Abstract
Identifying the downstream targets of microRNAs (miRNAs) is essential to understanding cellular regulatory networks. We devised a direct biochemical method for miRNA target discovery that combined RNA-induced silencing complex (RISC) purification with microarray analysis of bound mRNAs. Because targets of miR-124a have been analyzed, we chose it as our model. We honed our approach both by examining the determinants of stable binding between RISC and synthetic target RNAs in vitro and by determining the dependency of both repression and RISC coimmunoprecipitation on miR-124a seed sites in two of its well characterized targets in vivo. Examining the complete spectrum of miR-124 targets in 293 cells yielded both a set that were down-regulated at the mRNA level, as previously observed, and a set whose mRNA levels were unaffected by miR-124a. Reporter assays validated both classes, extending the spectrum of mRNA targets that can be experimentally linked to the miRNA pathway.
Keywords: gene regulation, RNAi, RISC, immunoprecipitation
MicroRNAs (miRNAs) are a widely distributed class of noncoding RNAs that play an integral role in gene regulation (1). Hundreds of microRNA species have been discovered in animals and plants, many of which exhibit temporally and spatially controlled expression. While the list is rapidly growing, clear examples of microRNA function have been shown in development, differentiation, proliferation, apoptosis, metabolism, and tumor initiation and progression (reviewed in ref. 2).
Mature miRNAs function in stable complexes with proteins of the Argonaute family, the core of the RNA-induced silencing complex (RISC) (3, 4). In animals, the 21- to 22-nt miRNA targets RISC to mRNAs with partial sequence complementarity. By using this flexible recognition mechanism, an average miRNA is estimated to affect expression of hundreds of mRNAs. The RISC–mRNA interaction results in translational repression that may also be accompanied by mRNA destruction (5–9). However, the precise factors that determine the extent to which mRNA decay versus translational repression contributes to silencing are not currently well understood.
A basic set of rules governing the recognition of the target by the miRNA has emerged from natural and artificial miRNA-target pairs. Complementarity to the target message in the 5′ region of the miRNA, particularly nucleotides 2–8 (the “seed” region), is the strongest indicator of functional interaction (1). However, extensive complementarity at the 3′ end of the miRNA can compensate for a nonideal seed interaction (10). Additionally, the sequence context outside of the miRNA binding site can impact regulation (11). Endogenous miRNA targets often have multiple complementary sites, and artificial constructs have shown that the presence of several sites improves regulation (12). Finally, synergistic activity of multiple miRNAs on the same mRNA has been demonstrated using reporters and has been postulated for endogenous targets (13, 14).
Steady progress is being made in genetically probing the functions of miRNAs themselves. However, identifying the targets that mediate such functions has proven more challenging. Currently, two approaches are widely used. One relies on measuring reductions in target mRNA levels caused by an exogenously added miRNA (15). In the seminal study, 174 and 96 potential targets of miR-124a and miR-1, respectively, were identified by miRNA delivery to HeLa cells. With this approach, targets whose stability is not affected appear as false negatives, and mRNAs that are down-regulated through secondary effects score as false positives. Multiple computer prediction algorithms have also been developed that use established miRNA–mRNA interaction rules, some of which have been trained on existing microarray data, to identify miRNA targets (16–19). To maximize their fidelity, the current methods require a fully complementary seed sequence in the 3′ UTR and conservation of the site across several species, thus potentially missing targets that do not conform to these rules. In both cases, potential targets are typically validated by using luciferase sensors containing the target 3′ UTR.
Results
Ago2 Coimmunoprecipitates mRNA Targets.
