Abstract
The regulated clearance of mitochondria is a well recognized but poorly understood aspect of cellular homeostasis, and defects in this process have been linked to aging, degenerative diseases, and cancer. Mitochondria are recycled through an autophagy-related process, and reticulocytes, which completely eliminate their mitochondria during maturation, provide a physiological model to study this phenomenon. Here, we show that mitochondrial clearance in reticulocytes requires the BCL2-related protein NIX (BNIP3L). Mitochondrial clearance does not require BAX, BAK, BCL-XL, BIM, or PUMA, indicating that NIX does not function through established proapoptotic pathways. Similarly, NIX is not required for the induction of autophagy during terminal erythroid differentiation. NIX is required for the selective elimination of mitochondria, however, because mitochondrial clearance, in the absence of NIX, is arrested at the stage of mitochondrial incorporation into autophagosomes and autophagosome maturation. These results yield insight into the mechanism of mitochondrial clearance in higher eukaryotes. Furthermore, they show a BAX- and BAK-independent role for a BCL2-related protein in development.
Keywords: autophagy, mitochondria, BCL2 family
BCL2-related proteins play essential roles in the regulation of programmed cell death. Members of the BCL2 family are divided into subgroups based on the presence of one or more BCL2 homology domains (BH1–BH4) (1). BCL2-related proteins possessing a single BH3 domain (BH3-only proteins) are activated by diverse death-inducing stimuli including DNA damage, glucocorticoids, and growth factor deprivation, and their signals are integrated at the mitochondria by the multidomain proapoptotic proteins BAX and BAK (2). BH3-only proteins activate BAX and BAK either directly or indirectly, through binding to and inhibiting the function of antiapoptotic BCL2-related proteins (3–5). BAX or BAK activation in turn causes cytochrome c release, caspase activation, and apoptosis (6, 7).
BNIP3 and NIX (also known as BNIP3L) are related proteins with limited homology to BH3-only proteins in a BH3-like domain (8–10). BNIP3 and NIX have uncertain biological function. BNIP3 and NIX localize to the mitochondria when overexpressed, induce cytochrome c release, and cause apoptosis (11–13), however, BNIP3 also causes necrosis-like cell death (14). Hypoxia induces and retinoblastoma protein represses BNIP3 expression through HIF-1α and E2F binding sites in the BNIP3 promoter, respectively (15–17). In contrast, NIX is induced by Gq-coupled hypertrophic agonists in neonatal rat cardiomyocytes, by p53 in U2OS osteosarcoma cells, and by differentiation of human erythroid cells (18–20). Accordingly, NIX functions as an effector of Gq-dependent cardiomyopathy and negatively regulates tumor growth in nude mice injected with U2OS osteosarcoma cells (19, 21). NIX has a role in erythroid development, because Nix−/− mice exhibit anemia and erythroid hyperplasia (22).
Results and Discussion
Defective Erythropoiesis in Nix−/− Mice.
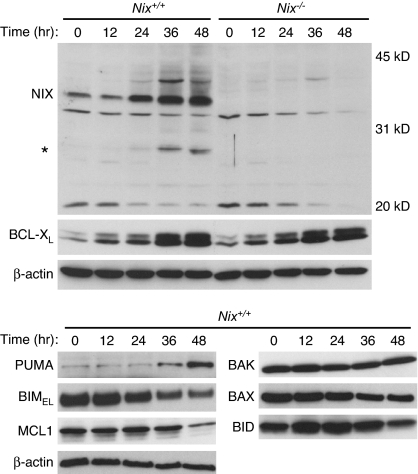
NIX, BCL-XL, and PUMA are up-regulated during terminal erythroid differentiation (Fig. 1 and ref. 20). To gain insight into the function of NIX, we generated mice with a targeted mutation of the Nix gene [supporting information (SI) Fig. 6]. Friend virus-infected erythroid cells (FVA cells) from Nix−/− mice did not express NIX protein, confirming that the targeted mutation created a null allele (Fig. 1). As previously reported (22), Nix−/− mice had a reduced erythrocyte count and reticulocytosis (SI Table 1). Furthermore, their blood smear showed increased polychromatophils and abnormally shaped erythrocytes, suggestive of a maturation defect (Fig. 2A). Nix−/− mice also had moderate splenomegaly (Nix+/+, 0.10 ± 0.01 g; Nix−/−, 0.32 ± 0.12 g) and increased numbers of splenic Ter119+, Gr1+, and Mac1+ cells (SI Fig. 7). Thus, NIX deficiency causes anemia, reticulocytosis, and erythroid-myeloid hyperplasia. We reproduced this phenotype in bone marrow transplant recipients, showing that the defect is intrinsic to the hematopoietic compartment (SI Table 2).
