Abstract
Interaction of monoglucosylated oligosaccharides with ER lectins (calnexin and/or calreticulin) facilitates glycoprotein folding but this interaction is not essential for cell viability under normal conditions. We obtained two distinct single Schizosaccharomyces pombe mutants deficient in either one of the two pathways leading to the formation of monoglucosylated oligosaccharides. The alg6 mutant does not glucosy- late lipid-linked oligosaccharides and transfers Man9GlcNAc2 to nascent polypeptide chains and the gpt1 mutant lacks UDP-Glc:glycoprotein glucosyltransferase (GT). Both single mutants grew normally at 28°C. On the other hand, gpt1/alg6 double-mutant cells grew very slowly and with a rounded morphology at 28°C and did not grow at 37°C. The wild-type phenotype was restored by transfection of the double mutant with a GT-encoding expression vector or by addition of 1 M sorbitol to the medium, indicating that the double mutant is affected in cell wall formation. It is suggested that facilitation of glycoprotein folding mediated by the interaction of monoglucosylated oligosaccharides with calnexin is essential for cell viability under conditions of extreme ER stress such as underglycosylation of proteins caused by the alg6 mutation and high temperature. In contrast, gls2/alg6 double-mutant cells that transfer Man9GlcNAc2 and that are unable to remove the glucose units added by GT as they lack glucosidase II (GII), grew at 37°C and had, when grown at 28°C, a phenotype of growth and morphology almost identical to that of wild-type cells. These results indicate that facilitation of glycoprotein folding mediated by the interaction of calnexin and monoglucosylated oligosaccharides does not necessarily require cycles of reglucosylation–deglucosylation catalyzed by GT and GII.
Keywords: glycoprotein folding, Schizosaccharomyces pombe, glucosylation, glucosyltransferase, endoplasmic reticulum
Membrane-bound calnexin and its soluble homologue calreticulin behave in mammalian cells as authentic chaperones of the ER. They bind monoglucosylated high mannose-type oligosaccharides present in both misfolded and properly folded glycoproteins (Rodan et al., 1996; Zapun et al., 1997). Monoglucosylated structures may be formed by partial deglucosylation of the oligosaccharide transferred to nascent polypeptide chains (Glc3Man9GlcNAc2) by glucosidases I (GI)1 and II (GII) or by reglucosylation of Man7-9GlcNAc2 by the UDP-Glc:glycoprotein glucosyltransferase (GT) (Sousa et al., 1992). GT behaves as a sensor of glycoprotein conformation since the glucose units removed from the monoglucosylated species by GII are added back by GT only when glycoproteins have not yet acquired their proper tertiary structures.
Binding of calnexin and calreticulin to monoglucosylated oligosaccharides of misfolded glycoproteins prevents their aggregation and premature degradation, inhibits formation of non-native disulfide bridges and retains misfolded species in the ER. The role of calnexin and calreticulin is therefore similar to that of classical chaperones of the Hsp70 and chaperonin families (see Helenius et al., 1997, and references therein).
The yeast Schizosaccharomyces pombe has the three elements required for the quality control of glycoprotein folding. This fission yeast has a calnexin-like molecule with a 40% amino acid identity with the mammalian lectin (Jannatipour and Rokeach, 1995; Parlati et al., 1995a ). The S. pombe calnexin is a transmembrane type I protein with a cytosolic tail and three highly conserved and one less conserved Ca2+-binding motifs, as in mammalian calnexin. Calnexin is essential for S. pombe viability and no calreticulin homologue was detected in this yeast genome (Jannatipour and Rokeach, 1995; Parlati et al., 1995a ). Moreover, it has been shown that conditions that lead to the accumulation of misfolded glycoproteins in the ER induce calnexin-encoding mRNA synthesis (Jannatipour and Rokeach, 1995; Parlati et al., 1995a ).
The oligosaccharide transferred to protein in S. pombe is Glc3Man9GlcNAc2, as in most eukaryotic cells. It is processed in the ER to Man9GlcNAc2, thus indicating the presence of both GI and GII activities although the genes encoding these glucosidases have not yet been identified (Ziegler et al., 1994).
We have previously characterized the S. pombe GT. The enzyme was purified to homogeneity and found to be even more stringent in its differential glucosylation of denatured and native glycoproteins than its rat liver counterpart (Fernández et al., 1994). We showed that synthesis of S. pombe GT-encoding mRNA was induced under stress conditions that promoted accumulation of misfolded glycoproteins in the ER lumen (Fernández et al., 1996). However, disruption of the GT-encoding gene showed that the enzyme is not essential for cell viability and no discernible phenotype was observed (Fernández et al., 1996). On the other hand, the yeast Saccharomyces cerevisiae lacks GT, a key element in the quality control of glycoprotein folding, and calreticulin and its calnexin-like molecule presents significant structural variations with respect to the mammalian protein (Fernández et al., 1994; Parlati et al., 1995 b; Jakob et al., 1998).
Facilitation of glycoprotein folding mediated by the interaction of calnexin/calreticulin with monoglucosylated oligosaccharides is dispensable for cell growth as shown by the viability of S. pombe mutant cells lacking GII (our unpublished results), the same as mammalian cells lacking the same enzyme (Reitman et al., 1982). No monoglucosylated oligosaccharides are formed in GII-deficient mutants. Facilitation of glycoprotein folding mediated by the ER lectins was therefore considered to be a back up system that the unique plasticity of the ER folding machinery could replace with alternative mechanisms.
Work reported here shows that: (a) facilitation of glycoprotein folding mediated by the interaction of calnexin and monoglucosylated oligosaccharides is essential for S. pombe viability under conditions of extreme ER stress such as glycoprotein underglycosylation and high temperature; (b) cycles of reglucosylation–deglucosylation catalyzed by GT and GII are apparently not essential for facilitation of glycoprotein folding; and (c) the formation of monoglucosylated oligosaccharides is not essential for S. pombe viability, thus indicating that calnexin has another essential role, independent of its recognition of monoglucosylated oligosaccharides. In addition, we have identified two amino acid residues that are apparently required for GT catalytic activity.
Materials and Methods
Strains and Media
The wild-type S. pombe strains used were Sp95 (h90, ura4-D18, ade6-M216, leu1-32) and Sp61 (h−, ura4-D18, ade6-M210, ade1, leu1-32). YE and minimal media were prepared as described (Moreno et al., 1991).
Construction of the alg6 Mutant
Degenerate oligonucleotide primers encoding three peptides that are conserved in known ALG6 sequences from other species (these sequence data are available from GenBank/EMBL/DDBJ under accession Nos. Z74910 and Z54342) were used as primers in PCR reactions: 5′-TAYTGGGGNTYNGAYTATCCNCC-3′ (Alg1′, sense), 5′-DATNGTYTTYTCRTGNACNYGRA-3′ (Alg4′, antisense), and 5′-CAYTGGATGGARATHAC-3′ (Alg6s, sense). The template was genomic S. pombe DNA. PCR reactions using primers Alg1′ and Alg4′ yielded an 812-bp fragment that was sequenced. The deduced amino acid sequence of this product was 82% and 80% homologous to the respective portions of S. cerevisiae ALG6 and putative Caenorhabditis elegans ALG6 gene products, respectively. An 876-bp fragment was obtained when primers Alg6s and Alg4′ were used. As expected, it contained the 812-bp fragment. The 812-bp fragment in pGEMT-Easy vector (Promega Corp., Madison, WI) was treated with BglII and NheI enzymes. A 102-bp segment was thus eliminated and replaced by the ura4 + gene (these sequence data are available from GenBank/EMBL/DDBJ under accession No. M36504; 1.8 kb) in an inverted position. The construct was liberated from the vector and used for transforming Sp95 cells. Ura4+ cells (alg6; Sp95A strain) were selected. Colony PCRs were performed using Alg6s (sense) and uraSC (sense, 5′-TGCTCCTACAACATTACC-3′; this primer is complementary to a portion of the 3′ end of the ura4 + gene). Selected colonies gave the expected 423-bp fragment. DNAs from wild-type (Sp95) and alg6 mutant cells were digested with BglII and NdeI (that do not cut the ura4 + gene) and analyzed by Southern blotting. A 603-bp fragment obtained by digestion of the 812-bp portion of the alg6 + gene with NheI and SphI was used as probe. Mutant and wild-type DNAs yielded 4.6-kb and 2.8-kb positive fragments, respectively. On the other hand, the mutant DNA 4.6-kb fragment gave a positive signal when a ura4 +-derived probe was used.
Construction of the gpt1/alg6 Double Mutant
Construction of the gpt1 Mutant.
The procedure used was similar to that previously described but Sp61 cells were used (Fernández et al., 1996). The resulting strain was named Sp61G.
Excision of the ura4+ Gene.