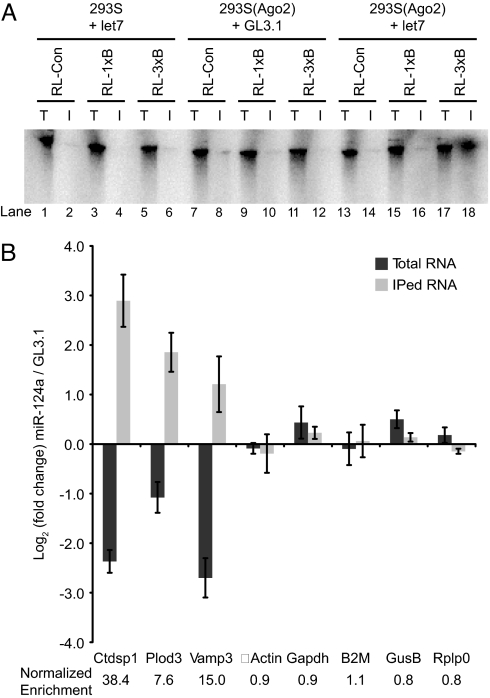
A detailed understanding of the biological function of any miRNA requires the identification of its direct mRNA targets. We sought to develop a biochemical method to isolate such targets, hypothesizing that an Ago2-coimmunoprecipitation approach might retain mRNAs containing miRNA binding sites. We initially combined biochemical systems, which we had previously used to study RISC effector mechanisms, with well established, artificial reporters of microRNA-mediated repression. 293S cells stably expressing c-myc-tagged Ago2 were transfected with either a let-7 siRNA or a control siRNA against firefly luciferase (GL3.1) and used to prepare extracts. In vitro-transcribed let-7 targets were prepared that contained zero, one or three bulged (miRNA-like) let-7 sites (9). These targets were incubated with lysates, and RISC was recovered by immunoprecipitation with anti-c-myc beads (Fig. 1A). Strikingly, c-myc-Ago2 programmed with let-7 was able to coimmunoprecipitate target RNAs containing three let-7 sites, but not the control RNA or the RNA with only a single site. Control extracts containing GL3.1-programmed RISC or let-7 programmed RISC that lacked a c-myc tag did not permit recovery of synthetic targets in immunoprecipitates (IPs). In similar studies, Ago1 performed analogously to Ago2 [supporting information (SI) Fig. 6]. Thus, retention of targets by RISC was observed in a miRNA- and binding site-dependent manner.
Fig. 1.
Programmed Ago2 coimmunoprecipitates miRNA targets. (A) Lysates from 293S or 293S(c-myc-Ago2) cells transfected with let-7 or GL3.1 siRNAs were incubated with radiolabeled RNAs containing zero, one or three let-7 binding sites, and immunoprecipitated with anti-c-myc beads. T, total lysate (1/10 of immunoprecipitated volume); I, immunoprecipitated complexes. (B) Retention of endogenous miR-124a targets. 293S(Ago2) cells were transfected with miR-124a or GL3.1 siRNAs, lysed, and immunoprecipitated. Transcript levels in total and immunoprecipitated fractions were quantified by RT-QPCR.
Building upon these in vitro results, we assessed the ability of RISC to retain endogenous miRNA targets. miR-124a served as an attractive model because of the availability of a large set of experimentally determined target candidates (15). Because miR-124a is absent in 293S cells, we were able to compare retention of specific mRNAs by miR-124a-programmed and unprogrammed RISC. Both total mRNAs and c-myc coimmunoprecipitated mRNAs from miR-124a- or GL3.1-transfected cells were analyzed by reverse transcription followed by quantitative PCR (RT-QPCR) using primers for several targets, including Ctdsp1/SCP1, Plod3, and Vamp3 (20, 21). Results were quantified as enrichment or depletion in the miR-124a sample relative to the GL3.1 control (Fig. 1B). Because no mRNA can be conclusively declared a non-target a priori, the QPCR data were not normalized to a particular control gene but rather were contrasted with a panel of housekeeping genes that were unlikely, in aggregate, to be miR-124a targets (14).
As previously reported, depletion of Ctdsp1, Plod3, and Vamp3 mRNAs in miR-124a-transfected cells was observed in total mRNA (Fig. 1B). Despite these reduced mRNA levels, all three targets were significantly enriched in the immunoprecipitates. Dividing the enrichment in the IP by the change in total mRNA level gave a Net IP enrichment (see below) of 7.6- to 38-fold for the targets, compared with 0.8- to 1.1-fold for housekeeping mRNAs (Fig. 1B). Thus, RISC programmed with miR-124a robustly and specifically coimmunoprecipitates endogenous mRNA targets.
miR-124a Seed Sites in the 3′ UTRs of Vamp3 and Ctdsp1 Are Required for Regulation and Coimmunoprecipitation.