Fig. 1.
Expression of BCL2-related proteins during terminal erythroid differentiation. Expression of NIX, BCL-XL, and other BCL2-related proteins during FVA cell differentiation. Wild-type and Nix−/− FVA cells were cultured for 48 h in the presence of erythropoietin. NIX monomer and a lower molecular mass form of NIX (asterisk) are shown. Molecular mass marks are shown on the right.
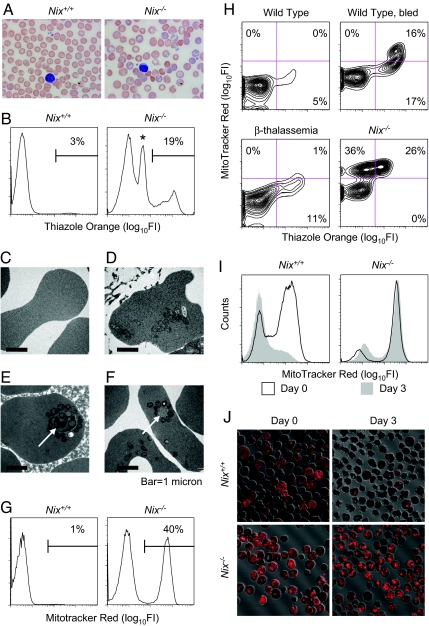
Fig. 2.
Defective mitochondrial clearance in Nix−/− reticulocytes. (A) Blood smears from Nix+/+ and Nix−/− mice. The Nix−/− blood smear shows polychromatophils (blue-tinged cells), spherocytes (absent central pallor), and abnormally shaped erythrocytes. The slides are stained with Wright–Giemsa. (B) Flow cytometry of Nix+/+ and Nix−/− blood stained with TO. The asterisk marks erythrocytes that failed to clear their mitochondria. (C–F) Ultrastructure of erythrocytes from the blood of Nix+/+ (C) and Nix−/− (D–F) mice. Nix−/− erythrocytes contain mitochondria and ribosomes (D) and double-membraned autophagosomes (E and F, arrows). (G) Flow cytometry of wild Nix+/+ and Nix−/− blood stained with MTR. (H) Flow cytometry of erythrocytes stained with TO and MTR. (I) Flow cytometry of Nix+/+ and Nix−/− reticulocytes stained with MTR at baseline (black lines) or after (gray shading) 3 days in culture. (J) Immunofluorescence imaging of cultured Nix+/+ and Nix−/− reticulocytes stained with MTR.
Mitochondrial Clearance Is Impaired in Nix−/− Reticulocytes.
Reticulocytes contain RNA, which can be stained with thiazole orange (TO) (23). Analysis of Nix−/− erythrocytes by TO staining and flow cytometry confirmed the presence of marked reticulocytosis (Fig. 2B and SI Table 1). Also, the presence of a discrete population of cells that stained weakly with TO provided evidence of a specific maturation defect and prompted further analysis (Fig. 2B, asterisk). Ultrastructurally, Nix−/− erythrocytes either were featureless, like wild-type erythrocytes (Fig. 2C); contained ribosomes and mitochondria, consistent with newly released reticulocytes (Fig. 2D); or had no ribosomes but contained mitochondria and undegraded autophagosomes (Fig. 2 E and F, arrows). By Mitotracker Red (MTR) staining and flow cytometry, approximately half of the circulating Nix−/− erythrocytes contained mitochondria (Fig. 2G and SI Table 1). Furthermore, consistent with the ultrastructural studies, Nix−/− erythrocytes contained three populations of cells (MTR−TO−, MTR+TO+, MTR+TOweak) (Fig. 2H). The MTR+TOweak population was not seen in wild-type cells, even under conditions of erythropoietic stress, or in hemolytic diseases like β-thalassemia. Thus, Nix−/− erythrocytes contain an abnormal subpopulation of cells that have cleared their ribosomes but retained their mitochondria. Freshly isolated Nix−/− reticulocytes retained their mitochondria after 3 days in culture, clearly showing that mitochondrial clearance is defective in the absence of NIX (Fig. 2 I and J). Mitochondrial clearance is regulated by 15-lipooxygenase (24); however, the effect of the lipooxygenase inhibitor eicosatetraynoic acid on mitochondrial clearance was modest compared with loss of NIX (data not shown), consequently, we explored other potential mechanisms of NIX activity.
