12, forms a complex with Mis13 and Mis14 (Hayashi et al. 2004; Obuse et al. 2004), but Mis12 is dispensable for CENP-A loading in fission yeast (Goshima et al. 1999, 2003). In vertebrate cells, the Mis12 complex has been proposed to form the core binding site within the kinetochore for the attachment of spindle microtubules (Cheeseman et al. 2006). Fission yeast cells bearing mutations in kinetochore complex components exhibit unequal chromosome segregation (Saitoh et al. 1997; Goshima et al. 1999; Takahashi et al. 2000; Pidoux et al. 2003). In the event that microtubules do not attach or tension on the spindle is relieved, the mitotic spindle checkpoint is activated to prevent the onset of anaphase, exit from mitosis, and initiation of cytokinesis (Lew and Burke 2003; Pinsky and Biggins 2005). Mis6 is required in fission yeast for loading of the mitotic spindle checkpoint protein Mad2 at the kinetochore (Saitoh et al. 2005).
While kinetochore protein structures are key elements in maintaining chromosome stability, the integrity of the centromeric chromatin, upon which kinetochores assemble, is critical as well. The function of mammalian centromeres relies not only on the histone variant CENP-A, but also on the histone H2A variant H2A.Z (Rangasamy et al. 2004). Recently, it has been shown that histone H2A.Z and CENP-A form distinct domains within the centromeric region (Greaves et al. 2007). While histone H2A.Z has not yet been localized in fission yeast, cells with a disruption in the pht1 gene encoding histone H2A.Z exhibit chromosome loss (Carr et al. 1994). In budding yeast, histone H2A.Z is found throughout the genome, specifically within one or two nucleosomes that flank nucleosome-free promoters, perhaps establishing marks for the activation of transcription (Guillemette et al. 2005; Li et al. 2005; Raisner et al. 2005; Zhang et al. 2005; Millar et al. 2006). A chromatin-remodeling complex containing the Swr1 protein facilitates the exchange of histone H2A.Z for the core histone H2A (Mizuguchi et al. 2004).
In this study, we characterize the phenotype of cells lacking the fission yeast gene msc1. Msc1 is required for chromosome stability and exhibits synthetic genetic interactions with at least two critical kinetochore components, Mis6 and Mis12. Multicopy expression of Msc1 suppresses loss of function of the centromere-specific histone H3 variant CENP-A encoded by the temperature-sensitive allele cnp1-1. In addition, localization of wild-type CENP-A to the centromere is compromised in cells lacking msc1. As Msc1 coprecipitates components of the Swr1 chromatin-remodeling complexes, we also tested whether Msc1 might act through histone H2A.Z. Indeed, the ability of Msc1 to rescue a chk1 mutant or a cnp1 mutant requires the pht1 gene. Given the role of the kinetochore in establishing proper spindle function, we tested whether suppression of chk1 by Msc1 might require the spindle checkpoint protein Mad2 and found that it does. Thus, we suggest that Msc1 promotes chromosome stability in fission yeast by facilitating deposition or retention of the histone H3 variant CENP-A at the centromere. Furthermore, genetic evidence suggests that the ability of Msc1 to maintain chromosome stability and allow for appropriate responses to cell cycle checkpoints requires the presence of the histone H2A variant H2A.Z.
MATERIALS AND METHODS
Strains and growth condition:
Strains are listed in Table 1. Standard genetic methods were utilized for strain construction (Moreno et al. 1991). Cells were grown at 30° unless otherwise indicated. Survival following UV treatment was determined as described previously (Walworth et al. 1993). For spotting assays, cells were grown to mid-log phase and 10-fold serial dilutions were made. Aliquots of 5 μl of each dilution were spotted on plates. For monitoring mitotic entry in the presence of a microtubule inhibitor, log-phase cdc25-22 cells were shifted to 36° for 3 hr to synchronize them in G2 and then shifted to 25° in the presence of 50 μg/ml thiobendazole (TBZ, Sigma, St. Louis). Samples were collected at 20-min intervals, fixed with 70% ethanol, and stained with DAPI and calcofluor before counting binucleate cells under the fluorescent microscope. Immunofluorescence studies were performed as described previously (Ahmed et al. 2004) with TAT-1 antibody to α-tubulin (a kind gift of Keith Gull) and DAPI staining to visualize nuclei. For determining the percentage of cells with lagging chromosomes, cells were grown at 18° and cells with elongated anaphase spindles as visualized with TAT-1 antibody were counted (Ekwall et al. 1995). The number of cells in anaphase with more than two DAPI staining spots were counted as having lagging chromosomes.
TABLE 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| SP6 | h−leu1-32 | Lab stock |
| NW309 | h+ leu1-32 nda3-KM311 | Lab stock |
| NW653 | h+ leu1-32 ura4D18 chk1∷ura4 cdc17-K42 ade6-216 | Lab stock |
| NW731 | h+ leu1-32 msc1∷kanR cdc25-22 | Lab stock |
| NW1469 | h−leu1-32 mis12-HA∷LEU2 | Mitsuhiro Yanagida |
| NW1525 | h−leu1-32 ura4D18 cnp1∷ura4 lys1+∷cnp1-1 | Mitsuhiro Yanagida |
| NW1526 | h−leu1-32 lys1+∷cnp1-GFP | Kohta Takahashi |
| NW1564 | h−leu1-32 msc1∷kanR | This study |
| NW1580 | h−leu1-32 mis12-375 | Mitsuhiro Yanagida |
| NW1601 | h−leu1-32 msc1∷kanR nda3-KM311 | This study |
| NW1608 | h−leu1-32 msc1-myc∷kanR | This study |
| NW1617 | h+ leu1-32 mis12-375 msc1∷kanR | This study |
| NW1698 | h−leu1-32 ura4D18 cdc25-22 | This study |
| NW1699 | h+ leu1-32 ura4D18 cdc25-22 msc1∷kanR | This study |
| NW1700 | h+ leu1-32 ura4D18 cdc25-22 mad2∷ura4 ade6-210 | Matthew O'Connell |
| NW1702 | h−leu1-32 mis6-302 | Mitsuhiro Yanagida |