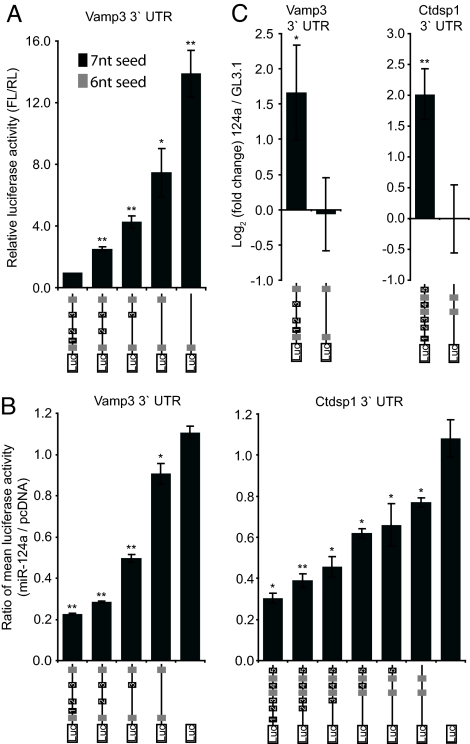
If mRNA retention on RISC related to microRNA-mediated regulation, it should depend upon miR-124a binding sites in the mRNA. To investigate the role of 3′ UTR seed matches in RISC association and regulation for direct targets, we created a series of seed site deletions in luciferase reporters bearing the UTRs of Ctdsp1 and Vamp3. When the Vamp3 reporter was tested in cortical neurons in the presence of endogenous miR-124a, a loss of repression was observed in constructs lacking miR-124a seed complementary sites (Fig. 2A). The seed sites seemed to act in a roughly additive manner, with the full set of sites necessary for complete regulation. A similar dependence of regulation on seed sites was observed when the reporters were tested in mouse kidney cells (TCMK1; Fig. 2B), or 293S(Ago2) cells transfected with miR-124a (SI Fig. 7). Importantly, both reporters showed the same dependency on seed sites when assayed for their association with RISC by coimmunoprecipitation (Fig. 2C), where a complete deletion of 7-mer seed sites reduced mRNA-RISC interactions to basal levels. We conclude that the predicted miR-124a binding sites in the 3′ UTRs of Ctdsp1 and Vamp3 are necessary both for regulation by miR-124a and for physical association with miR-124a-programmed Ago2.
Fig. 2.
Seed sites in the 3′ UTRs of Vamp3 and Ctdsp1 are important for miR-124a regulation and Ago2-coimmunoprecipitation. (A) Luciferase activity of sensor constructs bearing the Vamp3 3′UTR with sequential seed site deletions transfected into primary cortical neurons together with the RL expression vector. (B) Sensor constructs containing the Vamp3 and Ctdsp1 3′ UTRs with sequential deletions of the seed sequences were transfected into mouse kidney cells (TCMK1) together with either an expression vector for miR-124a or pcDNA and a vector expressing Renilla luciferase (RL). (C) Sensor constructs containing the Vamp3 and Ctdsp1 3′ UTRs, wild-type or lacking 7-mer seed sites were transfected into 293S(Ago2) cells along with miR-124a or GL3.1 siRNAs. Transcript levels in Ago2-coimmunoprecipitates were measured by RT-QPCR. Error bars represent standard deviation of three independent experiments (*, P ≤ 0.05; **, P ≤ 0.005; Student's t test).
Transcriptome-Wide Identification of miR-124a Targets.
To identify a comprehensive set of miR-124a targets, we used Ago2 coimmunoprecipitation followed by microarray hybridization. In parallel, total mRNA levels from the same samples were also measured on microarrays. For each mRNA, we envisioned a number of possible behaviors in the microarray studies. For all mRNAs, we expected the amount immunoprecipitated with Ago2, both for biologically relevant retention and for nonrelevant interactions, to depend, in part, on the starting levels of the mRNA. For non-target mRNAs (representing the majority of probes on the array), the immunoprecipitated amount would simply be proportional to mRNA levels in the miR-124a and control samples, as retention would be caused only by nonspecific association. Accordingly, an overall positive correlation between abundance in total mRNA and IP samples was observed (average correlation coefficient 0.81). In contrast, bona fide miRNA targets were predicted to show greater retention by Ago2, which would be superimposed on this basal trend.
