Abstract
Although great progress has been made in clarifying deep-level angiosperm relationships, several early nodes in the angiosperm branch of the Tree of Life have proved difficult to resolve. Perhaps the last great question remaining in basal angiosperm phylogeny involves the branching order among the five major clades of mesangiosperms (Ceratophyllum, Chloranthaceae, eudicots, magnoliids, and monocots). Previous analyses have found no consistent support for relationships among these clades. In an effort to resolve these relationships, we performed phylogenetic analyses of 61 plastid genes (≈42,000 bp) for 45 taxa, including members of all major basal angiosperm lineages. We also report the complete plastid genome sequence of Ceratophyllum demersum. Parsimony analyses of combined and partitioned data sets varied in the placement of several taxa, particularly Ceratophyllum, whereas maximum-likelihood (ML) trees were more topologically stable. Total evidence ML analyses recovered a clade of Chloranthaceae + magnoliids as sister to a well supported clade of monocots + (Ceratophyllum + eudicots). ML bootstrap and Bayesian support values for these relationships were generally high, although approximately unbiased topology tests could not reject several alternative topologies. The extremely short branches separating these five lineages imply a rapid diversification estimated to have occurred between 143.8 ± 4.8 and 140.3 ± 4.8 Mya.
Keywords: Ceratophyllum, molecular dating, phylogenetics, mesangiosperms
During the past decade, enormous progress has been made in clarifying relationships among the major lineages of angiosperms, which represent one of the largest branches of the Tree of Life (>250,000 species) (1, 2). The sequence of nodes at the base of the tree of extant angiosperms is now known with a reasonable degree of confidence. Numerous studies have revealed strong support for the successive sister relationships of Amborellaceae, Nymphaeales [sensu APGII (3), including Hydatellaceae], and Austrobaileyales to all other extant angiosperms (4–16), although some studies have placed Amborellaceae + Nymphaeales as sister to all other angiosperms (e.g., refs. 9 and 17).
Whereas the three basalmost angiosperm nodes are now well resolved and supported, relationships among the major lineages of Mesangiospermae [sensu (18)] have been more difficult to elucidate. Analyses of multigene data sets have provided strong support for the monophyly of each of the five major clades of mesangiosperms: Chloranthaceae, Magnoliidae [sensu (18), consisting of Laurales, Magnoliales, Canellales, and Piperales; for the rest of this paper, we will refer to this group as “magnoliids”], Ceratophyllum, monocots, and eudicots. However, the relationships among these five lineages remain unclear. For example, Ceratophyllum has been variously recovered as sister to eudicots (7, 16, 19) or monocots (20, 21). Bootstrap (BS) and Bayesian posterior probability values for these alternative relationships of Ceratophyllum and for the positions of these clades relative to magnoliids and Chloranthaceae have usually been low, even when as many as nine genes have been combined (2, 15). The phylogenetic position of monocots has also been problematic. In molecular phylogenetic studies, monocots have been recovered as sister to magnoliids, Ceratophyllum, or as part of a clade with magnoliids and Chloranthaceae, generally with low support (7, 12, 15, 17, 19–24). The unstable relationships exhibited among these five major lineages of angiosperms are likely due to a combination of the relatively ancient age of these taxa [at least four of which have fossil records that extend back >100 Mya to the Early Cretaceous (25–29)], the short evolutionary branches separating these lineages, and the relatively long branches leading to Ceratophyllum and to the basal lineages of monocots (2).
The increasing number of complete angiosperm plastid genome sequences presents an opportunity to explore whether character-rich data sets can resolve the relationships among these five major angiosperm lineages (13, 14, 30). Here we present phylogenetic analyses of 61 plastid protein-coding genes (≈42,000 bp of sequence data) derived from complete plastid genome sequences of 45 taxa, including at least one member of every major basal lineage of angiosperms. As part of this study, we also report the complete nucleotide sequence of the Ceratophyllum demersum plastid genome. Although topology tests do not exclude several alternative relationships, we find generally high support for a fully resolved topology of Mesangiospermae, including a clade of Ceratophyllum, eudicots, and monocots. We also provide a time frame for the likely rapid diversification of the five major lineages of mesangiosperms.
Results
Ceratophyllum Plastid Genome.
The Ceratophyllum plastid genome possesses the typical genome size and structure found in most angiosperms, with an inverted repeat region of ≈25 kb separating large and small single-copy regions (31, 32). The Ceratophyllum genome is unrearranged relative to Nicotiana (33), and the plastid gene content in Ceratophyllum is identical to that in most angiosperms (32) [supporting information (SI) Fig. 3]. General genome characteristics as well as 454 sequence assembly characteristics are available in SI Table 2.
Phylogenetic Analyses.
The total analyzed aligned length (total aligned length minus excluded base pairs) of the 61-gene combined data set was 42,519 bp, whereas the analyzed aligned lengths of the fast and slow gene partitions (see Materials and Methods) were 22,682 and 19,837 bp, respectively. Total aligned and analyzed aligned lengths for all partitions and genes are given in SI Table 3. The Akaike Information Criterion selected GTR + I + Γ as the optimal model for maximum likelihood (ML) and Bayesian searches for the 61-gene combined, fast, and slow gene data sets, and TVM + I + Γ for the 61-gene first/second codon position data set (although all analyses used GTR + I + Γ; see Materials and Methods).
