Abstract
Amt/MEP/Rh proteins are a family of integral membrane proteins implicated in the transport of NH3, CH2NH2, and CO2. Whereas Amt/MEP proteins are agreed to transport ammonia (NH3/NH4+), the primary substrate for Rh proteins has been controversial. Initial studies suggested that Rh proteins also transport ammonia, but more recent evidence suggests that they transport CO2. Here we report the first structure of an Rh family member, the Rh protein from the chemolithoautotrophic ammonia-oxidizing bacterium Nitrosomonas europaea. This Rh protein exhibits a number of similarities to its Amt cousins, including a trimeric oligomeric state, a central pore with an unusual twin-His site in the middle, and a Phe residue that blocks the channel for small-molecule transport. However, there are some significant differences, the most notable being the presence of an additional cytoplasmic C-terminal α-helix, an increased number of internal proline residues along the transmembrane helices, and a specific set of residues that appear to link the C-terminal helix to Phe blockage. This latter linkage suggests a mechanism in which binding of a partner protein to the C terminus could regulate channel opening. Another difference is the absence of the extracellular π-cation binding site conserved in Amt/Mep structures. Instead, CO2 pressurization experiments identify a CO2 binding site near the intracellular exit of the channel whose residues are highly conserved in all Rh proteins, except those belonging to the Rh30 subfamily. The implications of these findings on the functional role of the human Rh antigens are discussed.
Keywords: carbon dioxide, channel, proline kinks
Rhesus (Rh) proteins are best known for their role as a human blood factor and antigen (Rh+/Rh−) (1, 2). Functionally, Rh proteins appear to be membrane channels (based on their electroneutral transport properties and presumed hydrophobic selectivity filter), but their exact substrate has remained controversial (3, 4). Based in part on their homology to Amt/Mep proteins and their complementary organismal distribution, Rh proteins were initially proposed to transport ammonia (NH3/NH4+) (4–6). In support of this role, yeast mep mutants recovered growth at low ammonium after the introduction and expression of the human rhAG gene (6, 7). Subsequent studies on the transport of human Rh proteins in oocytes and yeast indicated that ammonia transport was unaffected by the electrochemical potential, indicating that if Rh proteins transport ammonia it is as NH3 (3, 8–10).
Physiological studies on the green alga Chlamydomonas reinhardtii, however, showed that its Rh1 protein was involved in CO2 metabolism, whereas its major Amt protein, Amt4, was involved in nitrogen metabolism. Current support for the function of Rh proteins in CO2 transport include (i) the high expression of Rh1 during high CO2-dependent growth of C. reinhardtii (11); (ii) the observation that RNA interference of C. reinhardtii rh1 mRNA results in a growth defect at high CO2 but does not affect methylamine uptake; (iii) the observation that RNA interference of C. reinhardtii rh1 during growth on acetate leads to failure to express genes under control of the carbon concentration mechanism (12); (iv) the lower CO2 transport rate of Rhnull cells as revealed by mass spectroscopic studies (13); and (v) phylogenetic analyses that clearly separate Rh and Amt/MEP proteins into different families with distinctive features (14, 15).
One of four bacterial Rh proteins identified by phylogenetic analyses (14) and subsequent sequence alignments is from Nitrosomonas europaea, an obligate chemolithoautotroph that utilizes CO2 as its principal carbon source and derives all reducing equivalents for energy and biosynthesis from the oxidation of ammonia to nitrite. This organism fixes CO2 through a type I ribulose bisphosphate carboxylase (Rubisco) located in the cytoplasm while the first step of ammonia oxidation takes place at the ammonia monooxygenase (AMO), an integral membrane protein (16, 17). Recent biochemical studies have implicated the N. europaea Rh protein in ammonia uptake based on Rh transcript analysis and 14C-labeled methylamine uptake assays on N. europaea cells grown under oxic and anoxic conditions (18). Analogous studies to explore its putative role in CO2 transport have been hindered by the lack of a reliable biochemical assay.
Given the clear phylogenetic differences between Rh and Amt proteins, a structural comparison of the two families could be illuminating. Although there have now been a number of structures of Amt/Mep proteins, structures of Rh proteins have lagged. We here present the structure of the N. europaea Rh protein and its structure in complex with CO2.
Results and Discussion
The Protein Fold.