Thus, for each mRNA, the change in overall abundance in the total mRNA sample was determined by comparison of miR-124a and GL3.1 control samples. The subset in which mRNA levels decreased specifically in the presence of miR-124a was defined as the Down-regulated set. Raw enrichment in the Ago2 IP (Raw IP enrichment) was also determined, and the ratio of these values yielded a Net IP enrichment that incorporated both mRNA abundance changes and specific binding to Ago2.
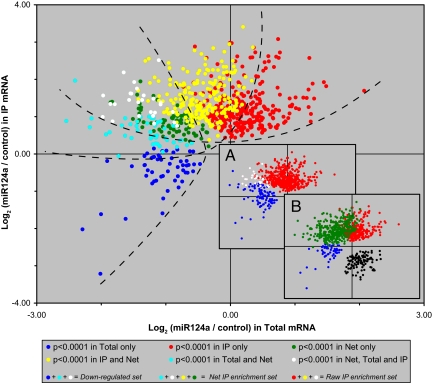
This analysis identified 126 probes that, upon miR-124a transfection, were significantly reduced (adj. P < 0.0001) in total mRNA (Down-regulated set; Fig. 3Inset A, blue) and 550 probes that showed significant Raw IP enrichment in RISC (red), with little overlap (white) between the two sets. Calculating the Net IP enrichment identified 370 probes that showed miR-124a-dependent association with Ago2 (Fig. 3 Inset B, green), with substantial overlap between those mRNAs that showed Raw IP enrichment and those in the down-regulated set (Fig. 3, colored to show the overlap). The converse analysis, looking for probes with Net IP enrichment in RISC immunoprecipitated from the GL3.1 control samples, identified only 105 probes (Fig. 3 Inset B, black), likely representing “off-targets” for the synthetic GL3.1 siRNA. For the identified probe sets, see SI Dataset 1
Fig. 3.
Probes that are significantly enriched or depleted in total RNA and miR-124a-programmed Ago2 IPs. (Inset A) Probes significantly down-regulated (adj. P < 0.0001) in total RNA (blue) and enriched in immunoprecipitated RNA (red) in miR-124a-transfected cells. The overlap is shown in white. (Inset B) As in Inset A, with probes identified by net miR-124a IP enrichment shown in green, and by converse, net GL3.1 IP enrichment shown in black. (Main figure) As in Inset B, but colored to show the overlap between blue and green (cyan), red and green (yellow), and blue and red (white). Dashed lines approximately delineate the Down-regulated, Net IP, and Raw IP sets for illustration purposes.
Thus, coimmunoprecipitation with Ago2 and decrease in total mRNA levels identified distinct, but overlapping, sets of possible miR-124a targets. The effectiveness and scope of these approaches was initially evaluated by searching the 3′ UTRs of the identified gene sets and their overlapping subsets for the presence of miR-124a 7-mer seed matches (Table 1). In agreement with previous studies (15), 109 genes corresponding to the 126 probes that show mRNA reduction upon miR-124a delivery (Down-regulated set; Fig. 3, blue, cyan, and white) were significantly enriched in 7-mer seed sites (0.56 sites per kilobase). The set identified by Raw IP enrichment (red, yellow, and white) also had a high prevalence of seeds (0.40 sites per kilobase). The magnitude of this signal was likely reduced by probes that were also up-regulated in total mRNA and were retained by nonspecific interactions (Fig. 3, red, 0.26 sites per kilobase). Importantly, the gene set showing Net IP enrichment (cyan, green, yellow, and white) showed an overall seed signal (0.55 sites per kilobase) similar to the Down-regulated set. Strikingly, the Net IP enrichment set overlapped with the Down-regulated gene set by only 63 genes (Fig. 3, cyan and white), and included an additional 231 genes that could not be identified solely by monitoring changes in mRNA levels. Importantly, this final class still exhibited a significant enrichment for seed sites (0.48 sites per kilobase; green and yellow). This large set of candidate miR-124 targets may represent those where regulation occurs primarily by translational repression without changes in mRNA stability (22).