In almost all analyses, Amborella, Nymphaeales, and Austrobaileyales (represented by Illicium) were successively sister to the remaining angiosperms with strong support regardless of partitioning strategy or phylogenetic optimality criterion (Figs. 1 and 2; SI Figs. 4–10). Only ML analyses of combined first and second codon positions for the 61-gene data set recovered a differing optimal topology; in the best ML tree from this analysis, a clade of Amborella + Nymphaeales was sister to remaining angiosperms (Fig. 1; SI Fig. 5).
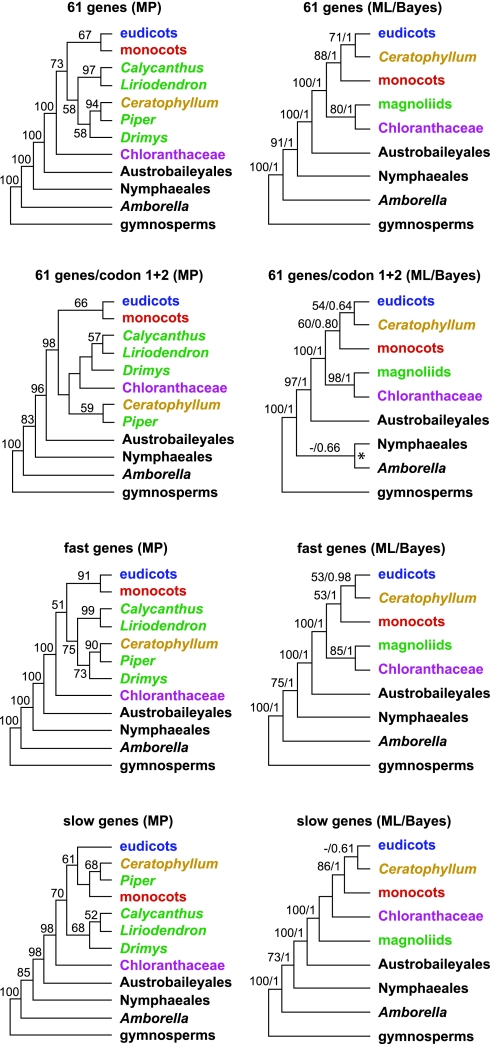
Fig. 1.
Comparison of simplified tree topologies among different partitions and phylogenetic optimization criteria. Numbers associated with branches in MP trees are MP BS support values >50%, whereas numbers associated with branches in ML trees are ML BS support values >50%/Bayesian posterior probabilities >0.5. Codon 1 + 2 refers to the combined first and second codon positions of the 61-gene combined analyses. The asterisk at the node uniting Amborella and Nymphaeales in the 61 genes/codons 1 and 2 ML/Bayes tree indicates that the GARLI ML BS analysis weakly favors a topology where Amborella is sister to all remaining angiosperms (BS = 54%).
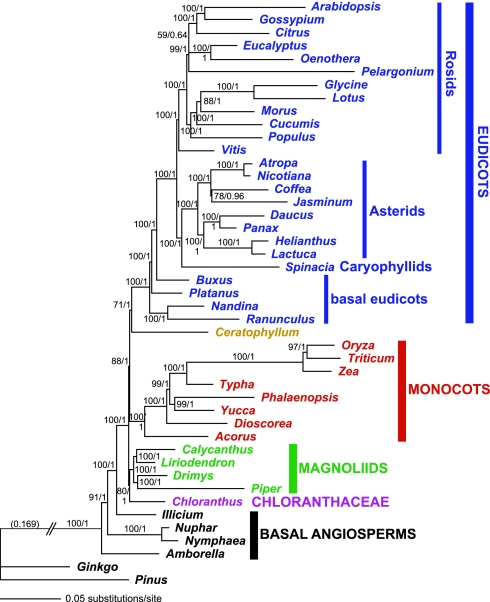
Fig. 2.
Phylogram of the best ML tree as determined by GARLI (−ln L = 460,654.151) for the 61-gene combined data set. Numbers associated with branches are ML BS values >50%/Bayesian posterior probabilities >0.5. The number in parentheses is the branch length separating outgroups from angiosperms.
The monophyly of mesangiosperms was also strongly supported in all analyses (Fig. 1), but the relationships among Chloranthaceae, magnoliids, Ceratophyllum, eudicots, and monocots varied depending on data partition and optimality criterion. Maximum parsimony (MP) recovered Chloranthaceae as sister to remaining mesangiosperms in the 61-gene combined, fast, and slow partitions, whereas in the 61-gene first/second codon position analysis, Chloranthaceae were sister to three of the four magnoliid taxa in the data set (Fig. 1). Magnoliids were never recovered as monophyletic with MP (Fig. 1). Instead, Piper was sister to Ceratophyllum in all partitions, with high BS support (94%) in the 61-gene combined analysis (Fig. 1). The position of these two taxa shifted between sister to monocots in the slow gene analysis to nested within magnoliids in the 61-gene and fast gene analyses (Fig. 1). Removing Piper from MP analyses resulted in the placement of Ceratophyllum as sister to monocots in the 61-gene combined data tree (BS support <50%) and slow gene partition tree (BS support = 76%), and as sister to Chloranthaceae in the fast gene partition tree (BS support = 58%) (SI Figs. 11–13). Likewise, removing Ceratophyllum resulted in a maximally supported monophyletic magnoliid clade that includes Piper (SI Fig. 14). Monocots and eudicots were sisters in the 61-gene, fast gene, and slow gene MP trees, with weak to moderate BS support (Fig. 1).