The pBluescript plasmid containing the entire gpt1 + gene (4,952 bp) (Fernández et al., 1996) was digested with BglII, thus eliminating 1,684 bp. The digestion product, containing the plasmid plus 612 bp of the 5′ end and 2,656 bp of the 3′ end of GT, was ligated. A 3,111-bp linear fragment obtained upon cutting the plasmid with PstI and MluI was used to transform gpt1 mutant cells. Colonies resistant to fluororotic acid (1 mg/ml) were analyzed by Southern blotting (see below). The strain was named Sp61G4.
Disruption of the alg6+ Gene.
The alg6 + gene was disrupted in the gpt1- D1684 mutant (Sp61G4 strain), as described above for wild-type cells. The resulting strain was named Sp61G4A.
Characterization of gpt1 and gpt1/alg6 Mutants
DNAs from wild-type cells (Sp61), and from the gpt1::ura4 + (Sp61G), the gpt1-D1684 (Sp61G4) and the gpt1/alg6 (Sp61G4A) mutant cells were cut with BglII and analyzed in Southern blots with a 1,200-bp probe corresponding to the 5′ end of the gpt1 + gene. Wild-type DNA yielded two positive bands as BglII cuts at 612, 1,750, and 2,290 bp from the 5′ end of the gpt1 + gene whereas gpt1::ura4 +, gpt1-D1684, and gpt1/alg6 DNAs only showed a positive signal with the smaller band as 1,684 bp (from 612 to 2,296) from the gpt1 + gene had been deleted for inserting the ura4 + gene (gpt1 mutant) or for constructing the gpt1-D1684 mutant. Wild-type DNA gave a 2.8-kb positive signal, the same as gpt1::ura4 + and gpt1-D1684 DNAs when the above mentioned 603-bp alg6 + gene probe was used. On the other hand, gpt1/alg6 DNA gave the expected 4.6-kb band.
Construction of the gls2/alg6 Double Mutant
Disruption of the GII-encoding gene (gls2+).
Primers (sense 5′-TGGAAYGAYATGAAYGARCC-3′, and antisense 5′-IGCRTGNGCICKRAARAA-3′) were designed according to peptides conserved in three known GII sequences from other species (these sequence data are available from GenBank/EMBL/DDBJ under accession Nos. Z36098, D42041, and Z700753). A 414-bp fragment of the homologue S. pombe gene (here designated gls2 +) was obtained by PCR using genomic wild-type DNA as template. The fragment was sequenced and an antisense primer (5′-CCACCGAACAAACAATTCAGC-3′) was designed corresponding to the sequence of positions 355–375 of the fragment. This primer and a sense primer derived from another sequence conserved among the three GIIs of other species (5′-AAYGCNGCNGANACNTGG-3′) were used to synthesize a 998-bp fragment by PCR. This fragment was cloned in the pGEMT-Easy vector (Promega Corp.) and sequenced. The deduced amino acid sequence of this fragment showed 49%, 43%, and 47% homologies with the corresponding respective fragments of human, S. cerevisiae and putative C. elegans GII sequences. The ura4 + marker gene was introduced in reverse orientation in position 569 (HindIII site) of the 998-bp fragment. Wild-type cells (Sp95) were transformed with the linear 2,766-bp NotI restriction DNA fragment (Sp95IIα strain).
Excision of the ura4+ Gene.
The vector containing the 998-bp fragment of gls2 + with the inserted ura4 + marker was treated with Bsu36I. A 4,565-bp fragment containing the vector plus 359 bp of the 5′ end of gls2 +, 776 bp of the 5′ end of the ura4 +, and 430 bp of the 3′ end of gls2 + genes was ligated. A 1,565-bp linear fragment was liberated from it by NotI digestion and used for transforming the gls2::ura4 + (Sp95IIα strain) mutant. Fluororotic acid (1 mg/ml) resistant colonies were characterized by PCR. Primers used were 5′-GTCGTCCGCTTCTCCCTC-3′ (GIIBs) and 5′-CCCCATAATGAATAGCAT-3′ (GIIBa). The primers are complementary to portions of the gls2 + gene fragment. PCR generated a 542-bp fragment in wild-type cells, a 2,306-bp fragment in gls2::ura4 + (Sp95IIα) cells, and a 1,108-bp fragment in gls2::ura4 (Sp95IIα2) cells.
Disruption of the alg6+ Gene.
Gls2::ura4 (Sp95IIα2) cells were then transformed with the linear fragment mentioned before having portions of the alg6 + gene interrupted with ura4 + (see above, construction of the alg6 mutant). Ura+ cells (gls2/alg6; Sp95IIα2A strain) were selected. Colony PCRs were performed using primers GIIBs and GIIBa or Alg6s and uraSC. Selected colonies gave the expected fragments of 1,108 bp with GIIBs and GIIBa primers and of 423 bp with the Alg6s and uraSC ones. It is worth mentioning that primer Alg6s was not present in the construct used for disrupting the alg6 + gene but was complementary to a portion of the gene located to the 5′ end.
Mutagenesis of gpt1+
The gpt1 + gene was removed from the pBluescript plasmid and ligated to pALTER1 (Promega Corp.). Mutagenesis reactions were carried out as described by the manufacturer (pAlter kit; Promega Corp.). The primers used were Ampicillin repair, Tetracycline knockout, CTTACGTGGATTAAAAGCTCACATTAGTGCATTATATG for the Y1312A mutation and CCCTGTCTAATTTAGCTCAAGATTTGCCAAATC for the D1352A mutation. AmpR and TetS cells were selected. Plasmid DNA (pALTER1-gpt1YD-AA) was isolated from those cells and the presence of new AluI sites in the gpt1 gene was assayed. The mutations introduced are expected to create two new sites. As the gpt1 + gene has 25 AluI sites, a 1,236-bp fragment of the COOH-terminal portion that contained the putatively mutated region was synthesized by PCR. The fragment coming from gpt1 + yielded, as expected, five fragments upon digestion with AluI, whereas seven fragments were obtained from that coming from the Y1312A, D1352A GT mutant (gpt1YD-AA). Similar results were obtained in all five independently PCR-synthesized 1,236-bp fragments.
Subcloning of gpt1YD-AA in pREP1
pALTER1-gpt1YD-AAwas digested with SphI, treated with T4 DNA polymerase for generating blunt ends, and then digested with BamHI. The gpt1YD-AA gene was isolated and ligated to the pREP1 expression vector (this vector contains the LEU2 S. cerevisiae gene that complements the leu1 S. pombe mutation) that had been cut with SmaI and BamHI. Resulting plasmid was called pREP1-gpt1YD-AA. S. pombe gpt1/alg6 cells were transformed with this construct and plated in minimal medium supplemented with adenine.
Subcloning of gpt1+ in pREP1 and pREP41
The gpt1 + gene was isolated from the pBluescript as described above and ligated to pREP1 or pREP41. Resulting expression vectors were called pREP1-gpt1 + or pREP41-gpt1 +. Gpt1 or gpt1/alg6 cells were then transformed with these constructs and plated in minimal medium supplemented with adenine.
DNA and RNA procedures
Standard DNA manipulations were carried out as described (Sambrook et al., 1989). S. pombe genomic DNA was prepared as previously described (Moreno et al., 1989). Hybridization conditions were as previously described (Sambrook et al., 1989). RNA was extracted from cells grown under normal conditions (YE medium at 28°C) in the exponential growth phase at the same OD and submitted to Northern blotting as previously performed (Fernández et al., 1996). The probe used was a 1,004-bp DNA fragment of the BiP-encoding gene synthesized by PCR using genomic DNA as template.
Materials
[14C]Glucose (250 Ci/mol) and EasyTag Express protein labeling mix (>1,000 Ci/mmol) were from New England Nuclear (Boston, MA). Jack bean α-mannosidase and EndoH were from Sigma (St. Louis, MO). UDP- [14C]Glc (250 Ci/mol) was prepared as described by Wright and Robbins (1965). Polyclonal antiserum to recombinant S. pombe calnexin was a generous gift from Dr. L. Rokeach (Département de Biochimie, Université de Montréal, Montréal, Canada).
Substrates and Standards
[glucose-14C]Glc1Man9GlcNAc, [glucose-14C]Glc3Man9GlcNAc, [glucose- 14C]Glc2Man9GlcNAc, [glucose-14C]Glc1Man4GlcNAc, [glucose-14C]Glc1 Man5GlcNAc, [14C]ManGlcNAc, [14C]Man9GlcNAc, and [14C]Man8 GlcNAc were prepared as described previously (Fernández et al., 1994).
In Vivo Labeling of S. pombe Cells with [14C]glucose
Pulse labeling of S. pombe wild type, alg6, gpt1/alg6, and gls2/alg6 cells with [14C]glucose was performed as described before for S. cerevisiae but 150 μCi of [14C]glucose was used (Fernández et al., 1994). Preincubations and incubations with the labeled precursor lasted for 15 min. Where indicated 5 mM 1-deoxynojirimycin (DNJ) was added. Isolation of Endo H–sensitive oligosaccharides was performed as already described (Fernández et al., 1994).