| NW1703 | h+ leu1-32 mis6-302 msc1∷kanR ade6-210 | This study |
| NW1704 | h+ leu1-32 ura4D18 mad2∷ura4 chk1∷ura4 cdc17-K42 ade6-216 | This study |
| NW1713 | h−leu1-32 ura4D18 mad2∷ura4 chk1∷ura4 ade6-216 | This study |
| NW1718 | h−leu1-32 ura4D18 mad2∷ura4 cdc17-K42 ade6-210 | This study |
| NW1736 | h+ leu1-32 ade6-210 ura4D18 msc1∷kanR cnp1∷ura4 lys1+∷cnp1-1 | This study |
| NW1737 | h−leu1-32 lys1+∷cnp1-GFP msc1∷kanR | This study |
| NW1800 | h+ leu1-32 ade6-210 msc1∷kanR | This study |
| NW1828 | h+ leu1-32 ade6-210 ura4D18 pht1∷ura4 | Antony Carr |
| NW1829 | h+ leu1-32 ade6-210 ura4D18 pht1∷ura4 msc1∷kanR | This study |
| NW1830 | h−leu1-32 ade6-210 ura4D18 mis6-302 pht1∷ura4 msc1∷kanR | This study |
| NW1831 | h−leu1-32 ura4D18 mis12-375 pht1∷ura4 | This study |
| NW1832 | h+ leu1-32 ura4D18 mis12-375 pht1∷ura4 msc1∷kanR | This study |
| NW1833 | h−leu1-32 ade6-210 ura4D18 pht1∷ura4 cdc17-K42 | This study |
| NW1834 | h+ leu1-32 ade6-210 ura4D18 arg3-D4 pht1∷ura4 chk1∷arg3 cdc17-K42 | This study |
| NW1835 | h+ leu1-32 ade6-210 arg3-D4 chk1∷arg3 cdc17-K42 | This study |
| NW1836 | h−leu1-32 ade6-210 ura4D18 mis6-302 cnp1∷ura4 lys1+∷cnp1-1 | This study |
| NW1837 | h+ leu1-32 ade6-210 ura4D18 pht1∷ura4 cnp1∷ura4 lys1+∷cnp1-1 | This study |
| NW1839 | h+ leu1-32 ade6-210 ura4D18 pht1∷ura4 msc1∷kanR cnp1∷ura4 lys1+∷cnp1-1 | This study |
| NW1854 | h+ ade6-210 (Ch16 ade6-216) leu1-32 | Chris Norbury |
| NW1855 | h+ ade6-210 (Ch16 ade6-216) msc1∷kanR leu1-32 | This study |
| NW1856 | h+ ade6-210 (Ch16 ade6-216) pht1∷ura4 leu1-32 ura4D18 | This study |
| NW1857 | h+ ade6-210 (Ch16 ade6-216) msc1∷kanR pht1∷ura4 leu1-32 ura4D18 | This study |
Chromosome loss assay:
The minichromosome Ch16 (Niwa et al. 1986) was introduced into msc1∷kanR and msc1+ backgrounds using standard genetic techniques. Cultures were grown to mid-log phase in medium lacking adenine at 30°. An aliquot was removed to determine the percentage of Ade− cells. Remaining cells were then grown for 24 hr at 30° in rich medium containing adenine, and samples were removed and plated on media with limiting adenine. The number of red colonies were counted and chromosome loss was calculated from the following formula: loss rate = 1 − (F/I)1/N, where F is the final percentage of white colonies and I is the initial percentage of white colonies, while N is the number of generations between I and F.
Chromatin immunoprecipitations:
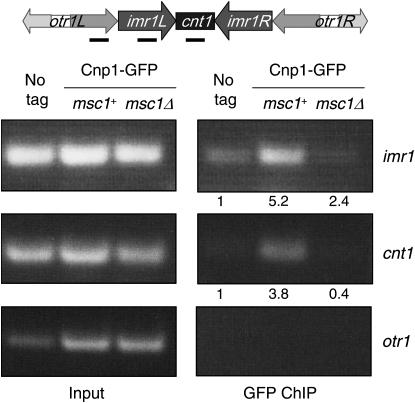
Strains with GFP-tagged cnp1 (Takahashi et al. 2000) were grown in 100-ml cultures to mid-log phase. Cells were pelleted and resuspended in 40 ml of YEA to which 1.4 ml of 37% formaldehyde was added. The samples were shaken gently at 25° for 15 min. Six milliliters of 1.25 m glycine was added, and the samples were transferred to 50-ml tubes and incubated overnight in the cold room. The following day the samples were washed twice with ice-cold TBS and resuspended in 400 μl of FALB (50 mm HEPES–potassium hydroxide, 150 mm NaCl, 1 mm EDTA, 1% Triton-X-100, 0.1% Na-deoxycholate) plus protease inhibitors (PI) (Ren et al. 2000). Cells were lysed with acid-washed glass beads in a Fastprep vortexing machine (Bio101). The lysate was centrifuged at 16,100 × g for 5 min at 4° and the supernatant was discarded. The pellets were resuspended in 800 μl of FALB+PI. These samples were then sonicated at a 30% duty cycle for 20 sec four times and for 10 sec once. The samples were pelleted in a microfuge at 16,100 × g for 10 min at 4°. The supernatants were harvested and brought to a final volume of 1 ml. A volume of 200 μl was set aside as the input fraction and the remaining material was precleared by incubating with washed sepharose beads in the cold room for 30 min. The samples were centrifuged at 4000 × g and the supernatant was transferred to a new tube and incubated with antibody overnight. The next day, 40 μl of sepharose bead volume was added to capture immune complexes and incubated in the cold room for 1 hr. The samples were pelleted and the beads were washed while rotating with the following buffers, each for 10 min in the cold room: FALB, FALB–500 mm NaCl, wash buffer (10 mm Tris–Cl, pH 8.0, 250 mm LiCl, 1 mm EDTA, 0.5% NP-40, 0.5% Na-deoxycholate), and TE. After the TE wash, the immunoprecipitate (IP) beads were resuspended in 400 μl of TE. The input samples were brought up to 400 μl as well. Five microliters of 10 mg/ml RNase A were added and incubated at 37° for 30 min. To reverse the crosslinks, 20 μl of 10% SDS was added and the samples were incubated at 65° for 20 hr. The next day, the samples were incubated with 10 μl of 10 mg/ml proteinase K for 6 hr at 55°. The samples were then purified using a DNA QIAGEN (Valencia, CA) column or by phenol–chloroform extraction and precipitated. One microliter of each IP sample and 1 μl of each input sample (diluted fivefold) was used in the PCR reaction. Samples were amplified for 28 cycles, which results in reaction products that are linear with respect to the amount of template DNA used in the reactions. PCR products were separated on an agarose gel and photographed with a digital camera. Photographs were scanned and analyzed with National Institutes of Health ImageJ for quantitation of signals, which were in the linear range of the detection program. Enrichments were calculated as the ratio of signal at cnt or imr relative to otr [where CENP-A is not expected to bind (Takahashi et al. 2000)] for the immunoprecipitates, normalized to the ratio of signals for the input DNA. Normalized enrichments, reported in Figure 4, were calculated as the ratio of E for the tagged strain vs. E for the untagged strain for the given PCR reaction.