Compared with MP, ML and Bayesian methods provided different but more stable topological results across our partitioning scheme. Regardless of partitioning strategy, ML and Bayesian methods recovered monocots as sister to a clade of Ceratophyllum + eudicots, with generally high support values (Fig. 1) in most cases despite the extremely short branch lengths separating these groups. A sister relationship of Ceratophyllum to eudicots received the highest support in the 61-gene combined data trees (ML BS = 71%; Bayesian posterior probability = 1.0) but was less well supported in fast and slow gene trees (Fig. 1). Chloranthaceae and magnoliids (including Piper) formed a clade that received moderate to high support values in the 61-gene and fast gene ML and Bayesian trees, but the slow gene analyses recovered magnoliids as sister to a clade of Chloranthaceae + Ceratophyllum/eudicots/monocots (Fig. 1). However, the latter relationship received ML BS support <50% and Bayesian posterior probability <0.5. Removing Piper from ML 61-gene combined data, fast gene, and slow gene analyses in no case altered the overall ML topology (SI Figs. 15–17).
After submission of this paper, the Ceratophyllum genome sequence data were added to an expanded 64-taxon (including extra eudicot and monocot taxa as well as a cycad outgroup), 81-gene (≈76,000-bp) plastid genome matrix in conjunction with Jansen et al. (34). The analyses of this data set are presented as supporting information figures 7–9 in ref. 34. ML analyses were topologically identical to those in the 61-gene combined analyses, with higher BS support for Ceratophyllum + eudicots (82%) but with lower support for the sister relationship of this clade to monocots (73%) as well as for the clade of Chloranthaceae and magnoliids (64%) (supporting informamtion figure 8 in ref. 34). MP analyses continued to unite Piper and Ceratophyllum with high support (supporting information figure 7 in ref. 34).
Topology Tests.
The approximately unbiased (AU) test failed to reject 17 of 104 alternative topologies involving Ceratophyllum, Chloranthaceae, eudicots, magnoliids, and monocots at the 0.05 significance level (SI Table 4). A strict consensus of these 17 trees and the best ML tree provided no resolution among these five lineages. An AU test of the 81-gene 65-taxon data set (including Ceratophyllum) jointly undertaken with Jansen et al. (34) also failed to resolve mesangiosperm relationships (supporting information figure 9 in ref. 34).
Molecular Dating.
Divergence time estimates varied little (<0.5% for all nodes) across the three fossil constraint schemes used in the penalized likelihood (PL) analyses (Table 1); the results of the unconstrained analysis are therefore reported here. The unconstrained PL analysis indicated that extant angiosperms began to diversify in the mid-Jurassic, ≈170 Mya, and that the five major mesangiosperm lineages diversified relatively rapidly in the earliest Cretaceous. The initial divergence of these five lineages was dated to 143.8 ± 4.8 Mya, and the youngest divergence (of Chloranthus and magnoliids) was dated to 140.3 ± 4.8 Mya (Table 1; SI Fig. 18). The origins of the extant crown groups of magnoliids, monocots, and eudicots were dated to somewhat later in the Cretaceous: 130.1 ± 4.4 Mya for magnoliids, 128.9 ± 4.9 Mya for monocots, and 124.8 ± 6.3 Mya for eudicots (Table 1; SI Fig. 18).
Table 1.
Divergence times and standard errors (in Mya) for deep-level angiosperm nodes as estimated by PL analyses
| Node | Unconstrained | Stem eud, 125 Mya | Crown eud, 125 Mya |
|---|---|---|---|
| Angiosperms | 169.6 (3.79) | 169.7 (3.46) | 169.8 (3.46) |
| Nymph + Ill + mesangiosperms | 163.3 (2.68) | 163.5 (2.63) | 163.5 (2.63) |
| Illicium + mesangiosperms | 154.7 (2.47) | 154.8 (2.53) | 154.9 (2.53) |
| Mesangiosperms | 143.8 (2.45) | 143.9 (2.67) | 144.0 (2.66) |
| Chloranthus + magnoliids | 140.3 (2.43) | 140.4 (2.54) | 140.5 (2.54) |
| Magnoliids | 130.1 (2.24) | 130.3 (2.20) | 130.3 (2.20) |
| Cerato + eud + mono | 143.1 (2.99) | 143.1 (3.18) | 143.0 (2.97) |
| Monocots | 128.9 (2.50) | 129.2 (2.69) | 129.1 (2.69) |
| Cerato + eud | 141.3 (2.72) | 141.4 (2.97) | 141.4 (3.18) |
| Eudicots | 124.8 (3.2) | 124.9 (3.43) | 125.0 (3.43) |
The first column gives results of the unconstrained analysis; the second and third columns give results when the minimum age of stem group or crown group eudicots is constrained to be 125 Mya. Standard errors are given in parentheses. Cerato, Ceratophyllum; eud, eudicots; mono, monocots; Ill, Illicium; Nymph, Nymphaeales.
Discussion
Resolving Deep-Level Angiosperm Relationships.