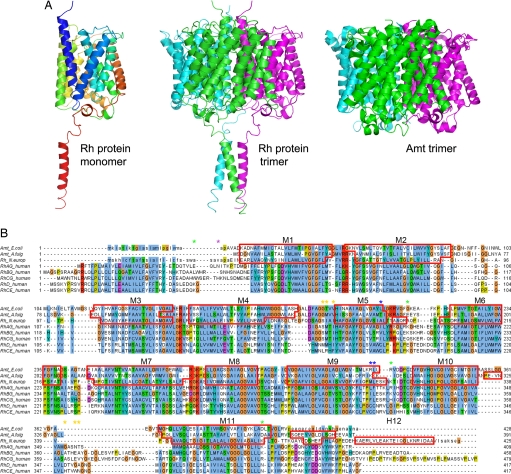
The structure of the N. europaea Rh protein was determined to 1.85-Å resolution [Fig. 1A and supporting information (SI) Table 1]. As observed in the previous Escherichia coli AmtB (19, 20) and Archaeoglobus fulgidus Amt1 structures (21), the N. europaea Rh protein structure reveals an α3 homotrimer generated by a crystallographic three-fold axis that passes through the center of the three subunits (Figs. 1A and 2A). The overall fold and membrane topology of each subunit are also similar. Structural alignment of the Rh protein to E. coli AmtB using the program DaliLite (22) yields an rmsd of 2.2 Å for 322 Cα atoms based on their overlapping residues (SI Fig. 3). Each Rh monomer comprises 11 transmembrane helices (Fig. 1A) with pseudo two-fold symmetry (helices 1–5 being related to helices 6–10) and contains a central channel that is presumed to mediate gas transport (Fig. 2B). The first 30 aa were absent and, based on computational and MALDI MS analysis, are likely cleaved by a signal peptidase around residue 27 (see SI Text). It should be mentioned that topology prediction and immunochemical studies have suggested that human Rh proteins have 12 transmembrane helices with both termini ending in the cytoplasm (23–25). Human Rh proteins have a longer N-terminal sequence than the Rh protein from N. europaea (Fig. 1B) and likely have an additional N-terminal transmembrane helix within this region.
Fig. 1.
Structure of the N. europaea Rh protein. (A Left) Ribbon diagram of N. europaea Rh protein subunit colored in rainbow from blue to red. (A Center) Ribbon diagram of N. europaea Rh protein trimer colored by subunit. (A Right) Ribbon diagram of E. coli AmtB colored by subunit. Note the additional C-terminal helix on the Rh protein. The diagram is oriented such that the periplasmic surface is on the top and the cytoplasmic face is on the bottom. (B) Sequence alignment of the N. europaea Rh protein to the E. coli AmtB and A. fulgidus Amt1 based on their respective structures and to the five human Rh proteins based on the program ClustalW (34). The residues not observed in the Rh protein structure are shown in lowercase. Segments assigned to α-helices are framed with red lines. Asterisks mark topologically relevant residues: glycosylation and protease sensitive sites on RhAG are indicated with purple and green asterisks, respectively; highly antigenic and potentially palmitoylated residues of RhD are labeled with orange and blue asterisks.
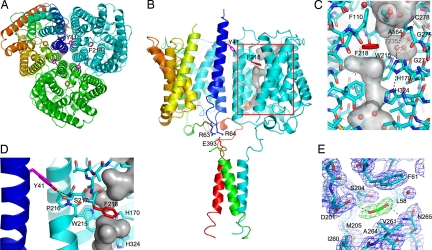
Fig. 2.
Functionally relevant sites within the N. europaea Rh protein. (A) Periplasmic view of the N. europaea Rh protein along the three-fold axis. Two key residues implicated in channel opening are colored in violet (Tyr-41) and red (Phe-218). Subunits are colored in cyan, green, and rainbow [from blue (N terminus) to red (C terminus)]. (B) Side view of the N. europaea Rh protein emphasizing the key residues implicated in partner protein-mediated channel gating. In the cyan subunit the surface of the channel is shown together with the location of the putative CO2 binding site. Tyr-41 on helix M1 from one subunit (in rainbow) is proposed to regulate the extent of kinking of transmembrane helix M6 on the adjacent subunit (in cyan). The position of Tyr-41 in turn is linked to the position of helix M1. Partner protein binding to the C-terminal α-helix is proposed to alter the position of the M1 helix via salt bridge interactions between Glu-393 on the C-terminal helix and Arg-63 and Arg-64 on helix M1. (C) Key channel residues on the periplasmic side of the channel. The protein is depicted in stick form with carbon atoms colored in cyan and the remaining atoms in CPK. Waters and the putative ethylene glycol are shown in ball-and-stick form. Potential cavities are highlighted with a transparent surface colored in gray. For the unique hydrophilic pocket adjacent to Phe-218, the hydrogen-bonding interactions of its three waters are depicted as black dotted lines. (D) Interactions between Tyr-41 and the M6 helix of the adjacent subunit. Potential hydrogen bonds between Tyr-41 and Ser-217 are shown as black dotted lines. (E) Weighted density maps surrounding the CO2 binding site, 2mFO–DFC (in black, contoured at 1.0σ) and mFO–DFC (in green, contoured at 3.0σ, and in red, contoured at −3.0σ) with terms as determined by REFMAC5 (36).