Figure 4.—
CENP-A localization to the inner centromeric region is reduced in the absence of Msc1. Cells with GFP-tagged Cnp1 (CENP-A) with wild-type or a null allele of msc1, or an untagged control, were subjected to chromatin immunoprecipitation with antibody to GFP as described in materials and methods. DNA from the lysates (input) or the immunoprecipitations were subject to PCR with primers specific to the central (cnt), inner (imr), and outer (otr) regions of the fission yeast centromere on chromosome I. Enrichment of imr1 and cnt1 was assessed compared to otr1 in immunoprecipitates relative to input DNA. This experiment was performed three times with similar results.
RESULTS
Msc1 is important for chromosome stability:
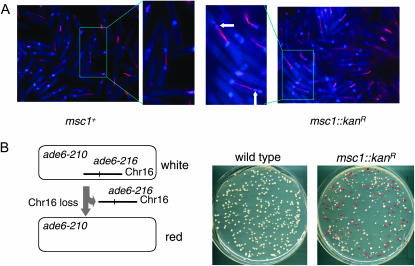
Previously we isolated Msc1 as a multicopy suppressor of a mutant defective for the checkpoint kinase Chk1. We showed that Msc1 associates with chromatin and coprecipitates a histone deacetylase activity (Ahmed et al. 2004). In the course of examining the msc1 null strain, we observed an unusual DAPI-staining pattern with individual cells often containing an extra DAPI-staining spot suggestive of lagging chromosomes. Therefore, we compared chromosome segregation in msc1∷kanR and msc1+ cells synchronized in G2 and released into mitosis. As shown in Figure 1A, cells lacking msc1 exhibit multiple DAPI spots along the mitotic spindle (white arrows), whereas this phenotype is not observed in the wild-type cells. To quantitate this phenotype, cells were grown at low temperature and the frequency of lagging chromosomes on late anaphase spindles was determined as described (Ekwall et al. 1995). While wild-type cells exhibit lagging chromosomes at a frequency of 2.0 ± 0.3% (SD), cells lacking msc1 show an elevated frequency of 12.6 ± 4.3% (SD).
Figure 1.—
Msc1 is required for chromosome stability. (A) Strains with a cdc25-22 mutant allele were synchronized in G2 by incubation at 36.5° and released to permissive temperature. After 90 min, cells were fixed with gluteraldehyde and processed for immunofluorescence with antitubulin antibody (red). Nuclei are stained with DAPI (blue). Portions of the micrographs in boxes are enlarged for better viewing of normal (left) and lagging (right) chromosomes (white arrows). (B) Schematic of chromosome loss assay. Ch16 is a minichromosome carrying the ade6-216 allele that complements in trans the ade6-210 allele (Niwa et al. 1986). Loss of Ch16 results in cells that solely express the ade6-210 allele, which confers a red color to the colonies. From the parent strains that are either msc1+ or msc1∷kanR, single white colonies were grown as described in materials and methods. Equal numbers of cells of the indicated strains were plated on plates with a limiting concentration of adenine.
Strains that exhibit the lagging-chromosome phenotype often exhibit an elevated rate of chromosome loss as well. To determine whether Msc1 is required for chromosome stability, we incorporated Ch16, a minichromosome derived from chromosome III (Niwa et al. 1986), in an msc1 knockout strain by genetic manipulation. This strain carries the ade6-210 mutation in the genome and the ade6-216 mutation on the minichromosome, making the strain phenotypically Ade+ and white in color because of trans-complementation of ade6-210 and ade6-216. We examined the loss of the minichromosome by growing cells under nonselective conditions for several generations and then scoring the number of Ade− red colonies produced after plating on media with limiting adenine. As shown in Figure 1B and quantified in Table 2, the rate of chromosome loss in the msc1∷kanR deletion strain is elevated ∼30-fold compared to an isogenic wild-type strain.
TABLE 2.
Frequency of chromosome loss
| Strain | Chromosome loss (rate/generation) | Fold difference |
|---|---|---|
| Wild type | 0.00024 | 1 |
| msc1Δ | 0.0083 | 35 |
| pht1Δ | 0.0036 | 15 |
| msc1Δ pht1Δ | 0.0078 | 33 |
Chromatin structure at the core centromere region shows a unique pattern after micrococcal nuclease (MNase) digestion (Polizzi and Clarke 1991; Takahashi et al. 1992). Since Msc1 has been implicated in modification of histones (Ahmed et al. 2004), and a dramatic change in chromatin structure at the centromere could influence attachment of chromosomes to the mitotic spindle, we examined the nucleosome pattern following MNase digestion of chromatin isolated from wild-type and from msc1 null cells. No differences were detected in the digestion pattern of centromeric chromatin when wild-type and msc1∷kanR cells were examined (supplemental data at http://www.genetics.org/supplemental/), indicating that no gross change in nucleosome organization results from the absence of msc1.
Msc1 exhibits genetic interactions with Mis12 and Mis6:
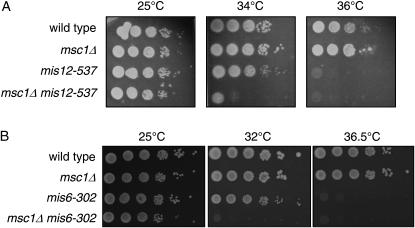
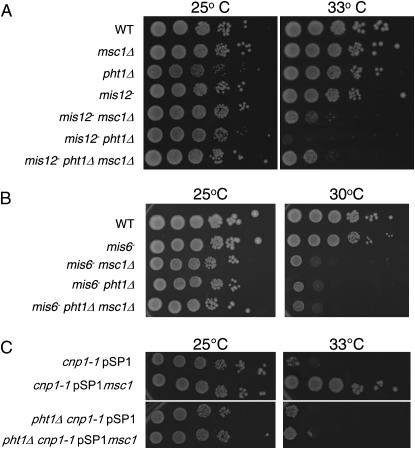
The lagging-chromosome phenotype is common in fission yeast mutants that affect chromosome segregation and kinetochore function. To evaluate whether Msc1 influences kinetochore function, double mutants with strains harboring temperature-sensitive mutations in the genes encoding Mis12 and Mis6 (Saitoh et al. 1997; Goshima et al. 1999) were constructed. Deletion of msc1 in the temperature-sensitive mis12-537 background results in a lower restrictive temperature as compared to mis12-537 alone (Figure 2A). While the mis12-537 mutant readily forms colonies at 34°, the double-mutant mis12-537 msc1∷kanR is compromised for growth at this temperature. Likewise, a mis6-302 msc1∷kanR mutant is compromised for growth at 32° whereas the mis6-302 strain is viable at this temperature (Figure 2B).