Following the diversification of Amborella, Nymphaeales, and Austrobaileyales, relationships among major basal angiosperm lineages have been enigmatic (reviewed in ref. 2). Our analyses of 61 plastid protein-coding genes reveal that even with ≈42,000 bp of sequence data, it is difficult to resolve the relationships among Ceratophyllum, Chloranthaceae, eudicots, magnoliids, and monocots with confidence. The combination of extremely short internal and relatively long terminal branches that characterize these lineages is almost certainly responsible for the topological differences among MP, ML, and Bayesian analyses (35–37). For example, MP recovers a clearly incorrect topology by uniting Ceratophyllum and Piper (Fig. 1). Strong molecular and morphological evidence supports a magnoliid clade that includes Piperales and excludes Ceratophyllum (21, 30, 38). The relatively long branches leading to Ceratophyllum and Piper, the occurrence of each of these taxa in different parts of the tree in the absence of the other taxon in MP analyses, the increasing support for the erroneous Ceratophyllum/Piper topology in MP with increasing sequence length, and the fact that ML never unites these taxa suggest this is almost certainly a case of long-branch attraction (35, 39, 40). Although breaking up the long branch to Ceratophyllum is impossible, the addition of unsampled Piperales may resolve this problem.
Despite the generally high ML support for a resolved basal angiosperm phylogeny in the 61-gene combined analyses (Fig. 2), the topology test results indicate that no statistically significant resolution of the relationships among Ceratophyllum, Chloranthaceae, eudicots, magnoliids, and monocots is possible with either the current data set (SI Table 4) or the 81-gene expanded data set (supporting information figure 9 in ref. 34). This difficulty in resolving mesangiosperm relationships may result from a number of phenomena, including the early and potentially rapid diversification of mesangiosperms in conjunction with the erosion of phylogenetic signal at more rapidly evolving sites. It is possible that increasing taxon sampling in several lineages (for example, magnoliids, monocots, and eudicots) in future analyses may alter the topology or support values by potentially reducing any phylogenetic error that arises from substitutional rate heterogeneity and mutational saturation (13, 14, 41–43). However, the problem of resolving mesangiosperm diversification may remain even with improved taxon sampling. More sophisticated analytical approaches may need to be developed before basal angiosperm branching order can be reconstructed confidently.
Monocot/Eudicot Relationships.
Systematists have long thought in terms of a major split in angiosperms between monocotyledons and dicotyledons. This longstanding view dates to Ray (44) and served until recently as a fundamental division in angiosperm classifications, with these two groups designated as distinct classes, Liliopsida and Magnoliopsida (45–47). Many earlier angiosperm systematists (e.g., refs. 45, 46, and 48) proposed that monocots formed a clade derived from “primitive” dicot ancestors, such as Nymphaeales. Early molecular phylogenetic analyses confirmed that monocots were derived from a paraphyletic grade of “dicots” but did not resolve their position with high support. Molecular analyses have variously placed monocots as sister to all remaining angiosperms after the Amborella–Nymphaeales–Austrobaileyales grade (11, 15, 17), as part of a clade with magnoliids and Chloranthaceae (7, 15), or as sister to the magnoliids (15, 24).
Our analyses of the plastid genome, although not conclusive, suggest that monocots may be sister to eudicots or part of a clade with Ceratophyllum + eudicots. A close relationship between monocots and eudicots is recovered in all ML analyses, with high support in several cases (Figs. 1 and 2). Likewise, of the 17 alternative topologies not rejected by the AU test, only five do not place monocots sister to eudicots or within a Ceratophyllum/eudicot/monocot clade (SI Table 4). Moreover, the P values of these five topologies fell just above the 0.05 significance level. Other recent analyses have also provided evidence of a monocot/eudicot sister relationship (24, 30, 49) or a monocot + Ceratophyllum/eudicot clade (16). Should this tentative support be validated in future analyses with additional data and/or taxa, it would suggest that after some initial evolutionary “experiments” (the Amborella–Nymphaeales–Austrobaileyales grade, magnoliids, Chloranthaceae), there was indeed a major split in angiosperms between monocots and eudicots + Ceratophyllum, which collectively represent 97% of extant flowering plants.
Mesangiosperm Radiation.
The PL divergence dates obtained for deep-level angiosperm diversification using the optimal 61-gene combined ML tree generally agree with those estimated in several previous studies. For example, several studies have documented a mid- to late-Jurassic age for extant angiosperms as well as Early Cretaceous ages for the divergences of Ceratophyllum, Chloranthaceae, eudicots, magnoliids, and monocots (50–52). As has been noted elsewhere, however, the age estimates for all of these basal angiosperm divergences antedate the earliest unambiguous fossil angiosperms, which are of Hauterivian Age, ≈136–130 Mya (53–56). A number of causes have been advanced to explain this discrepancy, ranging from missing fossil histories to the problems inherent in molecular-based dating techniques (51, 52, 57–59). Our dating analyses rely on a large and apparently internally consistent data set (as judged by the similar phylogenetic results among the various partitions under ML) and consequently should be less susceptible to phylogenetic sources of error (52, 57). The age estimated in the unconstrained PL analyses for the origin of eudicots (124.8 ± 6.3 mya) is also encouraging, because it is consistent with the earliest known appearance of fossil eudicot pollen (125 Mya; refs. 26 and 54). However, the data set used here contains relatively sparse taxon sampling in several major angiosperm lineages (e.g., Nymphaeales, magnoliids, and monocots) despite the fact that it contains exemplar taxa from all of the major basal lineages of angiosperms. Thus we cannot rule out other sources of error involving rate variation among lineages [the lineage effects of (57)] and the proper placement of fossil constraints. Adding key angiosperm taxa to our data set will therefore be important to correct such error and allow the exploration of the effects of more fossil constraints.