The structure of the Rh protein also reveals some clear differences from its AmtB cousins. Its most distinct feature is the presence of an additional cytoplasmic C-terminal α-helix directed along the crystallographic three-fold axis that leads away from the main oligomer (Fig. 1A). The three C-terminal helices from each monomer come together along the three-fold axis to form a left-handed three-helix bundle. Despite these interactions, the C-terminal helix appears fairly mobile based on its much higher average residue B factor (76 Å2) than the rest of the protein (20 Å2).
Two other short nontransmembrane spanning α-helices are located on the cytoplasmic face with orientations nearly parallel to the membrane plane. The first short α-helix (residues 186–190) is located between transmembrane helices M5 and M6. The second short α-helix (residues 380–384), which is also observed in the Amt1 and AmtB–GlnK structures (21, 26, 27), is part of the segment that leads from the transmembrane helix M11 to the novel C-terminal helix unique to the Rh structure.
Structural comparisons of the Rh and Amt structures reveal that the Rh periplasmic surface is smaller than Amt proteins because the Rh protein has much shorter loops between transmembrane helices M2 and M3 and helices M10 and M11. Although the structural consequences of these features are somewhat minor in this region, a narrower periplasmic face, a more important consequence is that it leads to a significant shift in the predicted sequence alignment to Amt proteins. The importance of this shifted alignment is most clearly manifested at the C terminus. Whereas the mature E. coli AmtB, A. fulgidus Amt1, and N. europaea Rh proteins are approximately the same size, the Rh protein has a much longer C terminus with 54 residues after the M11 helix, as compared with 25 and 24 for E. coli AmtB and A. fulgidus Amt1, respectively (Fig. 1B). It is this difference that results in the presence of the novel C-terminal helix.
Another distinct feature of the N. europaea Rh protein structure, as compared with the Amt structures, is a much higher occurrence of internal prolines within the transmembrane helices. Whereas only two such prolines are observed in each of the Amt/MEP protein structures, six internal prolines (Pro-82, Pro-140, Pro-216, Pro-229, Pro-306, and Pro-327) are present in the N. europaea Rh protein structure as illustrated in SI Fig. 4. The potential significance of this observation stems from the fact that such internal prolines can lead to distortions or kinks of the transmembrane helices (28). Thus, their presence can impact protein stability (e.g., helix packing) and play a critical role in function (e.g., dictating the hinge sites of a conformational change). Indeed, the potential functional importance of one such proline kink is discussed subsequently together with a proposed mechanism of channel opening.
The Extracellular Vestibule.
In their analysis of their E. coli AmtB structure, Stroud and coworkers (19) identified a site that they propose facilitates ammonium binding at the entrance to the channel on the extracellular face. This putative recruitment site was formed from aromatic residues Trp-148, Phe-103, and Phe-107 and appeared to bind a CH3NH3+ ion, implying recruitment via a π-cation interaction.
The Rh protein lacks this potential π-cation binding site. An aromatic residue corresponding to Trp-148 is absent in the Rh protein, and the helix on which it resides is oriented away from this binding site, so there is no corresponding residue. Phe-103 in E. coli AmtB is an isoleucine (Ile-106) in the N. europaea Rh. Only Phe-107 in AmtB is conserved, as Phe-110 in the Rh protein, but even this residue adopts a different orientation, as illustrated in SI Fig. 5.
This different conformation of Phe-110 may be linked to the binding of a small molecule based on the presence of strong connected density that occupies its location in the Amt structures. Attempts to model this connected density suggest an ethylene glycol molecule (perhaps originating as an impurity from PEG) based on the connected density and acceptable hydrogen-bonding interactions, but two waters, and/or two sets of waters at 50% occupancy, can also be used to fit the observed density (SI Fig. 6). Analysis of the residues surrounding this small-molecule site suggest that it is unlikely to serve as an alternate π-cation binding site because it is fairly hydrophobic with only one aromatic side chain, Phe-110. There are several carbonyl side chains that could help to stabilize ammonia binding, but CO2 binding would need to be stabilized by the hydrophobicity of the pocket and/or through interactions with neighboring waters in the channel.
The Phe Barrier.
Phe-110 and Phe-218 of N. europaea Rh are two highly conserved residues in the channels of Amt and Rh proteins that lie just under the extracellular vestibule. In Amt proteins, both conserved Phe residues adopt positions that block substrate passage through the pore (19–21). In their analysis of the E. coli AmtB structure, Stroud and coworkers (19) proposed a passive mechanism for opening of the channel in which the steric constriction provided by these Phe residues (Phe-107 and Phe-215) was removed by dynamic motion. This hypothesis was subsequently supported by Andrade et al. (21), who, in their analysis of the A. fulgidus Amt1 structure, noted that the more buried of the two conserved phenylalanines, Phe-204, had higher thermal parameters than the surrounding residues, suggesting that this Phe residue exhibits greater mobility.