Figure 2.—
Msc1 exhibits genetic interactions with Mis12 and Mis6. (A and B) The indicated strains were grown at 25° to mid-log phase. Tenfold serial dilutions were spotted on rich media plates and incubated at the indicated temperatures for 3–4 days.
Multicopy expression of msc1 suppresses loss of function of the histone H3 variant cnp1-1:
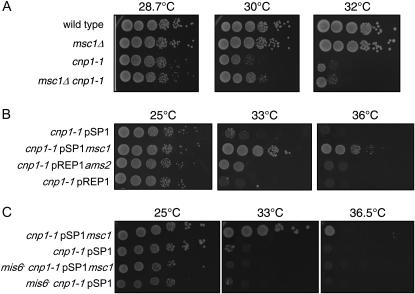
Chromosome stability relies not only on proper function of kinetochore proteins, but also on properly assembled centromeric chromatin. The histone H3 variant CENP-A, encoded by cnp1 in S. pombe, localizes to the cnt and imr regions of S. pombe centromeric chromatin (Kniola et al. 2001). Deletion of msc1 in a temperature-sensitive mutant cnp1-1 background does not exacerbate cnp1-1 temperature sensitivity; indeed, it seems to improve growth somewhat at 30° (Figure 3A), although this behavior is not apparent when single colonies are streaked on plates (data not shown). On the contrary, multicopy expression of msc1 suppresses the temperature sensitivity of cnp1-1 (Figure 3B), as does overexpression of ams2, a gene identified by virtue of this property. Ams2 is implicated in loading of CENP-A at the centromere (Chen et al. 2003). Mis6 is also required for CENP-A to localize to centromeres (Takahashi et al. 2000), and as shown in Figure 3C, the ability of multicopy msc1 to suppress cnp1-1 requires mis6 function.
Figure 3.—
An msc1 null is not synthetically sick with cnp1-1, but multicopy msc1 suppresses loss of function of CENP-A. (A) The indicated strains were grown at 25° to mid-log phase. Tenfold serial dilutions were spotted on rich media plates and incubated at the indicated temperatures for 3–4 days. (B and C) The cnp1-1 (B) or cnp1-1 mis6 (C) strain was transformed with the indicated plasmids. Cells were grown at 25° in minimal media to mid-log phase. Tenfold serial dilutions were spotted on minimal media plates and incubated at the indicated temperatures for 3–4 days.
To determine whether Msc1 affects localization of fission yeast CENP-A to the centromere, we performed chromatin immunoprecipitation experiments using GFP-tagged Cnp1. As shown in Figure 4, Cnp1-GFP localizes to the inner regions of the centromere (cnt and imr), but not to the outer region (otr) (Takahashi et al. 2000). In a mutant null for msc1, association of Cnp1-GFP to either cnt or imr is dramatically reduced.
Histone H2A.Z exhibits genetic interactions with mis6 and mis12 and is required for pmsc1 to suppress cnp1-1:
Msc1 associates with Swr1 and other components of the Swr1 chromatin-remodeling complex (X. Qiu, S. Ahmed and N. C. Walworth, unpublished results). Since Swr1 exchanges histone H2A.Z for histone H2A (Mizuguchi et al. 2004), we hypothesized that Msc1 might carry out its functions through histone H2A.Z. Consistent with this hypothesis, deletion of the H2A.Z-encoding gene in S. pombe, pht1, has been reported to result in elevated chromosome loss rates (Carr et al. 1994). In our hands, the loss rate for a pht1 null strain is elevated 15-fold above wild-type levels, whereas deletion of msc1 or simultaneous deletion of both pht1 and msc1 results in an ∼30-fold increase relative to wild-type cells (Table 2).
To determine if the pht1 deletion strain affects kinetochore function, crosses were carried out with mis12-537 and mis6-302. As shown in Figure 5, deletion of pht1 lowers the restrictive temperatures of mis12-537 even more so than does deletion of msc1 (Figure 5A). Like deletion of msc1, deletion of pht1 also reduces the restrictive temperature of mis6-302 (Figure 5B). A triple mutant of mis6 msc1Δ pht1Δ is as temperature sensitive as either mis6-302 msc1Δ or mis6-302 pht1Δ (Figure 5B). Paradoxically, the mis12 pht1Δ msc1Δ strain exhibits improved growth as compared to mis12 pht1Δ, equivalent to that seen for mis12 msc1Δ (Figure 5A).
Figure 5.—
A pht1 null mutant exhibits genetic interactions similar to msc1. (A and B) The indicated strains were grown at 25° to mid-log phase. Tenfold serial dilutions were spotted on rich media plates and incubated at the indicated temperatures for 3–4 days. (C) The cnp1-1 or cnp1-1 pht1Δ strain was transformed with the indicated plasmids. Cells were grown at 25° in minimal media to mid-log phase. Tenfold serial dilutions were spotted on minimal media plates and incubated at the indicated temperatures for 3–4 days.
To determine whether suppression of the temperature sensitivity of cnp1-1 by multicopy pmsc1 requires histone H2A.Z, a double mutant of pht1Δ and cnp1-1 was constructed and transformed with the pmsc1 plasmid. As shown in Figure 5C, pmsc1 is unable to rescue cnp1-1 in the absence of histone H2A.Z.
Histone H2A.Z and Mad2 are required for pmsc1 to suppress chk1:
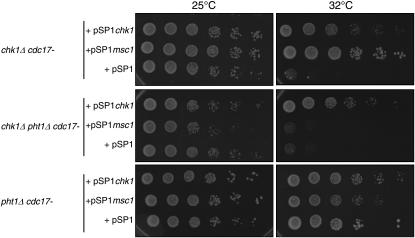
Msc1 was originally identified in fission yeast because of its ability to rescue a chk1-defective strain in which DNA ligase, encoded by cdc17, was mutated (Ahmed et al. 2004). We tested whether the ability of Msc1 to rescue a chk1Δ cdc17-K42 strain is also dependent on histone H2A.Z. As shown in Figure 6, while multicopy pmsc1 supports growth of a chk1Δ cdc17-K42 strain at 32°, it cannot do so when pht1 is deleted (middle). Deletion of pht1 alone in the cdc17-K42 strain does not compromise viability at 32° (bottom). Thus, the ability of pmsc1 to rescue loss of chk1 function requires the presence of H2A.Z.
Figure 6.—
The ability of multicopy Msc1 to restore survival of a chk1Δ strain requires histone H2A.Z. The indicated strains transformed with the indicated plasmids were grown in minimal media to mid-log phase at 25°. Serial dilutions (10-fold) were spotted on yeast nitrogen media plates lacking leucine and incubated at the indicated temperatures.