In Vivo Labeling of S. pombe Cells with [35S]Methionine and [35S]Cysteine
Wild type (Sp95), gls2, and alg6 mutant cells grown in YE medium were harvested, washed, and then suspended (2 × 108 cells) in 1.5 ml of 1% Yeast Nitrogen Base without amino acids (Difco Laboratories Inc., Detroit, MI) supplemented with adenine, leucine, and uracil. Samples were preincubated for 2 h at 28°C and incubated with 450 μCi of [35S]methionine plus [35S]cysteine (EasyTag Express protein labeling mix) for 5 min at the same temperature. Reactions were stopped by the addition of 10 mM sodium azide plus 10 mM iodoacetamide. Samples were then processed and calnexin was immunoprecipitated as described previously (Tabuchi et al., 1997). Samples were run on 9% SDS-PAGE and submitted to autoradiography.
Methods
Strong acid hydrolysis and treatment of oligosaccharides with Jack bean α-mannosidase were as described before (Fernández et al., 1994). Whatman 1 paper was used for chromatography. Solvents used were: (a) 1-propanol/nitromethane/water (5:2:4); (b) 1-butanol/pyridine/water (10:3:3); or (c) 1-butanol/pyridine/water (4:3:4). GII activity was assayed as described before using [glucose- 14C]Glc1Man9GlcNAc as substrate (Ugalde et al., 1979). GT was assayed using yeast cell microsomes as an enzyme source and 8 M urea-denatured thyroglobulin as acceptor, as described previously (Trombetta et al., 1989). S. pombe and rat liver microsomes were prepared as already described (Trombetta et al., 1992; Fernández et al., 1994). Endomannosidase was assayed in S. pombe microsomes with [glucose-14C]Glc1Man9GlcNAc as substrate as already described (Gañán et al., 1991). Rat liver microsomes with or without S. pombe microsomes were used as positive controls.
Results
Glycosylation and Oligosaccharide-processing Pathways
The glycosylation and oligosaccharide processing pathways occurring in the ER of wild type and mutant cells used in the work reported here are depicted in Fig. 1. Gpt1 +, gls2 +, and alg6 + code for GT, for GII and for the enzyme that transfers the first glucose unit from dolichol-P-Glc to Man9GlcNAc2-P-P-dolichol, respectively. Mutants were obtained and genetically characterized by Southern blotting and PCR analysis as described under Materials and Methods (Table I).
Figure 1.
Glycosylation and oligosaccharide processing pathways occurring in the ER of wild-type and mutant cells used in work reported here. D, dolichol; Pr, protein.
Table I.
S. pombe Strains Used in the Present Study
| Strain | Genotype | |
|---|---|---|
| Sp95 (wt) | h90, ura4-D18, ade6-M216, leu1-32 | |
| Sp61 (wt) | h−, ura4-D18, ade6-M210, ade1, leu1-32 | |
| Sp95A (alg6) | h90, ura4-D18, ade6-M216, leu1-32, alg6::ura4 + | |
| Sp61G (gpt1) | h−, ura4-D18, ade6-M210, ade1, leu1-32, gpt1::ura4 + | |
| Sp61G4 | h−, ura4-D18, ade6-M210, ade1, leu1-32, gpt1-D1684 | |
| Sp61G4A (gpt1/alg6) | h−, ura4-D18, ade6-M210, ade1, leu1-32, gpt1-D1684, alg6::ura4 + | |
| Sp95IIα (gls2) | h90, ura4-D18, ade6-M216, leu1-32, gls2::ura4 + | |
| Sp95IIα2 | h90, ura4-D18, ade6-M216, leu1-32, gls2::ura4 | |
| Sp95IIα2A (gls2/alg6) | h90, ura4-D18, ade6-M216, leu1-32, gls2::ura4, alg6::ura4 + |
Except for the first wild-type strains, all mutants were generated in the present study.
Biochemical Characterization of Mutants
The alg6 Mutation.
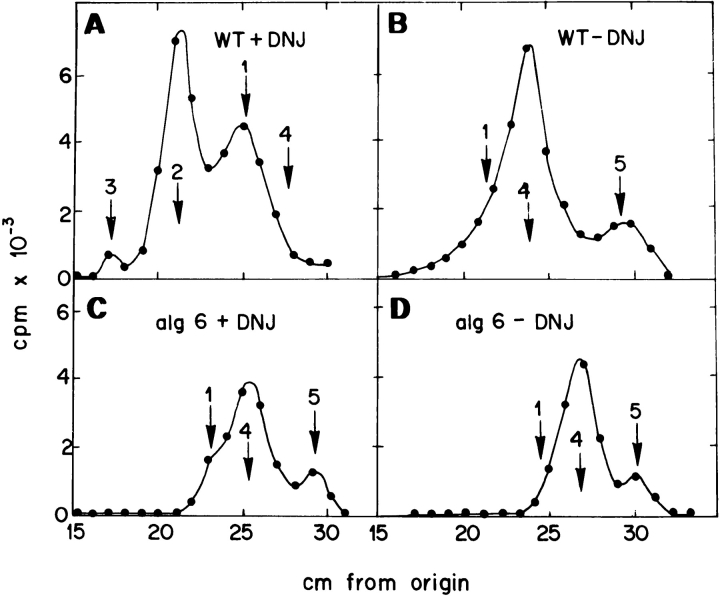
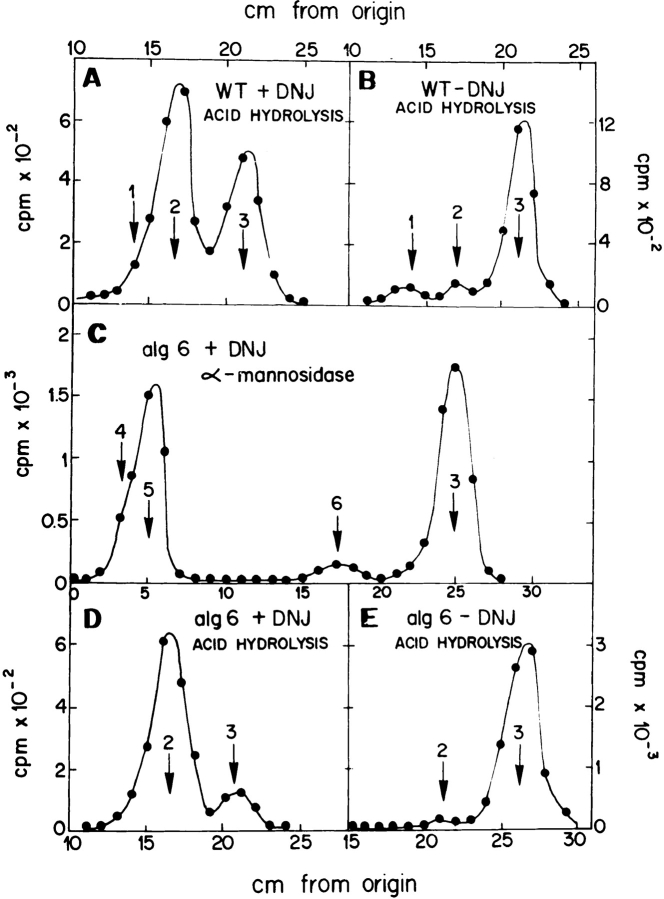
The gpt1 + gene encoding S. pombe GT has already been identified but as neither alg6 + nor gls2 + genes have yet been identified in S. pombe, it was necessary to characterize the alg6, gpt1/alg6, and gls2/alg6 mutants biochemically. For this purpose, the structure of protein-bound oligosaccharides synthesized in vivo by the wild-type and mutant cells in the presence or absence of the GI and GII inhibitor DNJ were compared. Incubation of wild-type cells with [14C]glucose for 15 min in the presence 5 mM DNJ yielded endo-β-N-acetylglucosaminidase H (Endo H)-sensitive, protein-linked oligosaccharides that migrated on paper chromatography as Glc3Man9GlcNAc, Glc2Man9GlcNAc, and Glc1Man9GlcNAc standards (Fig. 2 A). Strong acid hydrolysis of compounds that migrated as either one of the last two standards produced labeled glucose and mannose units (Fig. 3 A). Omission of DNJ mainly produced compounds that migrated as Man9GlcNAc and Man8GlcNAc standards but a trailing in the position of Glc1Man9GlcNAc could be observed (Fig. 2 B). Strong acid hydrolysis of compounds in this figure yielded mannose and very small amounts of glucose and galactose (Fig. 3 B). From these results it is concluded that DNJ only partially inhibited GI and GII activities.
Figure 2.
Characterization of alg6 mutant. Patterns of protein-linked oligosaccharides. Oligosaccharides synthesized in wild-type (Sp95) and alg6 mutant cells incubated for 15 min with [14C]glucose in the presence or absence of DNJ were obtained by Endo H treatment and subjected to paper chromatography with solvent A. Standards: 1, Glc1Man9GlcNAc; 2, Glc2Man9GlcNAc; 3, Glc3Man9GlcNAc; 4, Man9GlcNAc; and 5, Man8GlcNAc.
Figure 3.