In the N. europaea Rh structure, Phe-110 adopts an altered orientation, and thus only Phe-218 serves as a potential steric barrier. In examining the location of Phe-218, one notable feature with potential relevance to the mechanism of substrate transport is the presence of a neighboring proline (Pro-216) on the same α-helix (M6) as Phe-218. This proline leads to a helical kink whose bending angle dictates the position of Phe-218. This observation is significant in that it opens the possibility that helical kinking could be used to regulate the position of the Phe-218 side chain and, in turn, the open/closed state of the Rh protein channel.
The Twin-His Site.
Another feature observed in the Rh protein structure that is conserved in Amt proteins is the twin histidine (twin-His) site positioned just below Phe-218 (Fig. 2C). This site, which comprises His-170 and His-324, is unusual in that the two His residues adopt a coplanar orientation that may stabilize an H+ ion between them. This twin-His appears to be fairly hydrophobic based on the lack of bound water molecules in the refined structure in contrast to the rest of the channel, which is likely hydrophilic given the numerous water molecules.
The role of the twin-His site has been controversial. Stroud and coworkers (19) noted interactions of this site with ammonia in the AmtB structure and thus suggested that its role might be in ammonium ion deprotonation. This proposal was subsequently supported by combined structural and mutagenesis studies that linked the efficiency of transport with the conserved presence of the twin-His group (29). The hydrophobic twin-His site may also aid in selectivity by blocking the passage of small cations (such as Na+ and K+) while allowing the passage of neutral gas molecules such as NH3 and CO2 (19, 20).
The conservation of the twin-His site in the N. europaea Rh protein and other non-Rh30 Rh proteins may suggest a similar role(s). One difference with potential functional significance, however, is that, in the N. europaea Rh structure, the Nε2 atoms of both His-170 and His-324 form hydrogen bonds to neighboring water molecules. This is in contrast to the E. coli AmtB and A. fulgidus Amt1 structures, in which only one of the two histidines (corresponding to His-324 in N. europaea Rh) has a hydrogen-bonding partner.
The origin of this difference stems from a residue change in N. europaea Rh protein that is conserved for nearly the entire Rh protein family. Whereas in Amt proteins a conserved Thr provides steric hindrance that prevents water from accessing the more extracellular histidine, in N. europaea Rh this residue is replaced by a conserved Gly (Gly-275). The much smaller side chain of Gly-275 results in a hydrophilic pocket that is filled by three water molecules (Fig. 2C). The first water neighbors Phe-218 and forms hydrogen-bonding interactions with the side chain of His-170, the main-chain carbonyl of Ala-164, and the second water. The second water in turn has two additional hydrogen-bonding interactions with the side chain of Trp-215 and main-chain carbonyl of Gly-271, and a potential Van der Waals interaction with Gly-275. The third water hydrogen bonds to Thr-235, Cys-278, and the second water. The size and hydrophilicity of this pocket and its proximity to the Phe-218 steric barrier make this pocket an attractive target for further study.
A Cytoplasmic CO2 Binding Pocket.
As mentioned previously, there is a significant body of evidence in support of the role of Rh proteins in CO2 transport. To identify potential CO2 binding sites, native Rh protein crystals were pressurized under CO2 and then flash-cooled in liquid N2. These studies reveal a CO2 binding site within a deep cavity near the channel exit on the cytoplasmic side of the protein (Fig. 2 B and E). The putative binding site is formed by residues Leu-58, Phe-61, Asp-201, Ser-204, Met-205, Ile-260, Val-261, Ala-264, and Asn-265, which are fairly conserved in non-Rh30 Rh proteins.
Support for CO2 binding within this cavity comes from differences in the electron density maps of the native and CO2-pressurized crystals. Whereas in the native Rh protein structure these maps reveal positive density that appears to fit a water molecule, in the CO2-pressurized crystal structure the density is larger and elongated consistent with a linear triatomic species. Fitting and refinement of this density as CO2, formate, and two waters reveal that a CO2 molecule fits this density best. A formate ligand has negative FO–FC density at the bend of the formate carboxylate, whereas a model comprising two waters has positive density in between the waters. The final model based on occupancy refinement contains 73% bound CO2. The weighted 2mFO–DFC and mFO–DFC are shown in Fig. 2E and SI Fig. 7. Residues Asp-201, Ser-204, and Asn-265 form potential hydrogen bonds with the bound CO2 although the hydrogen bond with Asp 201 is slightly long (3.3 Å) and the geometry of the CO2 oxygen is not ideal for the Ser-204 interaction. Notably, Ser-204 appears more disordered in the native Rh structure, presumably because of its partial interaction with the second water in that structure.