Mitochondrial Clearance Does Not Require Established Proapoptotic Pathways.
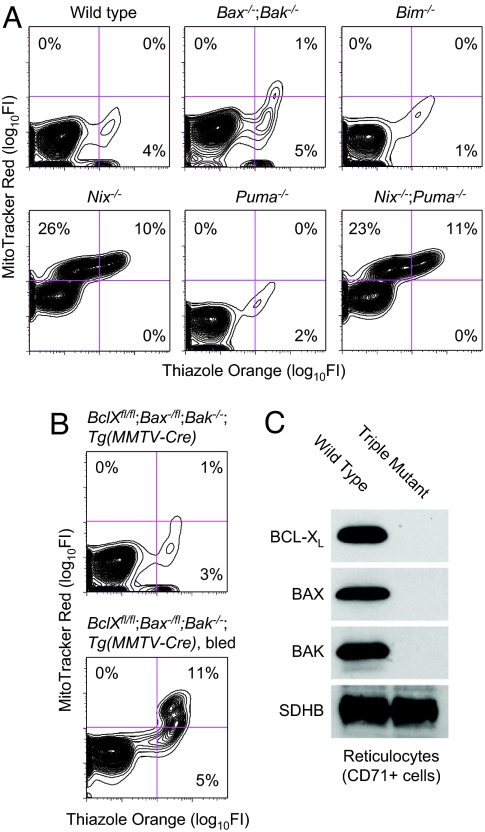
The proapoptotic activity of BH3-only proteins requires the downstream functions of BAX or BAK (2). Therefore, we tested whether NIX-dependent mitochondrial clearance was similarly dependent on these proteins. Bax−/−;Bak−/− mice were bred from compound heterozygotes and identified by genotyping and the presence of interdigital webbing (25, 26). Bax−/−;Bak−/− reticulocytes cleared their mitochondria normally (Fig. 3A). Additionally, mitochondrial clearance from wild-type reticulocytes was unaffected by chemical inhibitors of the mitochondrial permeability transition pore (MPTP), cyclosporin A, and bongkrekic acid (data not shown). Thus, NIX-dependent mitochondrial clearance is independent of BAX, BAK, and the MPTP.
Fig. 3.
Mitochondrial clearance is BAX-, BAK-, and BCL-XL-independent. (A and B) Flow cytometry of erythrocytes stained with TO and MTR. Genotypes are indicated. (C) Western blots of BCL-XL, BAX, BAK, and succinate dehydrogenase subunit B (SDHB). Reticulocytes were obtained from wild-type and BclXfl/fl;Bax−/fl;Bak−/−;Tg(MMTV-Cre) (triple mutant) phlebotomized mice by positive selection for CD71 expression.
BH3-only proteins can be categorized as activators or sensitizers depending on whether they activate BAX and BAK directly or indirectly (27). BIM, BID, and possibly PUMA are activators, whereas other BH3-only proteins are sensitizers. BIM, BID, and PUMA are expressed in late-stage erythroblasts, although BID is not present in its cleaved active form t-BID (Fig. 1); therefore, we examined the roles of BIM and PUMA. Reticulocytes from Bim−/− and Puma−/− mice cleared their mitochondria normally (Fig. 3A). Thus, mitochondrial clearance depends on NIX but not the activator BIM or the putative activator PUMA. Finally, the mitochondrial clearance defect in Nix−/− and Nix−/−;Puma−/− mice was equivalent, indicating a lack of redundancy between these proteins.