Recent results from others suggest that Chk1 may play a role not only in preventing the onset of mitosis when DNA is damaged, but also in slowing events within mitosis, particularly the metaphase-to-anaphase transition (Collura et al. 2005). Indeed, cells lacking the mitotic spindle checkpoint protein Mad2 are slightly sensitive to DNA-damaging agents, consistent with a possible back-up role for this checkpoint in promoting survival following DNA damage (Collura et al. 2005). Taking these observations into account, we tested the possibility that suppression of the chk1 defect by multicopy msc1 might require the function of Mad2. We constructed a strain with deletions of both chk1 and mad2 in the cdc17-K42 background. As shown in Figure 7A, the triple mutant cannot be suppressed by expression of pmsc1 (second panel), whereas a strain with the mad2 gene intact can be suppressed (top panel). Thus, suppression of the chk1 defect by multicopy msc1 when DNA damage is present requires an intact spindle checkpoint. As expected, a pchk1 plasmid was able to suppress the growth defect in the triple-mutant background (Figure 7A, third panel) by restoring DNA damage checkpoint function. Furthermore, the cdc17-K42 mad2Δ strain transformed with either the control vector plasmid or the pmsc1 plasmid is equally viable at 32° (Figure 7A, bottom panel), suggesting that Mad2 does not play a major role in promoting survival when DNA ligase function is compromised as long as Chk1 function is intact.
Figure 7.—
The ability of multicopy Msc1 to restore survival of a chk1Δ strain requires Mad2 function, although Msc1 is not required directly for the spindle checkpoint. (A) The indicated strains transformed with the indicated plasmids were grown in minimal media to mid-log phase at 25°. Serial dilutions (10-fold) were spotted on yeast nitrogen media plates lacking leucine and incubated at the indicated temperatures. (B) Cells were blocked in G2 by incubation at 36.5° and then released at permissive temperature in the presence of TBZ (50 μg/ml). At 20-min intervals cells were fixed with gluteraldehyde and processed for immunofluorescence with antitubulin antibody and stained with DAPI. For wild-type and msc1Δ cells, the 100-min time point is shown. For mad2Δ cells, the 60-min time point is shown.
Msc1 does not play a direct role in the spindle checkpoint:
To determine whether Msc1 might play a direct role in the mitotic spindle checkpoint, we compared the behavior of an msc1Δ strain to that of a mad2 null. The null mutations of msc1 and mad2 were constructed in a cdc25-22 background to allow synchronization of the cell population in G2. Cells arrested in G2 by incubation at 36.5° were released to 25° in the presence of the microtubule-destabilizing drug TBZ. Samples were collected at 20-min intervals and examined for progression through mitosis by visualizing nuclei with DAPI. As shown in Figure 7B, wild-type and msc1Δ cells remain arrested with condensed chromosomes in the presence of TBZ, even at 100 min. In contrast, even at 60 min, mad2Δ cells have already attempted to segregate their chromosomes exhibiting the cut phenotype and chromosome decondensation, indicating loss of spindle checkpoint function and progression through mitosis (He et al. 1997).
DISCUSSION
This study demonstrates a role for fission yeast Msc1 in chromosome stability. Genetic interactions with core components of the kinetochore and with the centromere-specific histone H3 variant CENP-A suggest that Msc1 may influence either directly or indirectly the structure or function of the site at which the mitotic spindle associates with the chromosome. We postulate that Msc1 acts through the histone H2A variant H2A.Z because the ability of multicopy Msc1 to elicit phenotypes is dependent on the presence of the H2A.Z gene, pht1. Furthermore, a mutant deficient in H2A.Z (pht1Δ) shares some phenotypes with a mutant deficient in Msc1 (msc1Δ). CENP-A and H2A.Z have been shown to bind within distinct centromeric domains in mammalian chromosomes (Okada et al. 2006; Greaves et al. 2007). CENP-A is a critical functional component of the fission yeast centromere (Takahashi et al. 2000) and the work described here provides genetic evidence supporting the possibility that histone H2A.Z could function within the fission yeast centromere as well. Detailed analysis of H2A.Z localization in fission yeast has not as yet been reported.
Msc1 was first identified in fission yeast as a protein that in multiple copies restores survival to a strain lacking Chk1 function (Ahmed et al. 2004). Two homologs of Msc1 exist in mammalian cells, one of which (PLU-1) is upregulated in breast cancer cells (Lu et al. 1999) and both of which (PLU-1 and RBP2) have been implicated in the control of gene expression by virtue of their associations with other proteins (Fattaey et al. 1993; Mao et al. 1997; Lu et al. 1999; Tan et al. 2003; Catteau et al. 2004). In fission yeast, cells lacking Msc1 are viable, but exhibit sensitivity to the histone deacetylase inhibitor TSA (Ahmed et al. 2004). Msc1, like its mammalian counterparts, possesses several motifs consistent with roles in chromatin modification and/or transcriptional control. These include three PHD fingers, JmjN, and JmjC domains (Ahmed et al. 2004). However, RBP2 and PLU-1 possess an ARID motif that does not appear to be conserved in Msc1. Published data for RBP2 and PLU-1 currently favor roles in transcriptional regulation. Whether fission yeast Msc1 influences transcriptional control is under investigation.
PHD domains have been suggested to have two disparate activities. The structural similarity of PHD domains to RING domains (Pascual et al. 2000; Capili et al. 2001) suggested that they might possess E3 ubiquitin ligase activity, which has been demonstrated for several PHD motifs found in non-nuclear proteins (Coscoy et al. 2001; Lu et al. 2002; Goto et al. 2003; Yonashiro et al. 2006). More recently, the PHD domains of at least two sets of proteins have been shown to behave as histone H3 di- or trimethyl-binding domains (Li et al. 2006; Martin et al. 2006; Pena et al. 2006; Shi et al. 2006; Wysocka et al. 2006). The PHD fingers of Msc1 do not appear to bind to histones, but do possess ubiquitin E3 ligase activity (Dul and Walworth 2007). Whether the corresponding PHD domains of RBP2 or PLU-1 possess either function remains to be determined.
JmjC domains have garnered attention recently as a subset of such domains have been shown to possess histone demethylase activity (Tsukada et al. 2006). Indeed, the JmjC domains of RBP2 and PLU-1 are reported to demethylate histone H3-K4 (Christensen et al. 2007; Klose et al. 2007; Yamane et al. 2007). However, many of the amino acid residues implicated in iron and α-ketoglutarate binding, which are thought to be essential for enzymatic activity of this domain, are not conserved in Msc1 (Klose et al. 2006). Thus, elucidation of the functional activity of the JmjC domain in Msc1 will require further investigation.