The role for this cytoplasmic CO2 binding site is currently unclear, but if the physiological substrate of Rh proteins is CO2, as has been proposed (4, 11, 12), one possibility could be as a secondary binding site to promote CO2 movement in and out of the central channel. One important property of CO2 is that, whereas the major form in solution is bicarbonate, a significant fraction will remain as dissolved CO2. In humans, for example, the dissolved CO2 concentrations varies between ≈5% and ≈10% depending on the pH (30). Dissolved CO2 is hydrophobic and would likely associate with the hydrophobic patch that lies near the twin-His site. As has been suggested for the Amt proteins, the hydrophobicity of the twin-His site suggests its role as part of the selectivity filter that distinguishes neutral molecules, such as CO2 and NH3, from water and ions, such as K+ and Na+ (19, 20). Unlike NH3, whose association within the hydrophobic twin-His site channel can be made unfavorable by protonation, CO2 removal may need to be assisted. The putative CO2 binding site could facilitate this removal by providing an intermediate binding site that binds CO2 more tightly, making its removal from the selectivity filter more favorable. Notably, the putative CO2 binding site lies along the pathway for CO2 egress but does not block the channel itself. Thus, even when this site is bound, other CO2 molecules could pass without binding. Alternatively, we note that the back of the CO2 binding site cavity is blocked from the cytoplasm by the side chain of a single residue, Met-205, whose directed movement could provide an alternative route for CO2 egress. If this movement could be linked to partner protein binding, it would provide a means to directly deliver the CO2 to a specific protein target.
A Proposed Gating Mechanism for Channel Opening.
The presence of the cytoplasmic C-terminal helix is highly suggestive of a protein–protein interaction relevant to function. The likelihood of this possibility is further enhanced by the recent structures of AmtB–GlnK complexes, which have confirmed that GlnK suppresses ammonia uptake by binding to the AmtB cytoplasmic face (26, 27). Binding of proteins to the C-terminal helix of the Rh protein could serve a similar role in blocking substrate transport, but there are a number of other possibilities. Two possibilities linked to the putative CO2 binding site include (i) facilitated substrate transfer mediated by partner protein binding and (ii) substrate-induced protein recruitment as a mechanism of sensing, as has been suggested for the AmtB of Azospirillum brasilense (31).
Our analysis of the N. europaea Rh protein structure, however, suggests that partner protein binding to the cytoplasmic C-terminal helix may be important for channel gating. Recall that one of the concerns regarding AmtB/Mep/Rh proteins discussed previously is the fact that the conserved Phe residues block the channel closing it for transport. Our analysis of the Rh protein suggests that the orientation of the key Phe could be regulated by partner protein binding. The key features and interactions that support this possibility are (i) the presence of the C-terminal helix and (ii) the fact that this helix has a glutamate that is salt-bridged to two arginines (Arg-63 and Arg-64) at the C terminus of helix M1. How the M1 helix could be linked to channel opening comes from the fact that Tyr-41 (on helix M1) is hydrogen-bonded to the main-chain nitrogen and side-chain hydroxyl of Ser-217 (on helix M6) of an adjacent subunit (Fig. 2D). Notably, Ser-217 lies directly adjacent to the internal proline (Pro-216) that induces a helical kink in the M6 helix and the phenylalanine (Phe-218) that is a potential steric hindrance for transport (Fig. 2 B–D). Thus, changes in the interactions between Tyr-41 and Ser-217 could influence the magnitude of helix M6 bending and, in turn, regulate channel opening (shift of Phe-218) or closing.
Based on our analysis of these interactions and the a priori requirement of there being an unhindered pathway for substrate passage, we propose a mechanism in which channel opening of the Rh protein is regulated by binding of a putative partner protein to the C-terminal helix. Possible partner proteins include carbonic anhydrase, Rubisco, or, if Rh transports NH3, AMO or glutamine synthetase, each of whose substrates could be metabolically linked to the N. europaea Rh protein.