Table 1.
Enrichment of miR-124a seed sites in 3' UTRs of gene sets identified by microarray analysis
| Gene sets | No. of genes | No. of sites | Total 3' UTR length | Sites per kilobase |
|---|---|---|---|---|
| Down-regulated | 109 | 71 | 126,051 | 0.5633 |
| Raw IP | 432 | 199 | 500,926 | 0.3973 |
| Net IP | 294 | 196 | 359,409 | 0.5453 |
| Subsets | ||||
| In Down-regulated only (blue) | 46 | 13 | 54,088 | 0.2403 |
| In Raw IP only (red) | 238 | 70 | 271,892 | 0.2575 |
| In Net IP only (green) | 60 | 37 | 83,437 | 0.4434 |
| In Down-regulated and Net IP only (cyan) | 40 | 30 | 46,938 | 0.6391 |
| In Raw and Net IP only (yellow) | 171 | 101 | 204,009 | 0.4951 |
| In Down-regulated, Raw, and Net IP (white) | 23 | 28 | 25,025 | 1.1189 |
| In Net IP, but not Down-regulated (green and yellow) | 231 | 138 | 287,446 | 0.4801 |
| In Net IP and Down-regulated (cyan and white) | 63 | 58 | 71,963 | 0.8060 |
| All other genes (background) | 21,948 | 2,312 | 26,445,994 | 0.087 |
Genes passing a P < 0.0001 cutoff for which a 3' UTR sequence could be obtained from public databases were analyzed.
Within the set of mRNAs down-regulated by miR-124a, those that were also in the Net IP enrichment set (cyan and white) showed a dramatically higher frequency of seed sites (0.81 sites per kilobase) than those that were not identified by Net IP enrichment (blue, 0.24 sites per kilobase). The blue set may represent genes that are down-regulated as an indirect consequence of miR-124a action. Thus, scoring for Net IP enrichment likely enhances the identification of bona fide miR-124a targets that are down-regulated in total mRNA.
Most Immunoprecipitated mRNAs Are Direct miR-124a Targets.
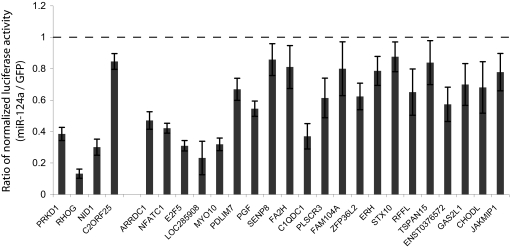
To probe the utility and specificity of the biochemical approach to target identification, we set out to validate a subset of the identified target candidates. We focused on the class of targets that did not change in total mRNA levels, as these genes would not be identified by existing experimental methods. Four genes identified by Net IP enrichment that were also in the Down-regulated set and 30 genes that were not significantly decreased in total mRNA, but identified by Raw IP enrichment, were randomly chosen. The 3′ UTR from each candidate was cloned into a firefly luciferase reporter plasmid and assayed in 293S(hAgo2) cells for response to transfected miR-124a. All four genes from the Net IP enrichment and Down-regulated sets, and 21 of 30 genes from the Raw IP enrichment set (but not Down-regulated) showed statistically significant regulation by miR-124a compared with a GFP siRNA control (P < 0.05, Student's t test) (Fig. 4).
Fig. 4.
Validation of targets identified by microarray analysis. 3′ UTRs were cloned into a firefly luciferase sensor construct and transfected into 293S(Ago2) cells along with a Renilla luciferase control and miR-124a or GFP siRNAs. Error bars represent 1 SD. (left 4 genes) Targets that are down-regulated in total mRNA levels. (right 21 genes) Targets that are not significantly down-regulated in total mRNA levels.
A Subset of miR-124a-Down-Regulated mRNAs Are Direct Targets.