Characterization of the alg6 mutant. Monosaccharide composition of oligosaccharides. (A) The compound migrating as the Glc1Man9GlcNAc standard in Fig. 2 A was submitted to strong acid hydrolysis. (B) All compounds depicted in Fig. 2 B were submitted to the same treatment. Resulting products in A and B were run on paper chromatography with solvent B. (C) Compounds depicted in Fig. 2 C that migrated as Glc1Man9 GlcNAc and Man9GlcNAc standards were treated with α-mannosidase and run on paper chromatography with solvent C. (D) Compounds migrating as Glc1Man5GlcNAc and Glc1Man4 GlcNAc standards in C were submitted to strong acid hydrolysis. (E) All compounds depicted in Fig. 2 D were submitted to strong acid hydrolysis. Resulting products in D and E were run on paper chromatography with solvent B. Standards: 1, galactose; 2, glucose; 3, mannose; 4, Glc1Man5GlcNAc; 5, Glc1Man4 GlcNAc; and 6, ManGlcNAc.
The alg6 mutant cells incubated in the presence of DNJ mainly produced compounds that migrated as Glc1Man9 GlcNAc, Man9GlcNAc, and Man8GlcNAc standards (Fig. 2 C). Treatment of compounds that migrated as the first two standards with Jack bean α-mannosidase yielded mannose, ManGlcNAc, and compounds that migrated as Glc1Man4GlcNAc and Glc1Man5GlcNAc standards (Fig. 3 C). Strong acid hydrolysis of these last two compounds produced labeled glucose and mannose units (Fig. 3 D). The α-mannosidase degradation products confirmed that a mixture of Man9GlcNAc and Glc1Man9GlcNAc was present in the main peak of Fig. 2 C. ManGlcNAc was produced upon degradation of the first compound and Glc1Man4,5 GlcNAc by that of the last one. Labeling of the mutant performed in the absence of DNJ only yielded Man9 GlcNAc and Man8GlcNAc (Fig. 2 D). Strong acid hydrolysis of those compounds only produced mannose units (Fig. 3 E).
As only partial inhibition of GII by DNJ was attained, there are two interpretations to results shown above: in alg6 mutants, Man9GlcNAc2 was transferred to proteins and this compound was either demannosylated to Man8 GlcNAc2 or glucosylated by GT to Glc1Man9GlcNAc2 or alternatively, the disrupted gene (alg6 +) corresponds to the enzyme that transfers not the first but the second glucosyl residue to Man9GlcNAc2-P-P-dolichol. According to this interpretation, Glc1Man9GlcNAc2 was transferred in the mutant obtained and this compound was deglucosylated and demannosylated to Man9GlcNAc2 and Man8 GlcNAc2. Characterization of the gpt1/alg6 double mutant discarded this last possibility.
The gpt1/alg6 Double Mutation.
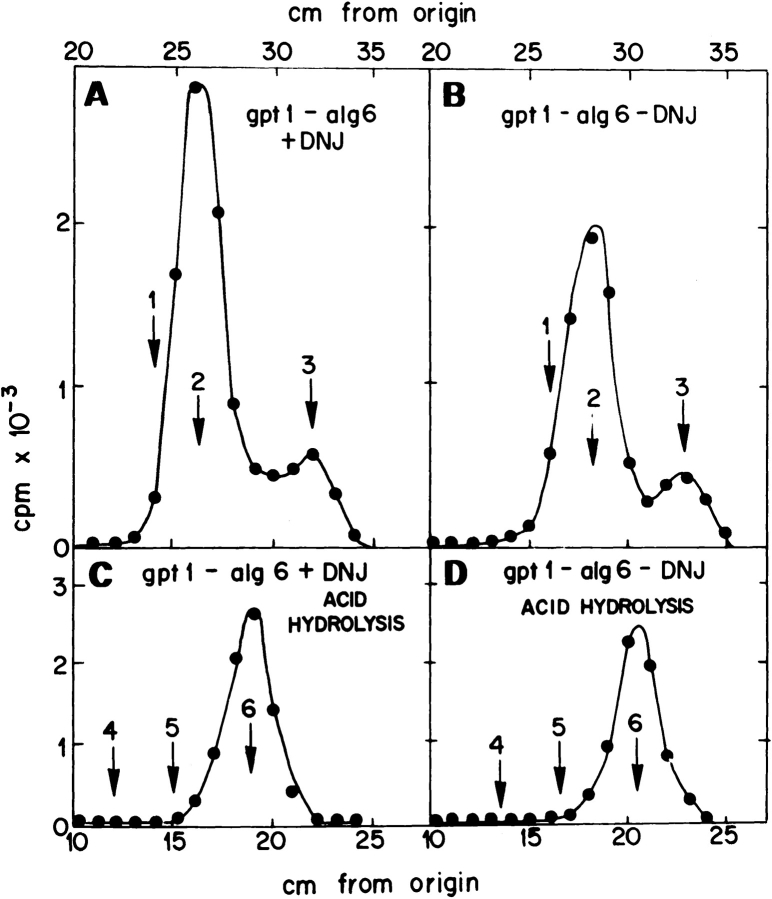
Incubation of gpt1/alg6 cells with [14C]glucose yielded protein-linked Man9GlcNAc and Man8GlcNAc both in the presence and absence of DNJ (Fig. 4, A and B). Strong acid hydrolysis of compounds obtained under both conditions only produced labeled mannose residues (Fig. 4, C and D). Furthermore, microsomes obtained from the double mutant were completely devoid of GT activity (not shown; the assay and results were as described before for the gpt1 mutant; Fernández et al., 1996). These results indicate that in gpt1/alg6 double-mutant cells Man9GlcNAc2 was transferred to nascent polypeptide chains and that this compound was not glucosylated. They further indicate that the same compound was transferred in the single alg6 mutant.
Figure 4.
Characterization of the gpt1/alg6 mutant. Patterns of oligosaccharides and monosaccharide compositions. (A and B) oligosaccharides synthesized upon incubation of gpt1/alg6 mutant cells with [14C]glucose, in the presence or absence of DNJ, were run on paper chromatography with solvent A. (C and D) Compounds synthesized in the presence or absence of DNJ were submitted to strong acid hydrolysis. Resulting products were run on paper chromatography with solvent B. Standards: 1, Glc1 Man9GlcNAc; 2, Man9GlcNAc; 3, Man8GlcNAc; 4, galactose; 5, glucose; and 6, mannose.
The gls2/alg6 Double Mutation.
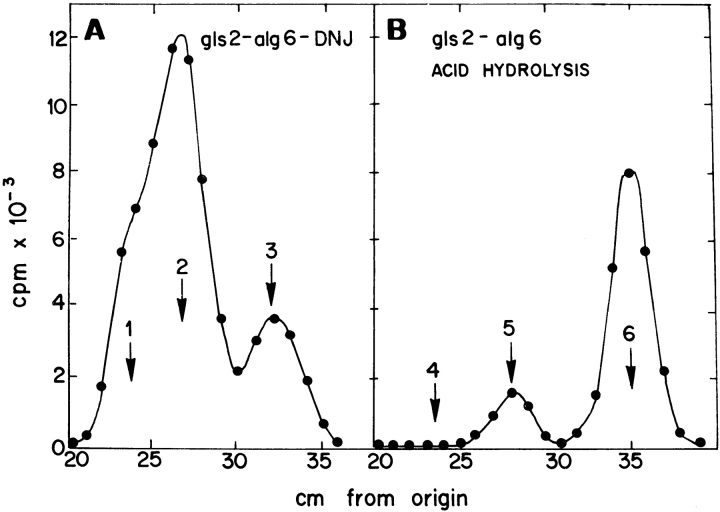
Incubation of gls2/alg6 mutant cells with [14C]glucose in the absence of DNJ yielded a mixture of Glc1Man9GlcNAc2, Man9GlcNAc2, and Man8GlcNAc2 (Fig. 5 A). The same result was obtained in the presence of the GII inhibitor (not shown). Strong acid degradation of above mentioned compounds yielded glucose and mannose (Fig. 5 B). No GII activity was detected in gls2/alg6 microsomes when assayed with [glucose-14C]Glc1Man9GlcNAc as substrate.
Figure 5.
Characterization of gls2/alg6 mutant. Patterns of oligosaccharides and monosaccharide composition. (A) Oligosaccharides synthesized by gls2/alg6 mutant cells upon incubation with [14C]glucose in the absence of DNJ were run on paper chromatography with solvent A. (B) Compounds synthesized were submitted to strong acid hydrolysis and resulting products run on paper chromatography with solvent B. Standards: 1, Glc1Man9 GlcNAc; 2, Man9GlcNAc; 3, Man8GlcNAc; 4, galactose; 5, glucose; and 6, mannose.