Regardless of the absolute divergence times of individual clades, the PL analyses indicate that mesangiosperms diversified rapidly, probably over just a few million years (Table 1; SI Fig. 18). The origin and relatively rapid rise of the angiosperms have long been considered enigmatic [e.g., Darwin's “abominable mystery” (60)]. Although the fossil record certainly supports the presence of many diverse lineages early in angiosperm evolution (56, 61–63), our analyses clearly indicate that the radiation responsible for nearly all extant angiosperm diversity was not associated with the origin of the angiosperms but occurred after the earlier diversification of Amborella, Nymphaeales, and Austrobaileyales.
Materials and Methods
Sequencing the Ceratophyllum Plastid Genome.
Fresh plant material of C. demersum was purchased from an aquarium supply store in Gainesville, FL; a voucher specimen (M. J. Moore 335) has been deposited in the herbarium of the Florida Museum of Natural History. Purified chloroplast DNA for genome sequencing was isolated by using sucrose gradient ultracentrifugation and amplified via rolling circle amplification (RCA) following the protocols of Moore et al. (64). The RCA product was sequenced at the University of Florida by using the Genome Sequencer 20 System (GS 20; 454 Life Sciences, Branford, CT) following the protocols in Moore et al. (64), with the exception that the sequencing run was conducted in a single region of a 70 × 75-mm PicoTiterPlate equipped with a four-region gasket. Gaps between the contigs derived from 454 sequence assembly were bridged by designing custom primers near the ends of the GS 20 contigs for PCR and conventional capillary-based sequencing. Two frame-shift errors in protein-coding sequence that were observed in the 454 sequence assembly were also corrected by using custom PCR and sequencing. The completed plastid genome was annotated by using DOGMA (65) and is available in GenBank (accession no. EF614270).
DNA Sequence Alignment.
The data set for phylogenetic analyses was composed of the nucleotide sequence of the 61 protein-coding genes (SI Table 3) that are present, with very few exceptions, in all angiosperm plastid genomes (32). We modified the 61-gene alignment of Cai et al. (30) by adding Ceratophyllum and several other recently sequenced chloroplast genomes as well as by reducing taxonomic coverage in Poaceae and Solanaceae, both of which have many available plastid genome sequences. The complete taxonomic sampling for the current analyses is given in SI Table 5. Manual realignment of some genes was necessary after the addition of new sequences. Several short regions that were difficult to align in the more quickly evolving genes (e.g., matK, ndhF, and rpoC2) were excluded from analyses, as were all sequence insertions present in only one taxon.
Phylogenetic Analyses.
MP, ML, and Bayesian searches were conducted on the combined 61-gene data set as well as on two partitions of the combined data set that were designed to test the influence of relative evolutionary rate on tree reconstruction. These partitions were created by first ranking the 61 genes based on the average pairwise distance across all taxa for each gene. A relatively large break in pairwise distances of 0.007 units between rps14 and atpA (SI Table 3) was then chosen to divide the genes into relatively more quickly and more slowly evolving groups (hereafter called fast and slow gene partitions) that contained roughly similar numbers of base pairs and genes. The genes included in each partition, along with sequence characteristics for each gene, are given in SI Table 3. The complete data set is available in SI Dataset 1.
We also investigated the effects of mutational saturation at third codon positions in the 61-gene combined data set. Uncorrected pairwise distances for transitions and transversions among all taxa in the data set were plotted against GTR + I + Γ distances to detect mutational saturation at first and second codon positions combined, as well as at third positions. Because third codon position transitions displayed the strongest evidence of mutational saturation (SI Fig. 19), we also performed phylogenetic analyses on combined first and second codon positions for the 61-gene data set.
MP heuristic searches were performed by using PAUP* 4.0 (66) with 1,000 random sequence addition replicates, TBR branch swapping and MULTREES, with gaps treated as missing data. Clade support under MP was assessed by using 1,000 BS replicates (67) with the same settings as for heuristic searches, except with 10 random sequence addition replicates per BS replicate. Trees were rooted with Pinus and Ginkgo.
ML and Bayesian searches were performed for the 61-gene combined data set and for all data partitions, incorporating the model selected as optimal by Modeltest Ver. 3.7 (68) by using the Akaike Information Criterion (AIC) (69) whenever possible. ML analyses were conducted by using the program GARLI, which uses a genetic algorithm to perform rapid heuristic ML searches (www.bio.utexas.edu/faculty/antisense/garli/Garli.html). Default parameters were used for the GARLI searches except that significanttopochange was set to 0.01. A total of 100 ML BS replicates was also performed by using GARLI. Bayesian searches were performed with MrBayes Ver. 3.1.2 (70). To ensure convergence on the appropriate posterior probability distribution, three replicate analyses were run for 6,000,000 generations each for all data sets except the 61-gene first/second codon position data set (1,500,000 generations each). Each replicate used four chains with default parameters. Trees were sampled every 1,000 generations, and the point of stationarity was determined by examining plots of the values of the estimated parameters against generation time and examining split parameters in the program AWTY (http://king2.scs.fsu.edu/CEBProjects/awty/awty_start.php). After ensuring that stationarity was reached in each run, the final 5,000 trees (the final 1,400 trees in the first/second codon position analyses) sampled from each replicate were combined to compute Bayesian majority-rule consensus trees. The AIC selected the TVM + I + Γ model for the 61-gene first/second codon position data set. Because neither MrBayes nor GARLI incorporates five-state models as analysis options, the model was set to GTR + I + Γ for this data partition.