For comparison with targets identified by direct biochemical methods, we used an independent experimental strategy to identify direct miR-124a targets in a previously published dataset. Mouse orthologs of the 174 candidate targets identified by Lim et al. (15) were interrogated in a series of assays: (i) down-regulation of the mRNA in mouse embryonic fibroblasts (MEFs) upon ectopic delivery of miR-124a, (ii) up-regulation of the mRNA in primary cortical neurons (CNs) upon inhibition of miR-124a with a 2′-O-methyl antisense oligonucleotide, and (iii) up-regulation of luciferase reporters bearing the 3′ UTR of the putative targets in CNs upon miR-124a inhibition (SI Fig. 8). Filtering the set based on the aforementioned assays identified a set of 22 genes that scored positively in all three tests, strongly suggesting that these genes were direct targets of miR-124a. Additionally, 62 genes that scored in assay 1 plus either assay 2 or assay 3 were classed as potential targets (SI Table 2). Forty-eight genes scored only in assay 1. These genes were classed as likely secondary or nonspecific targets as they do not show a response to endogenous miR-124a in a relevant cell type. Importantly, the 3′ UTRs of direct miR-124a targets contained significantly more 6-mer (positions 1–6 and 2–7 of miR-124a) and 7-mer seed matches than the potential targets, which were only slightly enriched compared with the nonspecific targets (Fig. 5A).
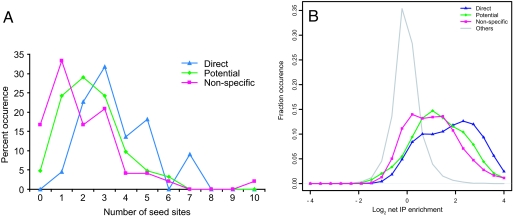
Fig. 5.
Direct miR-124a targets are enriched in miR-124a seed sites and show a higher degree of coimmunoprecipitation with Ago2. (A) Occurrence of 7-mer (positions 2–8 in miR-124a) and 6-mer (positions 1–6 and 2–7) seed sites in the 3′ UTRs of direct, potential, and nonspecific targets. (B) Histogram of net IP enrichment/depletion of direct, potential, and nonspecific targets of miR-124a.
To correlate these validated target sets with our biochemically identified targets, we analyzed the net IP enrichment of human orthologs of each gene set (Fig. 5B). The direct targets showed a markedly stronger Net IP enrichment with a mean fold change of 3.42. The potential target set was moderately enriched in the Net IP set (mean fold enrichment 2.58), and the non-targets showed the least Net IP enrichment (2.12-fold). This higher Net IP enrichment of direct targets reflected a combination of a somewhat stronger down-regulation in total mRNA (SI Fig. 9A) and a more pronounced enrichment in RISC immunoprecipitations (SI Fig. 9B). Therefore, Net IP enrichment of mRNAs in purified RISC preferentially identifies the direct targets of miR-124a, even though the experiment was performed in a model cell line.
Global Enrichment of miRNA Targets.
The immunoprecipitated RISC has the potential to contain the entire set of endogenously expressed miRNAs and their targets, not only targets of the transfected miR-124a or GL3.1. Because the targets of other microRNAs are found in RISC populations from cells transfected with both miR-124a and GL3.1, they would not be detected in the aforementioned analysis. To examine the possibility that endogenous miRNA targets might be recovered, we looked for overall enrichment of mRNAs in the IP as compared with total RNA, considering the miR-124a and GL3.1 samples together. This analysis identified 2,941 probes at a P value cutoff of 0.0001, which are related to 2,578 genes. Next, we considered hexamer seed density in the 3′ UTRs of these mRNAs, comparing it with density in the 3′ UTRs of a background set of genes that were not enriched in the IP. The seed density distribution for the 25 most abundant Ago2-associated miRNAs in 293T cells (23) was shifted toward the IP-enriched mRNAs, i.e., those seeds were more abundant in IP-enriched mRNAs. The distribution was significantly different from the density distribution of all possible hexamers between the two gene sets [P value = 0.002; Kolmogorov-Smirnov (KS) test]. Such a detectable shift is notable, considering that within the 2,578 IP-enriched genes, a given endogenous miRNA would be expected to have only tens to hundreds of targets contributing to its hexamer site enrichment. These observations raise the possibility that antagomir strategies might be used to identify miRNA targets in directly relevant cell types by their depletion from Ago2 IPs upon microRNA inhibition.