BCL-XL is up-regulated during late erythroid maturation in parallel with NIX (Fig. 1). We therefore considered whether the effect of NIX on mitochondrial clearance depends on BCL-XL. BCL-XL is essential for erythroid cell survival; consequently, we used a conditional model of BCL-XL deficiency (28). BclXfl/fl;Tg(MMTV-Cre) mice develop severe anemia and massive splenomegaly because of ineffective erythropoiesis (28, 29). To rescue the survival defect, we bred these mice with Bax and Bak mutant mice. BclXfl/fl;Bax−/fl;Bak−/−;Tg(MMTV-Cre) mice showed dramatic correction of the anemia and splenomegaly (SI Table 3). Furthermore, mitochondrial clearance was intact before or after erythropoietic stress induced by phlebotomy in BCL-XL/BAX/BAK triply deficient reticulocytes (Fig. 3B). Reticulocytes isolated from these mice had no detectable BCL-XL, BAX, or BAK protein but had equal levels of the mitochondrial marker protein succinate dehydrogenase subunit B used as an internal control (Fig. 3C). Therefore, BCL-XL does not have a BAX- or BAK-independent role in mitochondrial clearance from reticulocytes.
Ubiquitin-Like Conjugation Activity Is Induced Independently of NIX.
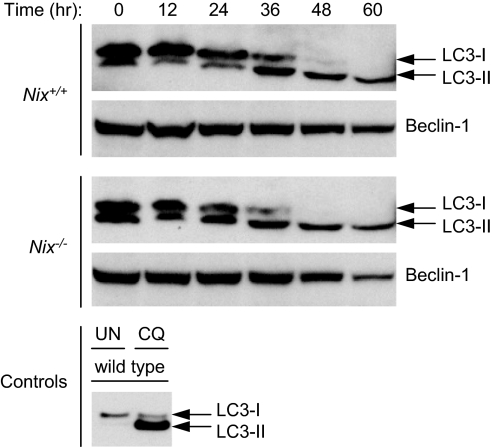
Mitochondrial clearance is thought to be regulated through an autophagy-related process, and autophagy is regulated by BCL2-related proteins (30, 31); therefore, we examined the induction of autophagy. Autophagy depends on two ubiquitin-like conjugation pathways (32). The final products of these pathways are a covalently linked form of the proteins Atg5 and Atg12 (Atg5-Atg12) and a phosphatidylethanolamine-linked form of the protein Atg8 (Atg8-PE). The mammalian homologues of Atg8 are γ2-GABA(A) receptor, golgi-associated enhancer of 16 kDa, and microtubule-associated protein-1 light chain-3 (LC3) (33, 34). Activation of autophagy converts LC3 from a slower migrating unconjugated form (LC3-I) to a faster migrating lipid-conjugated form (LC3-II). To assess the induction of autophagy during erythroid differentiation, and its regulation by NIX, we monitored LC3 conjugation by Western blotting of FVA cells. LC3-I was fully converted to LC3-II in Nix+/+ and Nix−/− FVA cells during terminal differentiation (Fig. 4). Therefore, autophagy is induced during terminal erythroid differentiation, and its induction is independent of NIX.
Fig. 4.
Ubiquitin-like conjugation activity is induced independently of NIX. Nix+/+ and Nix−/− FVA cells were cultured for 60 h in the presence of EPO. LC3-I is the unconjugated form of LC3, and LC3-II is the lipid-conjugated form of LC3. Beclin-1 levels are shown for comparison. Wild-type LC3-overexpressing murine embryonic fibroblasts untreated (UN) or treated with chloroquine (CQ), which causes accumulation of the lipid-modified LC3 (F.C.D. and J.L.C., unpublished results), were used to confirm the identity of LC3-I and LC3-II.
Mitochondrial Autophagy Is Defective in Nix−/− Reticulocytes.