Hypothesis Testing.
To assess whether alternative relationships among Ceratophyllum, Chloranthaceae, eudicots, magnoliids, and monocots could be statistically rejected, we performed AU tests (71) as implemented in CONSEL Ver. 0.1i (72). All 105 possible rooted alternative topologies (including the best ML tree) involving these five major lineages were tested, while holding all other relationships constant to those found in the best GARLI ML tree. Individual site likelihoods were estimated in PAUP* under the GTR + I + Γ model.
Molecular Dating Analyses.
A likelihood ratio test of rate constancy across lineages indicated that our data do not conform to a molecular clock model. Divergence times were therefore estimated under a relaxed molecular clock by using PL (73) as implemented in the program r8s (74). The smoothing parameter (λ) was determined by cross-validation. The best ML topology for the 61-gene combined data set as found by GARLI was used for divergence time analyses, but branch lengths and model parameters were reestimated in PAUP* by using a GTR + I + Γ model of sequence evolution, because GARLI does not fully optimize these parameters (although GARLI-estimated ML parameters are always extremely close to the fully optimized values). To quantify errors in our divergence time estimates, we used the nonparametric BS approach outlined by ref. 75.
Three PL analyses that varied in the application of fossil constraints were run. All analyses used root constraints of a maximum age of 310 Mya and a minimum age of 290 Mya as a conservative estimate of the age of crown group seed plants. The first PL analysis used no further age constraints (this will be referred to as the unconstrained analysis), whereas the second and third analyses used a minimum age of 125 Mya for crown and stem group eudicots, respectively. The latter two analyses also incorporated a number of other minimum age constraints across the tree. All fossil constraints are discussed in detail in SI Text.
Supplementary Material
Acknowledgments
We thank Bob Jansen for providing access to the Buxus, Chloranthus, Dioscorea, and Illicium plastid genomes. We are grateful to Jim Doyle, Terry Lott, and two anonymous reviewers for helpful comments. We also thank Rob Ferl and Beth Laughner for lab space and general lab help, Matt Gitzendanner for software help and general assistance, and D. L. Swofford for access to computational resources (National Science Foundation Grant EF 03-31495). This study was carried out as part of the Angiosperm Tree of Life Project (National Science Foundation Grant EF 04-31266, to D.E.S., P.S.S., W. Judd, S. Manchester, M. Sanderson, Y.-L. Qiu, C. Davis, K. Wurdack, R. Olmstead, K. Sytsma, K. Hilu, M. Donoghue, R. Beaman, N. Cellinese, and L. Hickey).
Abbreviations
- AU test
approximately unbiased test
- BS
bootstrap
- ML
maximum likelihood
- MP
maximum parsimony
- PL
penalized likelihood.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession no. EF614270).
This article contains supporting information online at www.pnas.org/cgi/content/full/0708072104/DC1.
References
- 1.Soltis PS, Soltis DE. Am J Bot. 2004;91:1614–1626. doi: 10.3732/ajb.91.10.1614. [DOI] [PubMed] [Google Scholar]
- 2.Soltis DE, Soltis PS, Endress PK, Chase MW. Phylogeny and Evolution of Angiosperms. Sunderland, MA: Sinauer; 2005. [Google Scholar]
- 3.Angiosperm Phylogeny Group. Bot J Linn Soc. 2003;141:399–436. [Google Scholar]
- 4.Mathews S, Donoghue MJ. Science. 1999;286:947–950. doi: 10.1126/science.286.5441.947. [DOI] [PubMed] [Google Scholar]
- 5.Parkinson CL, Adams KL, Palmer JD. Curr Biol. 1999;9:1485–1488. doi: 10.1016/s0960-9822(00)80119-0. [DOI] [PubMed] [Google Scholar]
- 6.Soltis PS, Soltis DE, Chase MW. Nature. 1999;402:402–404. doi: 10.1038/46528. [DOI] [PubMed] [Google Scholar]
- 7.Soltis PS, Soltis DE, Zanis MJ, Kim S. Int J Plant Sci. 2000;161:S97–S107. [Google Scholar]
- 8.Soltis DE, Soltis PS, Chase MW, Mort ME, Albach DC, Zanis M, Savolainen V, Hahn WH, Hoot SB, Fay MF, et al. Bot J Linn Soc. 2000;133:381–461. [Google Scholar]