Discussion
We present a method for comprehensive miRNA target identification by coimmunoprecipitation of messenger RNAs with miRNA-programmed Ago2 and show that this approach recapitulates the major characteristics of known miRNA–target interactions. Many natural and artificial targets, including the let-7 constructs used in this study (9), require multiple miRNA binding sites for efficient regulation. Accordingly, three let-7 sites were required for coimmunoprecipitation of artificial targets with Ago2, whereas a single site was insufficient. In natural targets of miR-124a, single sites, evolutionarily selected for their regulatory capacity, were often sufficient for retention. Importantly, in the cases where naturally functioning miR-124a miRNA binding sites have been mapped by mutagenesis (Ctdsp1 and Vamp3), the same sites were necessary both for repression of luciferase reporters and for retention by Ago2. The genes identified by coimmunoprecipitation with miR-124a-programmed Ago2 are significantly enriched in miR-124a seed complements in their 3′ UTRs, consistent with the notion that seed sites are strong first-order indicators of a miRNA–mRNA interaction.
Overall, Net IP enrichment is a highly specific and comprehensive predictor of consequential miRNA–mRNA interactions. This is likely the case for several reasons. First, the Down-regulated set contains both direct targets and mRNAs, whose abundance drops as a secondary consequence of miRNA action. The latter would not be enriched in the IP, thus distinguishing them from primary targets. The IP set contains not only bona fide targets but also those, which associate nonspecifically or bind Ago as a consequence of their interaction with an endogenous microRNA. Such mRNAs that are not miR-124a targets but that increase in abundance as a secondary consequence of miR-124a transfection would seem to be enriched in the IP. These mRNAs would score as false-positives in the Raw IP enrichment set. Therefore, taking into account both changes in abundance and IP retention focuses attention on the true target set. Importantly, the Ago IP identifies a large class of potential targets that are not decreased at the mRNA level and that would, therefore, be missed using current experimental approaches to target identification. For miR-124a, the set of targets identified through the procedures outlined here was ≈3-times as large as that identified by down-regulation alone.
A biochemical method for identifying microRNA targets holds the promise of deepening our understanding of the determinants of microRNA-mediated regulation, particularly by revealing targets that are repressed without changes in mRNA levels. Identification of this class of targets will provide an opportunity to glean sequence or structural features of these mRNAs that determine their regulatory fate. In this respect, a recent study by Sharp and coworkers (22) probed groups of mutations affecting the miRNA-target duplex that destine the reporter for mRNA degradation, translational repression, or a combination of both. The method described herein also provides an important balance to in silico methods of predicting microRNA targets, which, while growing in power, still fail to provide a complete and wholly precise picture of miRNA regulatory networks.
Materials and Methods
Coimmunoprecipitation with Ago2 and Analysis of Targets.