Presence of the Unfolded Protein Response in alg6, gpt1/alg6, and gls2/alg6 Mutants
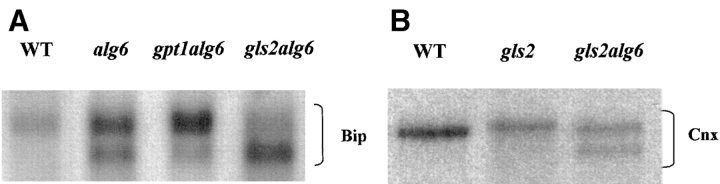
Accumulation of misfolded proteins in the ER leads to the so-called unfolded protein response, that is, to the induction of chaperones and other proteins that facilitate protein folding (BiP, calnexin, GT, etc.) (Pidoux and Armstrong, 1992; Jannatipour and Rokeach, 1995; Parlati et al., 1995a ; Fernández et al., 1996). Detection of this response is rather simple in S. pombe as under normal conditions a single BiP-encoding mRNA is synthesized. Accumulation of misfolded proteins leads to the induction of not only this mRNA species but also of a slightly smaller mRNA (Pidoux and Amstrong, 1992). Synthesis of both BiP-encoding mRNAs was detected in alg6, gpt1/alg6, and gls2/alg6 mutants but not in wild-type cells grown under normal conditions (Fig. 6 A). The most plausible explanation for induction of BiP-encoding mRNA in cells carrying the alg6 mutation might be that underglycosylation of glycoproteins occurs in them as has been described for S. cerevisiae mutants transferring Man9GlcNAc2 to proteins (Ballou et al., 1986). Cell-free assays have shown that in most eukaryotic cells Glc3Man9GlcNAc2 is transferred ∼10–25-fold more efficiently than Man9GlcNAc2 (Bosch et al., 1988). It has been reported that underglycosylation of glycoproteins triggers misfolding of glycoproteins (Marquardt and Helenius, 1992; Imperiali and Rickert, 1995). To confirm that underglucosylation of glycoproteins occurred in S. pombe mutants carrying the alg6 mutation, wild-type, gls2, and alg6 cells were incubated for 5 min with a mixture of [35S]methionine and [35S]cysteine, lysed in the presence of iodoacetamide and material immunoprecipitated by a calnexin antiserum run on SDS-PAGE under reducing conditions. As depicted in Fig. 6 B, whereas a single positive band appeared in wild-type cells, two positive signals were detected in the alg6 mutant. Their difference in size was of ∼2 kD, a value consistent with the presence of a single oligosaccharide in S. pombe calnexin (Jannatipour and Rokeach, 1995; Parlati et al., 1995a ). The gls2 mutant yielded a single positive signal that migrated slightly above that of wild-type cells. This result is consistent with the fact that in this mutant the oligosaccharide retains two glucose units.
Figure 6.
The unfolded protein response and underglycosylation of glycoproteins in S. pombe mutant cells. (A) Wild-type, alg6, gpt1/alg6, and gls2/alg6 cells were grown in YE medium at 28°C, mRNAs were extracted and submitted to Northern blotting. A fragment of BiP-encoding DNA was used as probe. (B) Wild-type, gls2, and alg6 mutant cells were incubated for 5 min with [35S]methionine and [35S]cysteine, and lysed in the presence of iodoacetamide. Material immunoprecipitated by antiserum to S. pombe calnexin was run on SDS-PAGE under reducing conditions and submitted to autoradiography.
Growth and Morphology of Wild Type, alg6, gpt1, and gpt1/alg6 cells at 28° and 37°C
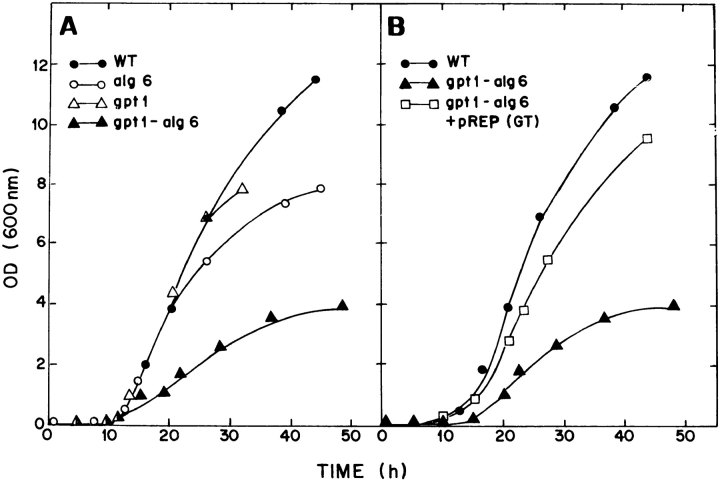
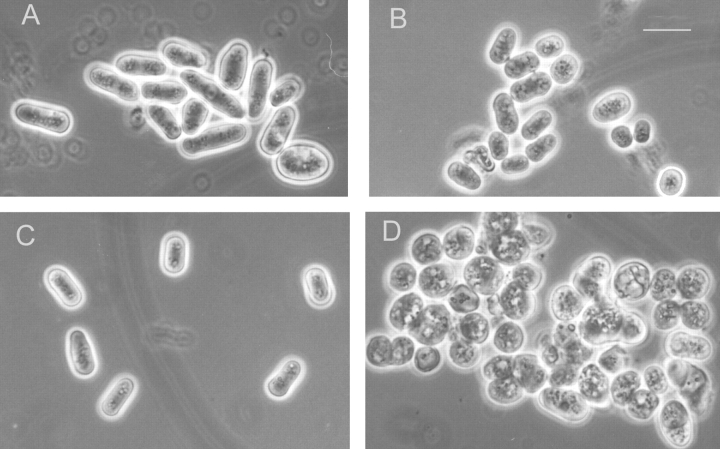
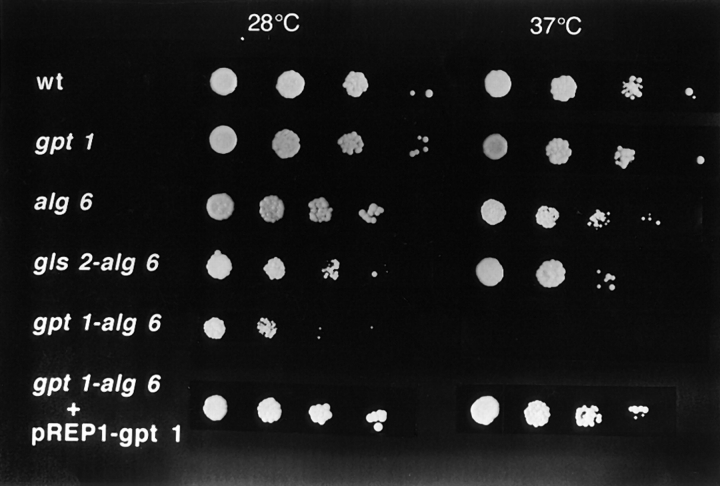
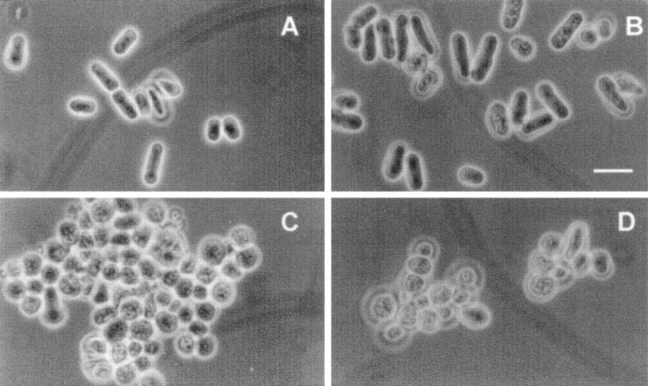
Alg6 and gpt1 cells grew at about the same rate as wild-type cells at 28°C (Fig. 7 A). Moreover, their short rod shapes were similar to those of wild-type cells (Fig. 8, A–C). On the other hand, gpt1/alg6 cells grew much more slowly than wild-type cells and growth stopped at a lower OD (Fig. 7 A). Gpt1/alg6 cells tended to clump and had a rounded shape with clearly visible inclusions (Fig. 8 D). All mutant cells grew at 37°C except for gpt1/alg6 that did not grow at this temperature (Fig. 9). Growth of gpt1 mutant cells at the high temperature was similar to that of wild-type cells, whereas alg6 cells grew somewhat less efficiently (Fig. 9). It is worth mentioning in experiments shown in Fig. 9 all strains were allowed to grow for 5 d except for gls2/alg6 and gpt1/alg6 that were incubated for 7 and 10 d, respectively. Both alg6 and gls2/alg6 mutants showed the so-called thermosensitive morphology at 37°C, characterized by swollen cells, displaying pear, lemon, and round shapes (Ishiguro et al., 1997).
Figure 7.
Growth of wild-type and mutant cells at 28°C. Minimal medium plus adenine was used. The medium was supplemented with leucine and uracil (wild type) or with leucine (gpt1, alg6, and gpt1/alg6). Sp95 or Sp61 wild-type cells yielded similar growth curves.
Figure 8.