- 9.Barkman TJ, Chenery G, McNeal JR, Lyons-Weiler J, Ellisens WJ, Moore G, Wolfe AD, dePamphilis CW. Proc Natl Acad Sci USA. 2000;97:13166–13171. doi: 10.1073/pnas.220427497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graham SW, Olmstead RG. Am J Bot. 2000;87:1712–1730. [PubMed] [Google Scholar]
- 11.Borsch T, Hilu KW, Quandt D, Wilde V, Neinhuis C, Barthlott W. J Evol Biol. 2003;16:558–576. doi: 10.1046/j.1420-9101.2003.00577.x. [DOI] [PubMed] [Google Scholar]
- 12.Hilu KW, Borsch T, Muller K, Soltis DE, Soltis PS, Savolainen V, Chase MW, Powell M, Alice L, Evans R, et al. Am J Bot. 2003;90:1758–1776. doi: 10.3732/ajb.90.12.1758. [DOI] [PubMed] [Google Scholar]
- 13.Leebens-Mack J, Raubeson LA, Cui L, Kuehl JV, Fourcade MH, Chumley TW, Boore JL, Jansen RK, dePamphilis CW. Mol Biol Evol. 2005;22:1948–1963. doi: 10.1093/molbev/msi191. [DOI] [PubMed] [Google Scholar]
- 14.Stefanovic S, Rice DW, Palmer JD. BMC Evol Biol. 2004;4:35. doi: 10.1186/1471-2148-4-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiu Y-L, Dombrovska O, Lee J, Li L, Whitlock BA, Bernasconi-Quadroni F, Rest JS, Davis CC, Borsch T, Hilu KW, et al. Int J Plant Sci. 2005;166:815–842. [Google Scholar]
- 16.Saarela JM, Rai HS, Doyle JA, Endress PK, Mathews S, Marchant AD, Briggs BG, Graham SW. Nature. 2007;446:312–315. doi: 10.1038/nature05612. [DOI] [PubMed] [Google Scholar]
- 17.Soltis DE, Gitzendanner MA, Soltis PS. Int J Plant Sci. 2007;168:137–157. [Google Scholar]
- 18.Cantino PD, Doyle JA, Graham SW, Judd WS, Olmstead RG, Soltis DE, Soltis PS, Donoghue MJ. Taxon. 2007;56:822–846. [Google Scholar]
- 19.Qiu YL, Lee J, Bernasconi-Quadroni F, Soltis DE, Soltis PS, Zanis M, Zimmer EA, Chen Z, Savolainen V, Chase MW. Nature. 1999;402:404–407. doi: 10.1038/46536. [DOI] [PubMed] [Google Scholar]
- 20.Zanis MJ, Soltis DE, Soltis PS, Mathews S, Donoghue MJ. Proc Natl Acad Sci USA. 2002;99:6848–6853. doi: 10.1073/pnas.092136399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zanis MJ, Soltis PS, Qiu Y-L, Zimmer E, Soltis DE. Ann Mo Bot Gard. 2003;90:129–150. [Google Scholar]
- 22.Chase MW, Soltis DE, Olmstead RG, Morgan D, Les DH, Mishler BD, Duvall MR, Price RA, Hills HG, Qiu Y-L, et al. Ann Mo Bot Gard. 1993;80:526–580. [Google Scholar]
- 23.Soltis DE, Soltis PS, Mort ME, Chase MW, Savolainen V, Hoot SB, Morton CM. Syst Biol. 1998;47:32–42. doi: 10.1080/106351598261012. [DOI] [PubMed] [Google Scholar]
- 24.Duvall MR, Mathews S, Mohammad N, Russell T. Aliso. 2006;22:79–90. [Google Scholar]
- 25.Dilcher DL, Crane PR. Ann Mo Bot Gard. 1984;71:351–383. [Google Scholar]
- 26.Doyle JA, Hotton CL. In: Pollen and Spores: Patterns of Diversification. Blackmore S, Barnes SH, editors. Oxford: Clarendon; 1991. pp. 169–195. [Google Scholar]
- 27.Bremer K. Proc Natl Acad Sci USA. 2000;97:4707–4711. doi: 10.1073/pnas.080421597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crepet WL, Nixon KC, Gandolfo MA. Am J Bot. 2004;91:1666–1682. doi: 10.3732/ajb.91.10.1666. [DOI] [PubMed] [Google Scholar]
- 29.Eklund H, Doyle JA, Herendeen PS. Int J Plant Sci. 2004;165:107–151. [Google Scholar]
- 30.Cai Z, Penaflor C, Kuehl JV, Leebens-Mack J, Carlson JE, dePamphilis CW, Boore JL, Jansen RK. BMC Evol Biol. 2006;6:77. doi: 10.1186/1471-2148-6-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palmer JD. In: Cell Culture and Somatic Cell Genetics of Plants. Hermann RG, editor. Vol. 7a. Vienna: Academic; 1991. pp. 5–53. [Google Scholar]
- 32.Raubeson LA, Jansen RK. In: Plant Diversity and Evolution: Genotypic and Phenotypic Variation in Higher Plants. Henry RJ, editor. Cambridge, MA: CABI; 2005. pp. 45–68. [Google Scholar]
- 33.Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K, et al. EMBO J. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jansen RK, Cai Z, Raubeson LA, Daniell H, dePamphilis CW, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, et al. Proc Natl Acad Sci USA. 2007;104:19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Felsenstein J. Syst Zool. 1978;27:401–410. [Google Scholar]
- 36.Huelsenbeck JP. Syst Biol. 1995;44:17–48. [Google Scholar]
- 37.Swofford DL, Waddell PJ, Huelsenbeck JP, Foster PG, Lewis PO, Rogers JS. Syst Biol. 2001;50:525–539. [PubMed] [Google Scholar]
- 38.Qiu Y-L, Lee J, Bernasconi-Quadroni F, Soltis DE, Soltis PS, Zanis M, Zimmer EA, Chen Z, Savolainen V, Chase MW. Int J Plant Sci. 2000;161:S3–S27. doi: 10.1038/46536. [DOI] [PubMed] [Google Scholar]