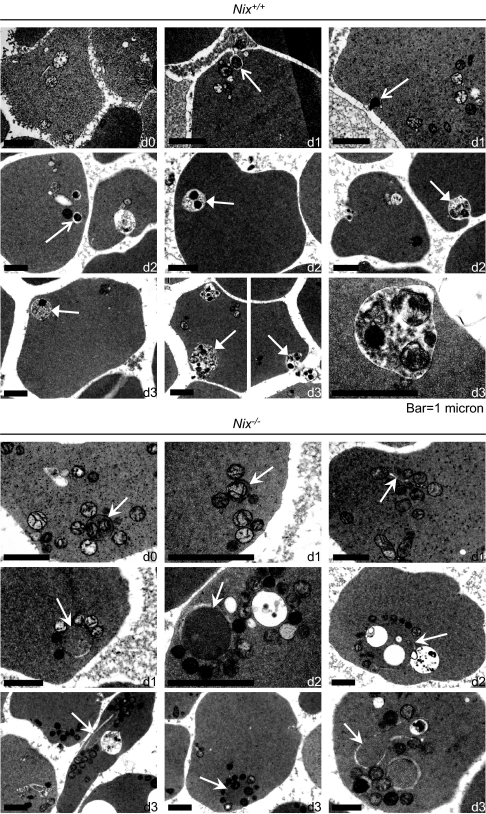
To determine the cellular processes affected by deficiency of NIX, we examined the ultrastructure of Nix+/+ and Nix−/− reticulocytes cultured in vitro for 3 days. In Nix+/+ reticulocytes, mitochondria showed an increase in electron density and loss of internal structure but did not show significant swelling or blebbing of the outer mitochondrial membrane (Fig. 5Upper). Mitochondria were found individually in small vacuoles (regular white arrows) and in multiples in mature autophagic vacuoles (closed white arrows). These structures were eliminated through exocytosis, and the number of mitochondria correspondingly decreased with time. The number of ribosomes also decreased with time, although their mode of elimination was not obvious. The ultrastructural appearance of Nix−/− reticulocytes was strikingly different (Fig. 5 Lower). Mitochondria in Nix−/− reticulocytes were intimately associated with autophagosomes and incompletely formed isolation membranes (barbed white arrows). Notably, although some mitochondria were located inside autophagosomes, the majority were associated with the cytoplasmic face of the outer autophagosomal membranes, and the contents of the autophagosomes were frequently undegraded. Thus, mitochondrial clearance depends on the incorporation of mitochondria into autophagosomes, and autophagosome maturation, and these processes depend on NIX.
Fig. 5.
Mitochondrial autophagy is defective in Nix−/− reticulocytes. Ultrastructure of Nix+/+ (Upper) and Nix−/− (Lower) reticulocytes. (Upper) Regular white arrows point to individual mitochondria in vacuoles, and closed white arrows point to mitochondria in mature autophagic vacuoles. Both were observed undergoing exocytosis. (Lower) Barbed white arrows point to isolation membranes and double-membraned autophagosomes. The length of time in culture (days) is indicated in the lower right corner of each panel as d0, d1, d2, or d3.
Collectively, these results show a critical role for NIX in programmed mitochondrial clearance during reticulocyte maturation. Recent studies show that the autophagy-inducing activity of beclin-1 is negatively regulated through interactions with multidomain antiapoptotic BCL2-related proteins and positively regulated through interactions with BH3-only proteins or the coiled-coil protein UVRAG (30, 31, 35). In the present context, however, NIX does not appear to function through beclin-1, because, as we have shown, NIX is dispensable for the induction of autophagy in erythroid cells. Indeed, the role of macroautophagy in mitochondrial clearance is not well established; Atg7−/− hepatocytes contain defective mitochondria, but Atg5−/− reticulocytes clear their mitochondria normally (36, 37). Instead, as recently described for two unrelated genes in yeast (38, 39), our results indicate that NIX is required for a selective form of autophagy that targets mitochondria for elimination.
NIX may function through the controlled release of cytochrome c or other mitochondrial effector molecules. In that regard, there are other examples of compartmentalized, caspase-dependent degradation in development (40, 41). However, to be consistent with our results, any pore-forming activity of NIX that mediates mitochondrial clearance must be independent of BAX and BAK, and the MPTP. Other mechanistic roles of NIX can be envisioned but remain to be tested. Mitochondrial clearance is important for the health of long-lived postmitotic cells, and defects in this process have been linked to aging, degenerative diseases, and cancer; therefore, the identification of NIX as a protein to regulate this process in higher eukaryotes, represents a significant advance.
Materials and Methods
Targeted Disruption of Nix.