Morphology of wild-type and mutant strains. (A) Wild type; (B) alg6; (C) gpt1; and (D) gpt1/alg6. Cells were grown at 28°C in minimal medium supplemented as in Fig. 7. Bar, 10 μm.
Figure 9.
Growth of wild-type and mutant strains at 28°C and 37°C. Cells from indicated strains were plated in minimal medium supplemented as indicated in Fig. 7. Absorbance of initial inocula (3 μl) was in all cases 0.3. Following inocula correspond to one-tenth serial dilutions. All strains were incubated for 5 d except for gls2/alg6 and gpt1/alg6 that were incubated for 7 and 10 d, respectively.
Rescue of the Wild-Type Phenotype in gpt1/alg6 Mutants
The rounded shape of gpt1/alg6 cells suggested that synthesis of one of the components of the cell wall was affected. A classical way to test this hypothesis is to grow cells in a hyperosmotic medium. As shown in Fig. 10 A, gpt1/alg6 cells recovered the wild-type morphology when grown in 1 M sorbitol. Double-mutant cells grew at 28°C at the same rate as wild-type ones in the presence of the above mentioned compound (not shown). To confirm that the phenotype observed in those double mutants was caused by the absence of GT, cells were transfected with two expression vectors, pREP1-gpt1 + and pREP41-gpt1 + both coding for GT. They respectively had strong and medium promoters and when transfected into gpt1 mutants they yielded cells displaying 4–10- and 1–2-fold the GT activity of wild-type cells. Gpt1/alg6 cells recovered the wild-type morphology (Fig. 10 B) and growth rate at 28°C (Fig. 7 B), and the ability to grow at 37°C (Fig. 9) when transfected with any of these expression vectors. The wild-type phenotype was not rescued by the vectors lacking the gpt1 + gene (Fig. 10 C).
Figure 10.
Rescue of wild-type phenotype in mutant strains. gpt1/alg6 cells were grown at 28°C in minimal medium supplemented as in Fig. 7. (A) 1 M sorbitol was added to the medium; (B) cells transformed with pREP1- gpt1 +; (C) cells transformed with pREP1; and (D) cells transformed with pREP1-gpt1YD-AA. Bar, 10 μm.
It was reported that under physiological conditions of pH and salt concentration, GT interacted with hydrophobic peptides and that this interaction was blocked by denatured glycoproteins. It was postulated that this interaction could be one of the recognition elements that determine the exclusive glucosylation of misfolded species (Sousa and Parodi, 1995). The in vivo interaction of GT with certain misfolded ER proteins under conditions of Ca2+ depletion has also been reported (Choudhury et al., 1997). To check if the rescue of the wild-type phenotype by the GT-expressing vectors was caused by the catalytic activity of the enzyme or by its capacity of binding hydrophobic domains, two point mutations were introduced at the COOH-terminal domain of GT. This domain is believed to harbor the catalytic activity as it is 68–70% identical among the four known GT sequences (Drosophila melanogaster, S. pombe, C. elegans, and rat liver) (Parker et al., 1995; Fernández et al., 1996; these sequence data are available from GenBank/EMBL/DDBJ under accession No. U28735; our unpublished results) and also has a limited but significant homology with bacterial glycosyltransferases that use UDP-Glc or UDP-Gal as sugar donor (Parker et al., 1995). The changes introduced were Y1312A and D1352A. Both amino acids are not only conserved in the above mentioned GTs but also in at least nine bacterial glycosyltransferases. The mutated GT in pREP1 (pREP1- gpt1YD-AA)was completely inactive when expressed in gpt1/ alg6 cells (Table II). The double-mutant cells expressed similar amounts of GT when transfected with pREP1- gpt1 + or with pREP1-gpt1YD-AA as judged by Coomassie brilliant blue staining of SDS-PAGE of the soluble content of microsomes. The high molecular weight of the enzyme and its absence in non-transformed cells eased its identification. The mutated enzyme did not revert the gpt1/alg6 phenotype (Fig. 10 D) and did not allow growth of the double mutant at 37°C (not shown). This result confirmed that the enzymatic activity of GT was responsible for the correction of the phenotype and strongly suggested that the correction observed was mediated by the interaction of monoglucosylated species with calnexin.
Table II.
Activity of Wild Type and Mutant GT Expressed in gpt1/alg6 Cells
| Addition | pREP1-gpt1 + | pREP1-gpt1YD-AA | pREP1 | |||
|---|---|---|---|---|---|---|
| -------- | 13,840 | 240 | 330 | |||
| Thyroglobulin | 98,550 | 1,080 | 900 |
Values expressed are cpm/mg of microsomal protein precipitated by 10% trichloroacetic acid.
Growth and Morphology of gls2/alg6 Cells
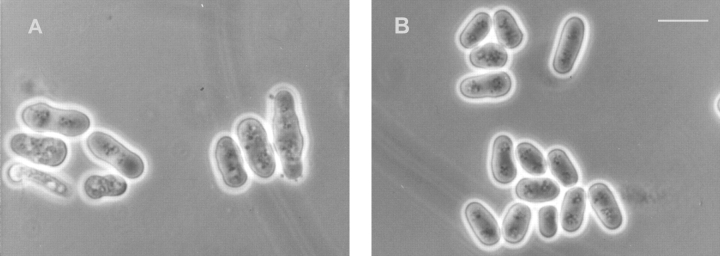
Gls2/alg6 cells grew at 28°C at almost the same rate as wild-type or alg6 cells (not shown). Moreover, the double-mutant cells displayed the same morphology as wild-type or alg6 cells when grown at 28°C (Fig. 11, A and B) and were able to grow at 37°C although not as efficiently as alg6 mutants (Fig. 9). As mentioned above, both alg6 and gls2/alg6 mutant cells showed the so called thermosensitive morphology at 37°C, characterized by swollen cells, displaying pear, lemon, and round shapes (Ishiguro et al., 1997). The fact that gls2/alg6 cells exhibited a phenotype of growth and morphology clearly different from that of gpt1/alg6 cells indicates that facilitation of glycoprotein folding mediated by the interaction of monoglucosylated oligosaccharides with calnexin does not necessarily require cycles of reglucosylation–deglucosylation catalyzed by the opposing activities of GT and GII.
Figure 11.
Morphology of wild-type and mutant strains. (A) Wild-type and (B) gls2/alg6 cells were grown at 28°C in minimal medium supplemented with adenine, uracil and leucine (wild type) and adenine and leucine (gls2/alg6). Bar, 10 μm.
Discussion
It has been reported that in wild-type S. pombe cells the Glc3Man9GlcNAc2 transferred is deglucosylated to Man9 GlcNAc2 in the ER, and then elongated by the addition of mannose and galactose residues in the Golgi (Ziegler et al., 1994). However, we have always obtained small amounts of Man8GlcNAc2. This compound was presumably not produced in the Golgi but in the ER as it was detected after very short incubations with the labeled precursor (5 min) or after long incubations (90 min) under conditions that prevented almost completely (addition of 5 mM DTT to the incubation medium) or completely (incubation at 39°C) exit of glycoproteins from the ER. Man8GlcNAc2 was not formed by an endomannosidase activity similar to that described previously in other cell types (Lubas and Spiro, 1987), as we have been unable to detect such activity in S. pombe microsomal membranes (see Materials and Methods). A phylogenetic survey has indicated a late evolutionary appearance of the enzyme (Dairaku and Spiro, 1997). Accordingly, no endomannosidase activity has been detected so far in unicellular organisms including protozoa and yeasts (Gañán et al., 1991; Gotz et al., 1991; Dairaku and Spiro, 1997).
Labeling of intact mutant cells with [14C]glucose for 15 min provided further evidence against the possibility that Man8GlcNAc2 could had been formed by an endomannosidase activity. As depicted in Fig. 4, Man8GlcNAc2 was produced in gpt1/alg6 mutant cells, in which no glucosylated oligosaccharides (the substrate for the endomannosidase) were synthesized. In addition, incubation of gls2 mutant cells (Sp95IIα strain; see Table I) with [14C]glucose led to the formation of protein-bound Glc2Man9GlcNAc2 and Glc2Man8GlcNAc2. No glucose-free compounds were detected, thus indicating that removal of the glucose units was not required for formation of oligosaccharides containing eight mannose residues.
In contrast to what was observed in mammalian, plant and protozoan cells incubated with [14C]glucose (see Parodi, 1993 for references), Glc1Man8GlcNAc2 was not detected in any of the S. pombe wild-type or mutant cells used in this study (Glc1Man8GlcNAc would have migrated on paper chromatography between Man9GlcNAc and Man8GlcNAc). The absence of Glc1Man8GlcNAc2 formation suggests that the mannosidase responsible for Man8 GlcNAc2 formation and GT might be located in different ER subcompartments. An alternative possibility is that the Man8GlcNAc2 isomer formed may lack the mannose unit to which the glucose residue is added by GT.