The key residues in this proposed mechanism are generally conserved in Rh proteins, with the exception of the Rh30 subfamily. There are, however, a limited number of non-Rh30 Rh proteins with shortened C-terminal sequences whose presence suggests variation in either the mechanism or the protein partner that interacts with the Rh protein in their respective organisms. This variation is perhaps most notable for the three other Rh proteins from bacteria: Nitrosospira multiformis, Acidobacteria bacterium Ellin 345, and Kueninia stuttgartiensis. The C-terminal helix of the N. multiformis Rh protein is approximately half the size of that of the N. europaea Rh protein but could still facilitate partner protein binding. The lengths of the C-terminal tails of A. bacterium and K. stuttgartiensis, however, indicate that the last helix is absent, suggesting that these organisms may use a distinct mechanism of Rh protein regulation. These differences could be linked to their different CO2 fixation pathways (S. Kustu and W. Inwood, personal permission confirmed communication): N. europaea and N. multiformis both fix CO2 via Rubisco; K. stuttgartiensis appears to convert CO2 to CO via an CO dehydrogenase/acetyl-CoA synthase based on studies of carbon isotopic fractionation in cells (32) and genomic analysis of possible carbon fixation pathways (33), and A. bacterium is presumed to incorporate CO2 via the reverse TCA cycle. All Amt/Mep proteins also have short C termini and differ in several key residues (e.g., Tyr-41 and Pro-216). Thus, if channel opening is gated in Amt/Mep proteins, it must also occur by an alternate mechanism.
Support for a Distinct Role for Rh30 Proteins.
One feature we noted in our structural analyses was the common divergence of the Rh30 subfamily. To evaluate this issue in greater detail, a multiple sequence alignment was calculated with the program ClustalW (34) using the 111 Rh proteins from 41 species identified by Huang and Peng (14). An alignment of the N. europaea Rh protein with the five human Rh proteins is included as part of Fig. 1B, and the overall alignment is provided in SI Table 2. Using this alignment we examined the key residues proposed to be involved in channel gating and CO2 binding. Perhaps the most significant outcome of this combined structural and sequence analysis is strong support for the view that the Rh30 proteins are a distinct subfamily of Rh proteins with a potentially different functional role in cells (14). This suggestion is consistent with the previous observation by Huang and Peng (14) that the Rh30 subfamily (RhD and RhCE) has undergone recent and rapid evolution based on its phylogenetic branch length.
One example that illustrates the divergence between Rh30 proteins and other Rh subfamilies is the conservation of the key residues comprising the central channel—namely, the pair of His residues (His-170 and His-324 in N. europaea Rh) forming the twin-His site and the pair of Phe residues (Phe-110 and Phe-218) that block the channel. Whereas the twin-His site is conserved in all 77 non-Rh30 Rh proteins, only five of the 24 Rh30 proteins retain this twin-His site. In 19 Rh30 proteins that have an altered twin-His site, each has a phenylalanine substituted for His-324, and five have a further substitution of tyrosine for His-170. A similar divergence was noted for the pair of Phe residues that block the central channel. In all Rh30 proteins, Phe-110 in N. europaea Rh is replaced by a smaller aliphatic amino acid (the most common being a methionine), whereas Phe-218 is replaced by an alternate amino acid in five of 24 Rh30 proteins. In the 77 other Rh proteins both Phe residues are highly conserved: Phe-110 is replaced by another residue in only seven instances, whereas Phe-218 is completely conserved.
Another site of potential functional significance is the putative CO2 binding site. In Rh30 proteins the residues forming this site are nearly all different from those observed in N. europaea Rh structure. In contrast, the CO2 binding site appears to be retained in all of the non-Rh30 Rh proteins with the residues forming the CO2 binding site being fairly conserved [residue in N. europaea Rh (percentage of conservation and primary amino acid): L58 (98% L), F61 (97% F), D201 (96% D), S204 (82% A), M205 (92% M), I260 (95% M), V261 (87% V), A264 (94% Q), and N265 (94% N)]. For the residues that are different, the replacement is generally by a residue with similar properties. This similarity is best exemplified for Asp-201 and Asn-265, the residues that form potential hydrogen-bonding interactions within the CO2-bound structure. In the five cases that exhibit replacement, all of the new residues are hydrogen-bonding residues of nearly the same size: Asp-201→His or Asn and Asn-265→Asp or Ser (twice).
One final illustration of the divergence of Rh30 and the other Rh proteins is the length of the C-terminal helix. Excluding the Rh30 subfamily, most Rh proteins (66 of 77) exhibit a long C terminus (≥15 residues past the conserved C-terminal proline, Pro-390 in N. europaea Rh), whereas all Rh30 proteins are short (<7 aa), as illustrated in SI Fig. 8.
Taken together these findings provide support for the view that Rh30 proteins are a distinct class of Rh protein with altered transport properties or function (11, 14). There are a number of possible origins for this evolutionary divergence, but two possibilities are suggested by their cell-type-specific expression in humans. Red blood cells solely express RhD, RhCE, and RhAG to form the Rh blood group substance. Given that the Rh blood group substance is critical for the structural integrity of red blood cells (2), one possibility is that the Rh30 proteins, RhD and RhCE, have adapted to serve this structural role (11, 14). The other possibility is that Rh30 proteins have evolved to serve an alternate transport role. Such an alternate transport function would be consistent with the observed residue differences in the channel and CO2 binding site.