The results of our phenotypic and genetic analysis of Msc1 lead us to hypothesize that the protein affects chromosome stability by directly or indirectly promoting kinetochore attachment to the mitotic spindle. The fact that multicopy Msc1 robustly suppresses a cnp1-1 mutant suggests that Msc1 might facilitate CENP-A incorporation or retention at the centromere, and indeed, CENP-A localization to the centromere is reduced in an msc1 null strain. However, the fact that cnp1 is an essential gene whereas msc1 is not indicates that a minimal level of CENP-A may be present at the centromere in the msc1 null strain. Indeed, the fact that centromeres in the msc1 null strain show a digestion pattern when exposed to nuclease (see supplemental Figure 2 at http://www.genetics.org/supplemental/) similar to that seen for wild-type cells suggests that CENP-A is present in sufficient amounts at the inner core to retain the chromatin structure characteristic of this region (Takahashi et al. 2000).
Histone H2A.Z must be present for multicopy pmsc1 to suppress the cnp1-1 mutant or to suppress loss of chk1 function. A molecular explanation for these requirements awaits characterization of the fission yeast Swr1 protein complex and determination of whether Swr1 promotes H2A.Z deposition as it does in budding yeast, as well as determination of the relationship of Msc1 to that complex. Notably, a clear homolog of Msc1 is absent from the budding yeast genome. It is possible that the mechanism by which Msc1 promotes cell survival in the absence of Chk1 may be through increasing the stability or segregation of damaged chromosomes. In support of this speculation, we note that suppression by pmsc1 of a chk1 mutant also requires an intact mitotic spindle checkpoint since suppression is lost in cells in which both chk1 and mad2 are compromised.
A relationship between various checkpoint pathways in fission yeast has begun to emerge. Sugimoto et al. (2004) reported that a hydroxyurea-induced delay to mitotic entry in cells lacking the protein kinase Cds1 requires the function of Mad2, just as it requires the function of Chk1 (Lindsay et al. 1998). In addition, Collura et al. (2005) have reported that Crb2, in conjunction with Chk1, is required to delay the metaphase-to-anaphase transition in cells treated with the topoisomerase I poison camptothecin in a Mad2-dependent fashion. The mechanism by which Chk1 influences the Mad2-dependent checkpoint pathway in fission yeast and the role that Msc1 plays in promoting checkpoint function remain to be determined.
Acknowledgments
We are grateful to Mitsuhiro Yanagida, Kohta Takahashi, Antony Carr, Matthew O'Connell, and Chris Norbury for providing numerous mutant strains and plasmids for these studies and Keith Gull for antitubulin antibody. We thank members of the Walworth lab for helpful discussions, Hui Rao for technical assistance, and Marc Gartenberg for critical reading of the manuscript. This work was supported by Public Health Service grant GM53194 from the National Institute of General Medical Sciences, National Institutes of Health.
References
- Abraham, R. T., 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15: 2177–2196. [DOI] [PubMed] [Google Scholar]
- Ahmed, S., C. Palermo, S. Wan and N. C. Walworth, 2004. A novel protein with similarities to Rb binding protein 2 compensates for loss of Chk1 function and affects histone modification in fission yeast. Mol. Cell. Biol. 24: 3660–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amon, A., 1999. The spindle checkpoint. Curr. Opin. Genet. Dev. 9: 69–75. [DOI] [PubMed] [Google Scholar]
- Amor, D. J., P. Kalitsis, H. Sumer and K. H. Choo, 2004. Building the centromere: from foundation proteins to 3D organization. Trends Cell Biol. 14: 359–368. [DOI] [PubMed] [Google Scholar]
- Benevolenskaya, E. V., H. L. Murray, P. Branton, R. A. Young and W. G. Kaelin, Jr., 2005. Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol. Cell 18: 623–635. [DOI] [PubMed] [Google Scholar]
- Capasso, H., C. Palermo, S. Wan, H. Rao, U. P. John et al., 2002. Phosphorylation activates Chk1 and is required for checkpoint-mediated cell cycle arrest. J. Cell Sci. 115: 4555–4564. [DOI] [PubMed] [Google Scholar]
- Capili, A. D., D. C. Schultz, F. J. Rauscher, III and K. L. B. Borden, 2001. Solution structure of the PHD domain from the KAP-1 corepressor: structural determinants for PHD, RING and LIM zinc-binding domains. EMBO J. 20: 165–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr, A. M., S. M. Dorrington, J. Hindley, G. A. Phear, S. J. Aves et al., 1994. Analysis of a histone H2A variant from fission yeast: evidence for a role in chromosome stability. Mol. Gen. Genet. 245: 628–635. [DOI] [PubMed] [Google Scholar]
- Catteau, A., I. Rosewell, E. Solomon and J. Taylor-Papadimitriou, 2004. A short region of the promoter of the breast cancer associated PLU-1 gene can regulate transcription in vitro and in vivo. Int. J. Oncol. 25: 5–16. [PubMed] [Google Scholar]
- Chan, S. W., and W. Hong, 2001. Retinoblastoma-binding protein 2 (Rbp2) potentiates nuclear hormone receptor-mediated transcription. J. Biol. Chem. 276: 28402–28412. [DOI] [PubMed] [Google Scholar]
- Cheeseman, I. M., J. S. Chappie, E. M. Wilson-Kubalek and A. Desai, 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127: 983–997. [DOI] [PubMed] [Google Scholar]
- Chen, E. S., S. Saitoh, M. Yanagida and K. Takahashi, 2003. A cell cycle-regulated GATA factor promotes centromeric localization of CENP-A in fission yeast. Mol. Cell 11: 175–187. [DOI] [PubMed] [Google Scholar]
- Christensen, J., K. Agger, P. A. Cloos, D. Pasini, S. Rose et al., 2007. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell 128: 1063–1076. [DOI] [PubMed] [Google Scholar]
- Cleveland, D. W., Y. Mao and K. F. Sullivan, 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112: 407–421. [DOI] [PubMed] [Google Scholar]
- Collura, A., J. Blaisonneau, G. Baldacci and S. Francesconi, 2005. The fission yeast Crb2/Chk1 pathway coordinates the DNA damage and spindle checkpoint in response to replication stress induced by topoisomerase I inhibitor. Mol. Cell. Biol. 25: 7889–7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coscoy, L., D. J. Sanchez and D. Ganem, 2001. A novel class of herpesvirus-encoded membrane-bound E3 ubiquitin ligases regulates endocytosis of proteins involved in immune recognition. J. Cell Biol. 155: 1265–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dul, B. E., and N. C. Walworth, 2007. The plant homeodomain fingers of fission yeast Msc1 exhibit E3 ubiquitin ligase activity. J. Biol. Chem. 282: 18397–18406. [DOI] [PubMed] [Google Scholar]