The Nix targeting construct was prepared by subcloning fragments from bacterial artificial chromosome 199e23 (Children's Hospital, Oakland Research Institute). Embryonic stem cell clones with homologous recombination were identified by Southern blot of genomic DNA digested with EcoRV. PCR primers used to generate the probe for hybridization were gcactgagtagtttgccattggcgt and ccaataaagcgtatcacaaagg. Chimeric mice were produced by the Transgenic Core Unit of St. Jude Children's Research Hospital (SJCRH).
Animal Studies.
Reticulocytosis was induced by phlebotomy of 0.4 ml of blood daily for 4 days with saline volume replacement through i.p. injection. On the fifth day, mice were anesthetized and terminally bled by cardiac puncture with a heparinized syringe and a 23-gauge needle. For bone marrow reconstitution studies, 5 × 106 Nix−/− or Nix+/+ bone marrow cells were injected into the tail vein of lethally irradiated (1,100 rads) C57BL6/J recipient mice. Bax and Bak mice were a gift of the Stanley Korsmeyer laboratory (Dana Farber Cancer Institute, Boston). BclXfl/fl;Tg(MMTV-Cre) mice were a gift of Lothar Hennighausen (National Institutes of Health, Bethesda) (28, 42). Puma mice were a gift of Gerard Zambetti (SJCRH) (43). Bim mice were purchased from The Jackson Laboratory (stock number 004525). All studies were performed under an animal protocol approved by the institutional animal care and use committee of SJCRH.
Flow Cytometry and Immunofluorescence.
Reticulocytes stained with MTR CMXRos (Molecular Probes) were analyzed on a BD FACSVantage SE DiVa cell sorter. Optimal separation was obtained with an argon/krypton laser excited at 568 nm, and a 610-nm-long pass filter. Reticulocytes stained with TO were analyzed with a 488-nm argon laser and a 525-nm band-pass filter. For some studies, reticulocytes were enriched by positive selection with magnetic beads for CD71 expression. For the imaging of mitochondria by immunofluorescence, reticulocytes were stained with MTR, transferred to a glass-bottom dish, and viewed with a Nikon TE2000-E inverted microscope equipped with a C1Si confocal system and a ×60 Plan Apo oil-immersion objective (N.A. 1.45).
Electron Microscopy.
Reticulocytes were fixed in 2.5% glutaraldehyde (Tousimis). The cell pellet was postfixed with osmium tetroxide, progressively dehydrated in ethanol, and embedded in low-viscosity resin. We cut 80-nm-thin sections, mounted the sections on copper grids, and stained them with uranyl acetate and lead citrate. Sections were examined with a Jeol JEM-1200EX II electron microscope and archived with a Gatan ES500W digital camera.
Protein Assays and Antibodies.
FVA cell culture was performed as described in ref. 44. Whole-cell extracts of FVA cells were made with RIPA buffer (150 mM NaCl, 1.0% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA, and 50 mM Tris·HCl, pH 8.0) with protease and phosphatase inhibitors. For the NIX Western blot, we resolved 50 μg of protein on an 11% SDS/PAGE gel with Laemmli running buffer and transferred to nitrocellulose. For the LC3 Western blot, we resolved 100 μg of protein on a 12% NuPAGE Bis-Tris gel (Invitrogen) with NuPAGE Mes SDS running buffer and transferred to PVDF membrane. We used the following primary antibodies: NIX (X1120P; Exalpha Biologicals), BCL-X (610209; BD Biosciences), PUMA (JM-3339–100; Medical and Biological Laboratories), BIM (AAP-330; Stressgen Bioreagents), MCL1 (600-401-394; Rockland Immunochemicals), BAK (06-536; Upstate Cell Signaling Solutions), β-actin monoclonal (A5441; Sigma–Aldrich), SDHB (sc-34150; Santa Cruz Biotechnology), LC3 monoclonal (M115–3; Medical and Biological Laboratories), and beclin-1 (sc-11427, Santa Cruz Biotechnology). BAX and BID antibodies were a gift of the Stanley Korsmeyer laboratory. Secondary antibodies were donkey anti-rabbit IgG-HRP conjugate, and sheep anti-mouse IgG-HRP conjugate (GE Healthcare Bio-Sciences).