Conclusions
Although this structure of the N. europaea Rh protein provides a framework to try to elucidate the physiological substrate of Rh proteins, it does not provide definitive support for either NH3 or CO2. Nevertheless, circumstantial evidence suggests that the physiological substrate is most likely CO2. This evidence includes (i) the fact that AMO, the primary enzyme that consumes ammonia, is membrane-bound, obviating the need for NH3 transport; (ii) the absence of the π-cation binding site proposed to recruit NH4+ in Amt proteins; and (iii) the identification of a possible CO2 binding site whose residues are primary conserved in Rh proteins, other than the Rh30 subfamily. Clearly other studies will be required to resolve this issue, but one intriguing is suggested by the Rh protein's C-terminal tail. If partner protein binding to this C-terminal tail leads to Rh protein activation or repression, then the identification of this partner protein could potentially resolve the identity of the true physiological substrate of the N. europaea Rh protein.
Materials and Methods
The structure of the Rh protein was determined by the Se-Met multiwavelength anomalous diffraction (MAD) method using data collected at the Stanford Synchrotron Radiation Laboratory. The details of the Rh protein overexpression and the structure determination are provided in SI Text. All of the structural figures presented in this article were prepared with the molecular display software package PYMOL (35).
Supplementary Material
Acknowledgments
TransMembrane Biosciences thanks Dr. Om Sahai of the National Science Foundation Small Business Innovation Research Program. This work was done partially at the Stanford Synchrotron Radiation Laboratory (SSRL), which is operated by the Department of Energy, Office of Basic Energy Sciences. The N. europaea Rh gene was kindly provided by Prof. Cheng-Han Huang (Laboratory of Biochemistry and Molecular Genetics, Lindsley F. Kimball Research Institute, New York Blood Center, New York). We thank Prof. Sydney Kustu for introducing us to this problem and for her numerous discussions, Bill Inwood for making us aware of the additional bacterial Rh protein sequences, Prof. Sunney Chan for his helpful insights, Yi Xiong for her assistance in preparaing pressurized crystals, and the staff at SSRL for the use of the SSRL gas pressure cell and for beamline assistance. The MAD script used to determine the phases was developed by Ana Gonzales and Qingyun Xu of SSRL. Methodologies to overexpress membrane proteins were developed at TransMembrane Biosciences with support from National Science Foundation Grant DMI0349777 and Department of Energy Grant ER84304 and with funding from the Defense Advanced Research Project Agency. This work was partially supported by National Institutes of Health Grant GM061796 (to M.K.C.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2B9Y and 2B9Z).
This article contains supporting information online at www.pnas.org/cgi/content/full/0709710104/DC1.
References
- 1.Avent ND, Reid ME. Blood. 2000;95:375–387. [PubMed] [Google Scholar]
- 2.Van Kim CL, Colin Y, Cartron JP. Blood Rev. 2006;20:93–110. doi: 10.1016/j.blre.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 3.Ludewig U. Transfus Clin Biol. 2006;13:111–116. doi: 10.1016/j.tracli.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 4.Kustu S, Inwood W. Transfus Clin Biol. 2006;13:103–110. doi: 10.1016/j.tracli.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Marini AM, Urrestarazu A, Beauwens R, Andre B. Trends Biochem Sci. 1997;22:460–461. doi: 10.1016/s0968-0004(97)01132-8. [DOI] [PubMed] [Google Scholar]
- 6.Westhoff CM. Transfusion. 2005;45:117S–121S. doi: 10.1111/j.1537-2995.2005.00535.x. [DOI] [PubMed] [Google Scholar]
- 7.Westhoff CM, Siegel DL, Burd CG, Foskett JK. J Biol Chem. 2004;279:17443–17448. doi: 10.1074/jbc.M311853200. [DOI] [PubMed] [Google Scholar]
- 8.Westhoff CM, Ferreri-Jacobia M, Mak DO, Foskett JK. J Biol Chem. 2002;277:12499–12502. doi: 10.1074/jbc.C200060200. [DOI] [PubMed] [Google Scholar]