- 39.Anderson FE, Swofford DL. Mol Phylogenet Evol. 2004;33:440–451. doi: 10.1016/j.ympev.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 40.Bergsten J. Cladistics. 2005;21:163–193. doi: 10.1111/j.1096-0031.2005.00059.x. [DOI] [PubMed] [Google Scholar]
- 41.Graybeal A. Syst Biol. 1998;47:9–17. doi: 10.1080/106351598260996. [DOI] [PubMed] [Google Scholar]
- 42.Pollock DD, Zwickl DJ, McGuire JA, Hillis DM. Syst Biol. 2002;51:664–671. doi: 10.1080/10635150290102357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hillis DM, Pollock DD, McGuire JA, Zwickl DJ. Syst Biol. 2003;52:124–126. doi: 10.1080/10635150390132911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ray J. Methodus Plantarum, Emendata et Aucta. London: Smith and Walford; 1703. [Google Scholar]
- 45.Cronquist A. An Integrated System of Classification of Flowering Plants. New York: Columbia Univ Press; 1981. [Google Scholar]
- 46.Takhtajan A. Systema Magnoliophytorum. Moscow: Nauka; 1987. [Google Scholar]
- 47.Thorne RF. Bot Rev. 1992;58:225–348. [Google Scholar]
- 48.Thorne RF. Evol Biol. 1976;9:35–106. [Google Scholar]
- 49.Jansen RK, Kaittanis C, Saski C, Lee SB, Tomkins J, Alverson AJ, Daniell H. BMC Evol Biol. 2006;6:32. doi: 10.1186/1471-2148-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Janssen T, Bremer K. Bot J Linn Soc. 2004;146:385–398. [Google Scholar]
- 51.Sanderson MJ, Thorne JL, Wikstrom N, Bremer K. Am J Bot. 2004;91:1656–1665. doi: 10.3732/ajb.91.10.1656. [DOI] [PubMed] [Google Scholar]
- 52.Magallon SA, Sanderson MJ. Evolution (Lawrence, Kans) 2005;59:1653–1670. doi: 10.1554/04-565.1. [DOI] [PubMed] [Google Scholar]
- 53.Hughes NF, McDougall AB, Chapman JL. J Micropalaeontol. 1991;10:75–82. [Google Scholar]
- 54.Hughes NF. The Enigma of Angiosperm Origins. Cambridge, UK: Cambridge Univ Press; 1994. [Google Scholar]
- 55.Brenner GJ. In: Flowering Plant Origin, Evolution and Phylogeny. Taylor DW, Hickey LJ, editors. New York: Chapman & Hall; 1996. pp. 91–115. [Google Scholar]
- 56.Friis EM, Pedersen KR, Crane PR. Palaeogeogr, Palaeoclimatol, Palaeoecol. 2006;232:251–293. [Google Scholar]
- 57.Sanderson MJ, Doyle JA. Am J Bot. 2001;88:1499–1516. [PubMed] [Google Scholar]
- 58.Soltis PS, Soltis DE, Savolainen V, Crane PR, Barraclough TG. Proc Natl Acad Sci USA. 2002;99:4430–4435. doi: 10.1073/pnas.032087199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feild TS, Arens NC, Doyle JA, Dawson TE, Donoghue MJ. Paleobiology. 2004;30:82–107. [Google Scholar]
- 60.Darwin F, Seward AC, editors. More Letters of Charles Darwin. Vol. 2. London: John Murray; 1903. [Google Scholar]
- 61.Crane PR, Friis EM, Pedersen KR. Nature. 1995;374:27–33. [Google Scholar]
- 62.Friis EM. Int J Plant Sci. 2000;161:S169–S182. doi: 10.1086/314248. [DOI] [PubMed] [Google Scholar]
- 63.Friis EM, Pedersen KR, Crane PR. Curr Opin Plant Biol. 2005;8:5–12. doi: 10.1016/j.pbi.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 64.Moore MJ, Dhingra A, Soltis PS, Shaw R, Farmerie WG, Folta KM, Soltis DE. BMC Plant Biol. 2006;6:17. doi: 10.1186/1471-2229-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wyman SK, Jansen RK, Boore JL. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- 66.Swofford DL. PAUP* Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer; 2000. Ver 4. [Google Scholar]
- 67.Felsenstein J. Evolution (Lawrence, Kans) 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 68.Posada D, Crandall KA. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 69.Posada D, Buckley TR. Syst Biol. 2004;53:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- 70.Ronquist F, Huelsenbeck JP. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 71.Shimodaira H. Syst Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- 72.Shimodaira H, Hasegawa M. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- 73.Sanderson MJ. Mol Biol Evol. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- 74.Sanderson MJ. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- 75.Baldwin BG, Sanderson MJ. Proc Natl Acad Sci USA. 1998;95:9402–9406. doi: 10.1073/pnas.95.16.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.