- Ekwall, K., J. P. Javerzat, A. Lorentz, H. Schmidt, G. Cranston et al., 1995. The chromodomain protein Swi6: a key component at fission yeast centromeres. Science 269: 1429–1431. [DOI] [PubMed] [Google Scholar]
- Fattaey, A. R., K. Helin, M. S. Dembski, N. Dyson, E. Harlow et al., 1993. Characterization of the retinoblastoma binding proteins RBP1 and RBP2. Oncogene 8: 3149–3156. [PubMed] [Google Scholar]
- Goshima, G., S. Saitoh and M. Yanagida, 1999. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev. 13: 1664–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., T. Kiyomitsu, K. Yoda and M. Yanagida, 2003. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 160: 25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto, E., S. Ishido, Y. Sato, S. Ohgimoto, K. Ohgimoto et al., 2003. c-MIR, a human E3 ubiquitin ligase, is a functional homolog of herpesvirus proteins MIR1 and MIR2 and has similar activity. J. Biol. Chem. 278: 14657–14668. [DOI] [PubMed] [Google Scholar]
- Gould, K. L., and P. Nurse, 1989. Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature 342: 39–45. [DOI] [PubMed] [Google Scholar]
- Greaves, I. K., D. Rangasamy, P. Ridgway and D. J. Tremethick, 2007. H2A.Z contributes to the unique 3D structure of the centromere. Proc. Natl. Acad. Sci. USA 104: 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemette, B., A. R. Bataille, N. Gevry, M. Adam, M. Blanchette et al., 2005. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 3: e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Z., A. Kumagai, S. X. Wang and W. G. Dunphy, 2000. Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 14: 2745–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell, L. H., and M. B. Kastan, 1994. Cell cycle control and cancer. Science 266: 1821–1828. [DOI] [PubMed] [Google Scholar]
- Hartwell, L. H., and T. A. Weinert, 1989. Checkpoints: controls that ensure the order of cell cycle events. Science 246: 629–634. [DOI] [PubMed] [Google Scholar]
- Hayashi, T., Y. Fujita, O. Iwasaki, Y. Adachi, K. Takahashi et al., 2004. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 118: 715–729. [DOI] [PubMed] [Google Scholar]
- He, X., T. E. Patterson and S. Sazer, 1997. The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc. Natl. Acad. Sci. USA 94: 7965–7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose, R. J., E. M. Kallin and Y. Zhang, 2006. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 7: 715–727. [DOI] [PubMed] [Google Scholar]
- Klose, R. J., Q. Yan, Z. Tothova, K. Yamane, H. Erdjument-Bromage et al., 2007. The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell 128: 889–900. [DOI] [PubMed] [Google Scholar]
- Kniola, B., E. O'Toole, J. R. McIntosh, B. Mellone, R. Allshire et al., 2001. The domain structure of centromeres is conserved from fission yeast to humans. Mol. Biol. Cell 12: 2767–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew, D. J., and D. J. Burke, 2003. The spindle assembly and spindle position checkpoints. Annu. Rev. Genet. 37: 251–282. [DOI] [PubMed] [Google Scholar]
- Li, B., S. G. Pattenden, D. Lee, J. Gutierrez, J. Chen et al., 2005. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc. Natl. Acad. Sci. USA 102: 18385–18390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., S. Ilin, W. Wang, E. M. Duncan, J. Wysocka et al., 2006. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442: 91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay, H. D., D. J. F. Griffiths, R. J. Edwards, P. U. Christensen, J. M. Murray et al., 1998. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 12: 382–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q., S. Guntuku, X.-S. Cui, S. Matsuoka, D. Cortez et al., 2000. Chk1 is an essential kinase that is regulated by Atr and required for the G2/M DNA damage checkpoint. Genes Dev. 14: 1448–1459. [PMC free article] [PubMed] [Google Scholar]
- Lopez-Girona, A., K. Tanaka, X. B. Chen, B. A. Baber, C. H. McGowan et al., 2001. Serine-345 is required for Rad3-dependent phosphorylation and function of checkpoint kinase Chk1 in fission yeast. Proc. Natl. Acad. Sci. USA 98: 11289–11294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, F. J., K. Sundquist, D. Baeckstrom, R. Poulsom, A. Hanby et al., 1999. A novel gene (PLU-1) containing highly conserved putative DNA/chromatin binding motifs is sepcifcally up-regulated in breast cancer. J. Biol. Chem. 274: 15633–15645. [DOI] [PubMed] [Google Scholar]
- Lu, Z., S. Xu, C. Joazeiro, M. H. Cobb and T. Hunter, 2002. The PHD domain of MEKK1 acts as an E3 ubiquitin ligase and mediates ubiquitination and degradation of ERK1/2. Mol. Cell 9: 945–956. [DOI] [PubMed] [Google Scholar]
- Lundgren, K., N. Walworth, R. Booher, M. Dembski, M. Kirschner et al., 1991. mik1 and wee1 cooperate in the inhibitory tyrosine phosphorylation of cdc2. Cell 64: 1111–1122. [DOI] [PubMed] [Google Scholar]
- Madsen, B., B. Spencer-Dene, R. Poulsom, D. Hall, P. J. Lu et al., 2002. Characterisation and developmental expression of mouse Plu-1, a homologue of a human nuclear protein (PLU-1) which is specifically up-regulated in breast cancer. Gene Expr. Patterns 2: 275–282. [DOI] [PubMed] [Google Scholar]
- Mao, S., G. A. Neale and R. M. Goorha, 1997. T-cell oncogene rhombotin-2 interacts with retinoblastoma-binding protein 2. Oncogene 14: 1531–1539. [DOI] [PubMed] [Google Scholar]
- Martin, D. G., K. Baetz, X. Shi, K. L. Walter, V. E. MacDonald et al., 2006. The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Mol. Cell. Biol. 26: 7871–7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo, J., and D. Toczyski, 2002. A unified view of the DNA-damage checkpoint. Curr. Opin. Cell Biol. 14: 237–245. [DOI] [PubMed] [Google Scholar]
- Millar, C. B., F. Xu, K. Zhang and M. Grunstein, 2006. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 20: 711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, J. B. A., C. H. McGowan, G. Lenaers, R. Jones and P. Russell, 1991. p80cdc25 mitotic inducer is the tyrosine phosphatase that activates p34cdc2 kinase in fission yeast. EMBO J. 10: 4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi, G., X. Shen, J. Landry, W. H. Wu, S. Sen et al., 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303: 343–348. [DOI] [PubMed] [Google Scholar]