Where indicated, 293S(Ago2) or the parental 293S cells were transfected with the appropriate plasmid constructs. Cells were harvested 48 h after transfection and washed in PBS followed by hypotonic lysis buffer [10 mM Tris, pH 7.5, 10 mM KCl, 2 mM MgCl2, 5 mM DTT, and 1 tablet per 10 ml of protease inhibitors, EDTA-free (Roche)]. Cells were incubated in lysis buffer for 15 min and lysed by douncing. Immediately after douncing, the lysates were supplemented with 5× ATP depletion mix [4 units/μl RNaseIn (Promega), 100 mM glucose, 0.5 units/μl hexokinase (Sigma), 1 mg/ml yeast tRNA (Invitrogen), 450 mM KCl] to a final concentration of 1×. The lysates were cleared by centrifugation at 16,000 × g for 30 min at 4°C. Capped and radiolabeled let-7 targets were incubated with lysates at 30°C for 1 h at this stage. Aliquots of total RNA (1/10 for radiolabeled samples and 1/20 for other) were taken and extracted with TRIzol. Before immunoprecipitation, anti-cmyc beads (Sigma) were preblocked for 30 min in wash buffer [0.5% Nonidet P-40, 150 mM NaCl, 2 mM MgCl2, 2 mM CaCl2, 20 mM Tris, pH 7.5, 5 mM DTT, and 1 tablet per 10 ml of protease inhibitors, EDTA-free (Roche)] supplemented with 1 mg/ml yeast tRNA and 1 mg/ml BSA, followed by a wash in wash buffer. One volume of wash buffer was added to the lysates, and cmyc-Ago2 was immunoprecipitated with the preblocked beads for 4 h at 4°C. The beads were washed once with wash buffer and twice in wash buffer containing 650 mM NaCl, transferring the slurry to a fresh tube on the last wash, and bound RNA was extracted with TRIzol. For RT-QPCR, the samples were treated with DNase I (amplification grade; Invitrogen), reverse-transcribed with gene-specific reverse primers (MessageSensor RT; Ambion), and amplified by using SYBR Green PCR Master mix (Applied Biosystems). Six replicate experiments (both total RNA and immunoprecipitated RNA), along with reference RNA, were amplified by using the MessageAmp II aRNA kit (Ambion) with direct incorporation of Cy3- and Cy5-labeled UTP. Samples were hybridized to Agilent Whole Human Genome 4 × 44K chips.
Overexpression and Depletion of miR-124a in MEFs and Cortical Neurons.
For overexpression of miR-124a, MEFs were plated at 1 × 105 cells per 6-cm dish and transfected by calcium phosphate precipitation as described in ref. 24. For depletion of miR-124a, embryonic day 15.5 primary cortical neurons at 4 days in vitro (DIV) were transfected with 500 nM 2′O-methyl (2′OMe) oligoribonucleotides (IDT) using Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. After the indicated time, relative abundance of selected mRNAs was determined by quantitative real-time PCR as described for cortical progenitors using a relative standard curve generated from 10-fold serial dilutions of MEF cDNA.
Luciferase Sensor Assays.
293S(Ago2), MEF, cortical neuron, or TCMK1 cells were transfected with the firefly luciferase reporter and pRL-TK plasmids and miR-124a, miR-124a 2′-OMe inhibitors, or GFP siRNA. Luciferase activity was measured by using the Dual Luciferase reporter system (Promega).
Detailed methods are presented in SI Materials and Methods. Oligonucleotides used in the study are listed in SI Table 3.
Supplementary Material
Acknowledgments
We thank Ramesh Pillai and Witold Filipowicz (Friedrich Miescher Institute for Biomedical Research, Basel, Switzerland) for plasmid constructs and Fabiola Rivas and Jong-Jin Han for experimental advice and help. F.V.K. is supported by American Cancer Society Postdoctoral Fellowship PF-07-058-01-GMC. This work was supported in part by National Institutes of Health (NIH) Grant HG001696 (to Z.X.), by NIH grants to G.M. and G.J.H., and by a kind gift from Kathryn W. Davis (to G.J.H.). G.M. and G.J.H. are supported by the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709971104/DC1.
References
- 1.Bartel DP. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Kloosterman WP, Plasterk RH. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Hammond SM, Bernstein E, Beach D, Hannon GJ. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 4.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 5.Wightman B, Ha I, Ruvkun G. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 6.Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 7.Parker R, Sheth U. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 8.Petersen CP, Bordeleau ME, Pelletier J, Sharp PA. Mol Cell. 2006;21:533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 9.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 10.Brennecke J, Stark A, Russell RB, Cohen SM. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Didiano D, Hobert O. Nat Struct Mol Biol. 2006;13:849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- 12.Doench JG, Petersen CP, Sharp PA. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doench JG, Sharp PA. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 15.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 16.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 18.Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z, Hatzigeorgiou A. Genes Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis BP, Burge CB, Bartel DP. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 20.Conaco C, Otto S, Han JJ, Mandel G. Proc Natl Acad Sci USA. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aleman LM, Doench J, Sharp PA. RNA. 2007;13:385–395. doi: 10.1261/rna.352507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.