Glycoproteins synthesized in S. pombe mutants that transfer Man9GlcNAc2 to proteins (alg6 mutants) were found to be underglycosylated. It is known that in cell-free assays the oligosaccharyltransferase transfers Glc3Man9 GlcNAc2 about 10–25-fold faster than Man9GlcNAc2 (Bosch et al., 1988). Glycoproteins synthesized in S. cerevisiae mutants that transfer the last compound were also found to be underglycosylated (Ballou et al., 1986). It is not surprising, therefore, that induction of BiP-encoding mRNA was detected in all S. pombe strains having the alg6 mutation. The extreme plasticity of the ER folding machinery compensated folding problems caused by glycoprotein underglycosylation by increased synthesis of chaperones and other folding-assisting proteins (BiP, GT, calnexin, etc.) and no discernable phenotype could be observed in those mutants at 28°C.
Facilitation of glycoprotein folding mediated by the interaction between monoglucosylated oligosaccharides and calnexin prevented appearance of a discernable phenotype in alg6 mutants because gpt1/alg6 double-mutant cells, in which no monoglucosylated oligosaccharides were formed, grew very slowly and with an altered morphology at 28°C and were unable to grow at 37°C. A wild-type phenotype was restored upon transfection with a GT-encoding plasmid. Two amino acid changes in the proposed catalytic domain of GT that abolished its glucosylating activity resulted in the non restoration of the wild-type phenotype. This result confirmed that the interaction of monoglucosylated compounds created by GT with calnexin was responsible for the rescue of gpt1/alg6 cells.
Single gpt1 mutants grown under normal conditions also showed an induction of BiP-encoding mRNA (not shown). This fact, as well as the already known induction of GT-encoding mRNA in cells submitted to conditions that promote accumulation of misfolded proteins in the ER (Fernández et al., 1996), confirmed the involvement of monoglucosylated oligosaccharides created by GT in facilitation of glycoprotein folding. Moreover, the fact that Glc1Man9GlcNAc2 could only be detected in alg6 mutants when DNJ had been added to the medium or when the gls2 + gene had been disrupted confirmed that S. pombe GT is involved, together with GII, in glucosylation–deglucosylation cycles, like its mammalian counterpart, and not in the synthesis of cellular polysaccharides. Single gpt1 mutants did not show a discernable phenotype as in them no underglycosylation of glycoproteins is expected to occur and probably increased synthesis of BiP and other folding-assisting proteins compensate for the absence of glycoprotein reglucosylation.
As mentioned above, many glycoproteins are expected to have folding problems in gpt1/alg6 mutant cells. However, not all glycoproteins are affected in the same manner as the requirement for the presence of all oligosaccharides for correct folding varies among different glycoproteins (see Helenius, 1994, and references therein). The one (or ones) that is responsible for the phenotype observed is probably somehow involved in cell wall formation as normal growth and morphology could be restored in a hyperosmotic medium (addition of 1 M sorbitol). It is worth mentioning that S. pombe mutant cells affected in the galactomannan content displayed a rounded morphology similar to that of gpt1/alg6 cells (Ribas et al., 1991). Moreover, the morphological alteration in those mutants was corrected when grown in 1 M sorbitol the same as with the double mutants described in the present report. Synthesis of galactomannan is initiated in the ER upon transfer of Glc3Man9GlcNAc2 to a protein core.
This is the first report showing that protein reglucosylation by GT is required for cell viability under conditions of extreme ER stress such as glycoprotein underglycosylation and high temperature.
It has been reported that proper folding was arrested upon preventing the dissociation of influenza hemagglutinin (a glycoprotein having seven N-linked oligosaccharides) with calnexin/calreticulin by the addition of glucosidase inhibitors to an in vitro translation system (Hebert et al., 1996). It was concluded that on-and-off cycles of interaction between the glycoprotein and the lectins catalyzed by GT and GII were required for folding facilitation. This would be analogous to what happens with chaperones of the Hsp70 family. Mutations in the NH2-terminal portion that abolish the ATPase activity and thus delay dissociation of the chaperones and misfolded proteins, prevent proper folding (Hendrick and Hartl, 1993). Apparently cycles of reglucosylation–deglucosylation are not essential for facilitation of proper folding of the glycoprotein(s) responsible for the observed phenotype in gpt1/alg6 cells as gls2/alg6 double-mutant cells that transferred Man9 GlcNAc2 and were unable to remove the glucose units added by GT as they lacked GII, had a phenotype of growth and morphology very similar to that of wild-type cells at 28°C and were able to grow at 37°C. It has been reported that at least two oligosaccharides in RNase B were required to immunoprecipitate the glycoprotein with anticalnexin antibodies (Rodan et al., 1996). This indicates that the affinity of calnexin for monoglucosylated oligosaccharides is not as high as those of classical plant lectins. This was confirmed by the fact that isolated monoglucosylated oligosaccharides could be washed out with buffer from an immobilized calnexin column (Ware et al., 1995). It may be speculated, therefore, that even in the absence of GII association–dissociation cycles of binding of calnexin with the glycoprotein(s) responsible for the altered phenotype of gpt1/alg6 cells could occur if the glycoprotein had a single (or few) oligosaccharide.
As mentioned above, calnexin is essential for S. pombe viability (Jannatipour and Rokeach, 1995; Parlati et al., 1995a ). The fact that gpt1/alg6 mutant cells grew, albeit at a slower rate, at 28°C indicates that calnexin has another role, besides that of binding monoglucosylated oligosaccharides in this yeast. The calnexin homologue in S. cerevisiae is not essential for survival, and although mammalian cells devoid of calnexin are viable it is unknown whether cells lacking both calnexin and calreticulin can survive (Parlati et al., 1995 b; Scott and Dawson, 1995).
It is worth mentioning that S. cerevisiae alg5 or alg6 mutants are similar to the gpt1/alg6 double mutant in S. pombe as Man9GlcNAc2 is transferred to proteins in them and there is no GT in S. cerevisiae. Nevertheless, no thermosensitive phenotype has been observed in S. cerevisiae alg5 or alg6 mutants (Jakob et al., 1998).
Trypanosomatid protozoa are similar to alg6 mutants in S. pombe as in them unglucosylated oligosaccharides are transferred to proteins and they have GT (Parodi, 1993). However, in this case no underglycosylation of glycoproteins is expected to occur as the protozoan oligosaccharyltransferase transfers, in cell-free assays, Glc1-3Man9 GlcNAc2 and Man7-9GlcNAc2 at the same rate (Bosch et al., 1988). It may be speculated that the specificity of the oligosaccharyltransferase in these protozoa is an evolutionary consequence of the loss of the ability to form Glc3Man9GlcNAc2 to prevent underglycosylation of proteins and thus a potential source of stress.
Abbreviations used in this paper
- BiP
binding protein
- DNJ
1-deoxynojirimycin
- Endo H
endo-β-N-acetylglucosaminidase H
- GI
glucosidase I
- GII
glucosidase II
- GT
UDP-Glc:glycoprotein glucosyltransferase
Footnotes
This work has received financial support from the United States Public Health Service (Grant GM 44500), from the Howard Hughes Medical Institute (Grant 75197-553502), from the University of Buenos Aires, and from the National Research Council (Argentina).