- 9.Ludewig U. J Physiol. 2004;559:751–759. doi: 10.1113/jphysiol.2004.067728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mayer M, Schaaf G, Mouro I, Lopez C, Colin Y, Neumann P, Cartron JP, Ludewig U. J Gen Physiol. 2006;127:133–144. doi: 10.1085/jgp.200509369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soupene E, King N, Feild E, Liu P, Niyogi KK, Huang CH, Kustu S. Proc Natl Acad Sci USA. 2002;99:7769–7773. doi: 10.1073/pnas.112225599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soupene E, Inwood W, Kustu S. Proc Natl Acad Sci USA. 2004;101:7787–7792. doi: 10.1073/pnas.0401809101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Endeward V, Cartron JP, Ripoche P, Gros G. Transfus Clin Biol. 2006;13:123–127. doi: 10.1016/j.tracli.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 14.Huang C-H, Peng J. Proc Natl Acad Sci USA. 2005;102:15512–15517. doi: 10.1073/pnas.0507886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng J, Huang CH. Transfus Clin Biol. 2006;13:85–94. doi: 10.1016/j.tracli.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Whittaker M, Bergmann D, Arciero D, Hooper AB. Biochim Biophys Acta. 2000;1459:346–355. doi: 10.1016/s0005-2728(00)00171-7. [DOI] [PubMed] [Google Scholar]
- 17.Chain P, Lamerdin J, Larimer F, Regala W, Lao V, Land M, Hauser L, Hooper A, Klotz M, Norton J, et al. J Bacteriol. 2003;185:2759–2773. doi: 10.1128/JB.185.9.2759-2773.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weidinger K, Neuhauser B, Gilch S, Ludewig U, Meyer O, Schmidt I. FEMS Microbiol Lett. 2007;273:260–267. doi: 10.1111/j.1574-6968.2007.00805.x. [DOI] [PubMed] [Google Scholar]
- 19.Khademi S, O'Connell J, III, Remis J, Robles-Colmenares Y, Miercke LJ, Stroud RM. Science. 2004;305:1587–1594. doi: 10.1126/science.1101952. [DOI] [PubMed] [Google Scholar]
- 20.Zheng L, Kostrewa D, Berneche S, Winkler FK, Li X-D. Proc Natl Acad Sci USA. 2004;101:17090–17095. doi: 10.1073/pnas.0406475101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andrade SLA, Dickmanns A, Ficner R, Einsle O. Proc Natl Acad Sci USA. 2005;102:14994–14999. doi: 10.1073/pnas.0506254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holm L, Park J. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 23.Avent ND, Ridgwell K, Tanner MJ, Anstee DJ. Biochem J. 1990;271:821–825. doi: 10.1042/bj2710821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avent ND, Butcher SK, Liu W, Mawby WJ, Mallinson G, Parsons SF, Anstee DJ, Tanner MJ. J Biol Chem. 1992;267:15134–15139. [PubMed] [Google Scholar]
- 25.Eyers SA, Ridgwell K, Mawby WJ, Tanner MJ. J Biol Chem. 1994;269:6417–6423. [PubMed] [Google Scholar]
- 26.Conroy MJ, Durand A, Lupo D, Li XD, Bullough PA, Winkler FK, Merrick M. Proc Natl Acad Sci USA. 2007;104:1213–1218. doi: 10.1073/pnas.0610348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gruswitz F, O'Connell J, III, Stroud RM. Proc Natl Acad Sci USA. 2007;104:42–47. doi: 10.1073/pnas.0609796104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chamberlain AK, Faham S, Yohannan S, Bowie JU. Adv Protein Chem. 2003;63:19–46. doi: 10.1016/s0065-3233(03)63002-0. [DOI] [PubMed] [Google Scholar]
- 29.Merrick M, Javelle A, Durand A, Severi E, Thornton J, Avent ND, Conroy MJ, Bullough PA. Transfus Clin Biol. 2006;13:97–102. doi: 10.1016/j.tracli.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 30.Geers C, Gros G. Physiol Rev. 2000;80:681–715. doi: 10.1152/physrev.2000.80.2.681. [DOI] [PubMed] [Google Scholar]
- 31.Huergo LF, Souza EM, Araujo MS, Pedrosa FO, Chubatsu LS, Steffens MB, Merrick M. Mol Microbiol. 2006;59:326–337. doi: 10.1111/j.1365-2958.2005.04944.x. [DOI] [PubMed] [Google Scholar]
- 32.Schouten S, Strous M, Kuypers MM, Rijpstra WI, Baas M, Schubert CJ, Jetten MS, Sinninghe Damste JS. Appl Environ Microbiol. 2004;70:3785–3788. doi: 10.1128/AEM.70.6.3785-3788.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strous M, Pelletier E, Mangenot S, Rattei T, Lehner A, Taylor MW, Horn M, Daims H, Bartol-Mavel D, Wincker P, et al. Nature. 2006;440:790–794. doi: 10.1038/nature04647. [DOI] [PubMed] [Google Scholar]
- 34.Thompson JD, Higgins DG, Gibson TJ. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeLano WL. The PyMol Molecular Graphics System. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]
- 36.Murshudov G, Vagin A, Dodson E. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.