- Moreno, S., A. Klar and P. Nurse, 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194: 795–823. [DOI] [PubMed] [Google Scholar]
- Morgan, D. O., 1997. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 13: 261–291. [DOI] [PubMed] [Google Scholar]
- Murray, A. W., 1992. Creative blocks: cell-cycle checkpoints and feedback controls. Nature 359: 599–604. [DOI] [PubMed] [Google Scholar]
- Nishihashi, A., T. Haraguchi, Y. Hiraoka, T. Ikemura, V. Regnier et al., 2002. CENP-I is essential for centromere function in vertebrate cells. Dev. Cell 2: 463–476. [DOI] [PubMed] [Google Scholar]
- Niwa, O., T. Matsumoto and M. Yanagida, 1986. Construction of a mini chromosome by deletion and its mitotic and meiotic behaviour in fission yeast. Mol. Genet. Genomics 203: 397–405. [Google Scholar]
- Obuse, C., O. Iwasaki, T. Kiyomitsu, G. Goshima, Y. Toyoda et al., 2004. A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat. Cell Biol. 6: 1135–1141. [DOI] [PubMed] [Google Scholar]
- O'Connell, M. J., N. C. Walworth and A. M. Carr, 2000. The G2-phase DNA-damage checkpoint. Trends Cell Biol. 10: 296–303. [DOI] [PubMed] [Google Scholar]
- Okada, M., I. M. Cheeseman, T. Hori, K. Okawa, I. X. McLeod et al., 2006. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 8: 446–457. [DOI] [PubMed] [Google Scholar]
- Pascual, J., M. Martinez-Yamout, H. J. Dyson and P. E. Wright, 2000. Structure of the PHD zinc finger from human Williams-Beuren syndrome transcription factor. J. Mol. Biol. 304: 723–729. [DOI] [PubMed] [Google Scholar]
- Pena, P. V., F. Davrazou, X. Shi, K. L. Walter, V. V. Verkhusha et al., 2006. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 442: 100–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux, A. L., W. Richardson and R. C. Allshire, 2003. Sim4: a novel fission yeast kinetochore protein required for centromeric silencing and chromosome segregation. J. Cell Biol. 161: 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky, B. A., and S. Biggins, 2005. The spindle checkpoint: tension versus attachment. Trends Cell Biol. 15: 486–493. [DOI] [PubMed] [Google Scholar]
- Polizzi, C., and L. Clarke, 1991. The chromatin structure of centromeres from fission yeast: differentiation of the central core that correlates with function. J. Cell Biol. 112: 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner, R. M., P. D. Hartley, M. D. Meneghini, M. Z. Bao, C. L. Liu et al., 2005. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123: 233–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raleigh, J. M., and M. J. O'Connell, 2000. The G2 DNA damage checkpoint targets both Wee1 and Cdc25. J. Cell Sci. 113: 1727–1736. [DOI] [PubMed] [Google Scholar]
- Rangasamy, D., I. Greaves and D. J. Tremethick, 2004. RNA interference demonstrates a novel role for H2A.Z in chromosome segregation. Nat. Struct. Mol. Biol. 11: 650–655. [DOI] [PubMed] [Google Scholar]
- Ren, B., F. Robert, J. J. Wyrick, O. Aparicio, E. G. Jennings et al., 2000. Genome-wide location and function of DNA binding proteins. Science 290: 2306–2309. [DOI] [PubMed] [Google Scholar]
- Rhind, N., B. Furnari and P. Russell, 1997. Cdc2 tyrosine phosphorylation is required for the DNA damage checkpoint in fission yeast. Genes Dev. 11: 504–511. [DOI] [PubMed] [Google Scholar]
- Saitoh, S., K. Takahashi and M. Yanagida, 1997. Mis6, a fission yeast inner centromere protein, acts during G1/S and forms specialized chromatin required for equal segregation. Cell 90: 131–143. [DOI] [PubMed] [Google Scholar]
- Saitoh, S., K. Ishii, Y. Kobayashi and K. Takahashi, 2005. Spindle checkpoint signaling requires the mis6 kinetochore subcomplex, which interacts with mad2 and mitotic spindles. Mol. Biol. Cell 16: 3666–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, X., T. Hong, K. L. Walter, M. Ewalt, E. Michishita et al., 2006. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442: 96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh, Y., 2001. ATM and ATR: networking cellular responses to DNA damage. Curr. Opin. Genet. Dev. 11: 71–77. [DOI] [PubMed] [Google Scholar]
- Sugimoto, I., H. Murakami, Y. Tonami, A. Moriyama and M. Nakanishi, 2004. DNA replication checkpoint control mediated by the spindle checkpoint protein Mad2p in fission yeast. J. Biol. Chem. 279: 47372–47378. [DOI] [PubMed] [Google Scholar]
- Takahashi, K., S. Murakami, Y. Chikashige, H. Funabiki, O. Niwa et al., 1992. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol. Biol. Cell 3: 819–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, K., E. S. Chen and M. Yanagida, 2000. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science 288: 2215–2219. [DOI] [PubMed] [Google Scholar]
- Tan, K., A. L. Shaw, B. Madsen, K. Jensen, J. Taylor-Papadimitriou et al., 2003. Human PLU-1 has transcriptional repression properties and interacts with the developmental transcription factors BF-1 and PAX9. J. Biol. Chem. 278: 20507–20513. [DOI] [PubMed] [Google Scholar]
- Tsukada, Y., J. Fang, H. Erdjument-Bromage, M. E. Warren, C. H. Borchers et al., 2006. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439: 811–816. [DOI] [PubMed] [Google Scholar]
- Walworth, N. C., and R. Bernards, 1996. rad-dependent response of the chk1-encoded protein kinase at the DNA damage checkpoint. Science 271: 353–356. [DOI] [PubMed] [Google Scholar]
- Walworth, N., S. Davey and D. Beach, 1993. Fission yeast chk1 protein kinase links the rad checkpoint pathway to cdc2. Nature 363: 368–371. [DOI] [PubMed] [Google Scholar]
- Wysocka, J., T. Swigut, H. Xiao, T. A. Milne, S. Y. Kwon et al., 2006. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442: 86–90. [DOI] [PubMed] [Google Scholar]
- Yamane, K., K. Tateishi, R. J. Klose, J. Fang, L. A. Fabrizio et al., 2007. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol. Cell 25: 801–812. [DOI] [PubMed] [Google Scholar]
- Yonashiro, R., S. Ishido, S. Kyo, T. Fukuda, E. Goto et al., 2006. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J. 25: 3618–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., D. N. Roberts and B. R. Cairns, 2005. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123: 219–231. [DOI] [PMC free article] [PubMed] [Google Scholar]