References
- Ballou L, Gopal P, Krummel B, Tammi M, Ballou C. A mutation that prevents glucosylation of the lipid-linked oligosaccharide precursor leads to underglycosylation of secreted yeast invertase. Proc Natl Acad Sci USA. 1986;83:3081–3085. doi: 10.1073/pnas.83.10.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M, Trombetta S, Engstrom U, Parodi AJ. Characterization of dolichol diphosphate oligosaccharide:protein oligosaccharyltransferase and of glycoprotein processing glucosidases occurring in trypanosomatids. J Biol Chem. 1988;263:17360–17365. [PubMed] [Google Scholar]
- Choudhury P, Liu Y, Bick RJ, Sifers RN. Intracellular association between UDP-glucose:glycoprotein glucosyltransferase and an incompletely folded variant of α1-antitrypsin. J Biol Chem. 1997;272:13446–13451. doi: 10.1074/jbc.272.20.13446. [DOI] [PubMed] [Google Scholar]
- Dairaku K, Spiro RG. Phylogenetic survey of endomannosidase indicates late evolutionary appearance of this N-linked oligosaccharide processing enzyme. Glycobiology. 1997;4:579–586. doi: 10.1093/glycob/7.4.579. [DOI] [PubMed] [Google Scholar]
- Fernández F, Jannatipour M, Hellman U, Rokeach L, Parodi AJ. A new stress protein: synthesis of Schizosaccharomyces pombeUDP-Glc: glycoprotein glucosyltransferase mRNA is induced under stress conditions but the enzyme is not essential for cell viability. EMBO (Eur Mol Biol Organ) J. 1996;15:705–713. [PMC free article] [PubMed] [Google Scholar]
- Fernández F, Trombetta SE, Hellman U, Parodi AJ. Purification to homogeneity of UDP-Glc:glycoprotein glucosyltransferase from Schizosaccharomyces pombe and apparent absence of the enzyme from Saccharomyces cerevisiae. . J Biol Chem. 1994;269:30701–30706. [PubMed] [Google Scholar]
- Gañán S, Cazzulo JJ, Parodi AJ. A major proportion of N-glycoproteins is transiently glucosylated in the endoplasmic reticulum. Biochemistry. 1991;30:3098–3104. doi: 10.1021/bi00226a017. [DOI] [PubMed] [Google Scholar]
- Gotz G, Gañán S, Parodi AJ. Glucosylation of glycoproteins in Crithidia fasciculata. . Mol Biochem Parasitol. 1991;45:265–274. doi: 10.1016/0166-6851(91)90094-m. [DOI] [PubMed] [Google Scholar]
- Hebert DN, Foellmer B, Helenius A. Glucose trimming and reglucosylation determine glycoprotein association with calnexin in the endoplasmic reticulum. Cell. 1995;81:425–433. doi: 10.1016/0092-8674(95)90395-x. [DOI] [PubMed] [Google Scholar]
- Hebert DN, Foellmer B, Helenius A. Calnexin and calreticulin promote folding, delay oligomerization and suppress degradation of influenza hemagglutinin in microsomes. EMBO (Eur Mol Biol Organ) J. 1996;15:2961–2968. [PMC free article] [PubMed] [Google Scholar]
- Helenius A. How N-linked oligosaccharides affect glycoprotein folding in the endoplasmic reticulum. Mol Biol Cell. 1994;5:253–265. doi: 10.1091/mbc.5.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A, Trombetta ES, Hebert DN, Simons JF. Calnexin, calreticulin and the folding of glycoproteins. Trends Cell Biol. 1997;7:193–200. doi: 10.1016/S0962-8924(97)01032-5. [DOI] [PubMed] [Google Scholar]
- Hendrick JP, Hartl FU. Molecular chaperone functions of heat-shock proteins. Annu Rev Biochem. 1993;62:349–384. doi: 10.1146/annurev.bi.62.070193.002025. [DOI] [PubMed] [Google Scholar]
- Imperiali B, Rickert KW. Conformational implications of asparagine-linked glycosylation. Proc Natl Acad Sci USA. 1995;80:7466–7470. doi: 10.1073/pnas.92.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro J, Saitou A, Durán A, Ribas JC. cps1 +, a Schizosaccharomyces pombe gene homolog of Saccharomyces cerevisiae FKSgenes whose mutation confers hypersensitivity to cyclosporin A and papulacandin B. J Bacteriol. 1997;179:7653–7662. doi: 10.1128/jb.179.24.7653-7662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob CA, Burda P, te Heesen S, Aebi M, Roth J. Genetic tailoring of N-linked oligosaccharides: the role of glucose residues in glycoprotein processing in Saccharomyces cerevisiae in vivo. . Glycobiology. 1998;8:155–164. doi: 10.1093/glycob/8.2.155. [DOI] [PubMed] [Google Scholar]
- Jannatipour M, Rokeach LA. The Schizosaccharomyces pombehomologue of the chaperone calnexin is essential for viability. J Biol Chem. 1995;270:4845–4853. doi: 10.1074/jbc.270.9.4845. [DOI] [PubMed] [Google Scholar]
- Lubas WA, Spiro RG. Golgi endo-α-mannosidase from rat liver, a novel N-linked carbohydrate unit processing enzyme. J Biol Chem. 1987;262:3775–3781. [PubMed] [Google Scholar]
- Marquardt T, Helenius A. Misfolding and aggregation of newly synthesized proteins in the endoplasmic reticulum. J Cell Biol. 1992;117:505–513. doi: 10.1083/jcb.117.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. . Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Parker CG, Fessler LI, Nelson RE, Fessler JH. DrosophilaUDP-Glc:glycoprotein glucosyltransferase: sequence and characterization of an enzyme that distinguishes between denatured and native proteins. EMBO (Eur Mol Biol Organ) J. 1995;14:1294–1303. doi: 10.1002/j.1460-2075.1995.tb07115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlati F, Dignard D, Bergeron JJ, Thomas DY. The calnexin homologue cnx1 + in Schizosaccharomyces pombe, is an essential gene which can be complemented by its soluble ER domain. EMBO (Eur Mol Biol Organ) J. 1995a;14:3064–3072. doi: 10.1002/j.1460-2075.1995.tb07309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlati F, Dominguez M, Bergeron JJ, Thomas DY. b. Saccharomyces cerevisiae CNE1encodes an endoplasmic reticulum (ER) membrane protein with sequence similarity to calnexin and calreticulin and functions as a constituent of the ER quality control apparatus. J Biol Chem. 1995;270:244–253. doi: 10.1074/jbc.270.1.244. [DOI] [PubMed] [Google Scholar]
- Parodi AJ. N-glycosylation in trypanosomatid protozoa. Glycobiology. 1993;3:193–199. doi: 10.1093/glycob/3.3.193. [DOI] [PubMed] [Google Scholar]
- Pidoux AL, Armstrong J. Analysis of the BiP gene and identification of an ER retention signal in Schizosaccharomyces pombe. . EMBO (Eur Mol Biol Organ) J. 1992;11:1583–1591. doi: 10.1002/j.1460-2075.1992.tb05203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitman ML, Trowbridge IS, Kornfeld S. A lectin-resistant mouse lymphoma cell line is deficient in glucosidase II, a glycoprotein processing enzyme. J Biol Chem. 1982;257:10357–10363. [PubMed] [Google Scholar]
- Ribas JC, Roncero C, Rico H, Durán A. Characterization of a Schizosaccharomyces pombemorphological mutant altered in the galactomannan content. FEMS Microbiol Lett. 1991;79:263–268. doi: 10.1016/0378-1097(91)90096-s. [DOI] [PubMed] [Google Scholar]
- Rodan AR, Simons JF, Trombetta ES, Helenius A. N-linked oligosaccharides are necessary and sufficient for association of glycosylated forms of bovine RNase with calnexin and calreticulin. EMBO (Eur Mol Biol Organ) J. 1996;15:6921–6930. [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning. A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Scott JE, Dawson JR. MHC class I expression and transport in a calnexin-deficient cell line. J Immunol. 1995;155:143–148. [PubMed] [Google Scholar]
- Sousa M, Parodi AJ. The molecular basis for the recognition of misfolded glycoproteins by the UDP-Glc:glycoprotein glucosyltransferase. EMBO (Eur Mol Biol Organ) J. 1995;14:4196–4203. doi: 10.1002/j.1460-2075.1995.tb00093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa M, Ferrero-García M, Parodi AJ. Recognition of the oligosaccharide and protein moieties of glycoproteins by the UDP-Glc:glycoprotein glucosyltransferase. Biochemistry. 1992;31:97–105. doi: 10.1021/bi00116a015. [DOI] [PubMed] [Google Scholar]
- Tabuchi M, Iwaihara O, Ohtani Y, Ohuchi N, Sakurai J-I, Morita T, Iwahara S, Takegawa K. Vacuolar protein transport sorting in fission yeast: cloning, biosynthesis, transport, and processing of carboxypeptidase Y from Schizosaccharomyces pombe. . J Bacteriol. 1997;179:4179–4189. doi: 10.1128/jb.179.13.4179-4189.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta S, Bosch M, Parodi AJ. Glucosylation of glycoproteins by mammalian, plant, fungal and trypanosomatid protozoa microsomal proteins. Biochemistry. 1989;28:8108–8116. doi: 10.1021/bi00446a022. [DOI] [PubMed] [Google Scholar]
- Ugalde RA, Staneloni RJ, Leloir LF. Microsomal glucosidases acting on the saccharide moiety of the glucose-containing dolichyl diphosphate oligosaccharide. Biochem Biophys Res Commun. 1979;91:1174–1181. doi: 10.1016/0006-291x(79)92003-5. [DOI] [PubMed] [Google Scholar]
- Ware FE, Vassilakos A, Peterson PA, Jackson MR, Lehrman MA, Williams DB. The molecular chaperone calnexin binds Glc1Man9 GlcNAc2oligosaccharide as an initial step in recognizing unfolded glycoproteins. J Biol Chem. 1995;270:4697–4704. doi: 10.1074/jbc.270.9.4697. [DOI] [PubMed] [Google Scholar]
- Wright A, Robbins PW. The enzymatic synthesis of uridine diphosphate [14C]glucose. Biochim Biophys Acta. 1965;104:594–595. [Google Scholar]
- Zapun A, Petrescu SM, Rudd PM, Dwek RA, Thomas DY, Bergeron JJM. Conformation independent binding of monoglucosylated ribonuclease B to calnexin. Cell. 1997;88:29–38. doi: 10.1016/s0092-8674(00)81855-3. [DOI] [PubMed] [Google Scholar]
- Ziegler FD, Gemmil TR, Trimble RB. Glycoprotein synthesis in yeast. Early events in N-linked oligosaccharide processing in Schizosaccharomyces pombe. . J Biol Chem. 1994;268:12527–12535. [PubMed] [Google Scholar]