Supplementary Material
Acknowledgments
We thank Lothar Hennighausen (National Institutes of Health) for the BclX-floxed and the Tg(MMTV-Cre) mice, Stanley Korsmeyer's laboratory for the Bax and Bak mice, Gerard Zambetti for the Puma mice, Derek Persons (SJCRH) for the β-thalassemic mice, Maurice Bondurant and Mark Koury (Vanderbilt University, Nashville, TN) for help and reagents, and Janet F. Partridge and Paul K. Brindle for comments. The authors acknowledge the support of the Animal Resource Center, Flow Cytometry, Scientific Imaging, and Cell Microinjection facilities of SJCRH. This work was supported by the National Institutes of Health (P.A.N., J.L.C., and M.K.), the Burroughs Wellcome Fund (M.K.), the American Society of Hematology (M.K.), the Pew Charitable Trusts (J.T.O.), and the American, Lebanese, and Syrian Associated Charities (ALSAC) of St. Jude Children's Research Hospital.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0708818104/DC1.
References
- 1.Danial NN, Korsmeyer SJ. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 2.Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ, Roth KA, MacGregor GR, Thompson CB, Korsmeyer SJ. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim H, Rafiuddin-Shah M, Tu HC, Jeffers JR, Zambetti GP, Hsieh JJ, Cheng EH. Nat Cell Biol. 2006;8:1348–1358. doi: 10.1038/ncb1499. [DOI] [PubMed] [Google Scholar]
- 4.Certo M, Del Gaizo Moore V, Nishino M, Wei G, Korsmeyer S, Armstrong SA, Letai A. Cancer Cell. 2006;9:351–365. doi: 10.1016/j.ccr.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 5.Willis SN, Fletcher JI, Kaufmann T, van Delft MF, Chen L, Czabotar PE, Ierino H, Lee EF, Fairlie WD, Bouillet P, et al. Science. 2007;315:856–859. doi: 10.1126/science.1133289. [DOI] [PubMed] [Google Scholar]
- 6.Jurgensmeier JM, Xie Z, Deveraux Q, Ellerby L, Bredesen D, Reed JC. Proc Natl Acad Sci USA. 1998;95:4997–5002. doi: 10.1073/pnas.95.9.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosse T, Olivier R, Monney L, Rager M, Conus S, Fellay I, Jansen B, Borner C. Nature. 1998;391:496–499. doi: 10.1038/35160. [DOI] [PubMed] [Google Scholar]
- 8.Boyd JM, Malstrom S, Subramanian T, Venkatesh LK, Schaeper U, Elangovan B, Sa-Eipper C, Chinnadurai G. Cell. 1994;79:341–351. doi: 10.1016/0092-8674(94)90202-x. [DOI] [PubMed] [Google Scholar]
- 9.Matsushima M, Fujiwara T, Takahashi E, Minaguchi T, Eguchi Y, Tsujimoto Y, Suzumori K, Nakamura Y. Genes Chromosomes Cancer. 1998;21:230–235. [PubMed] [Google Scholar]
- 10.Chen G, Ray R, Dubik D, Shi L, Cizeau J, Bleackley RC, Saxena S, Gietz RD, Greenberg AH. J Exp Med. 1997;186:1975–1983. doi: 10.1084/jem.186.12.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen G, Cizeau J, Vande VC, Park JH, Bozek G, Bolton J, Shi L, Dubik D, Greenberg A. J Biol Chem. 1999;274:7–10. doi: 10.1074/jbc.274.1.7. [DOI] [PubMed] [Google Scholar]
- 12.Yasuda M, Theodorakis P, Subramanian T, Chinnadurai G. J Biol Chem. 1998;273:12415–12421. doi: 10.1074/jbc.273.20.12415. [DOI] [PubMed] [Google Scholar]
- 13.Imazu T, Shimizu S, Tagami S, Matsushima M, Nakamura Y, Miki T, Okuyama A, Tsujimoto Y. Oncogene. 1999;18:4523–4529. doi: 10.1038/sj.onc.1202722. [DOI] [PubMed] [Google Scholar]
- 14.Vande Velde C, Cizeau J, Dubik D, Alimonti J, Brown T, Israels S, Hakem R, Greenberg AH. Mol Cell Biol. 2000;20:5454–5468. doi: 10.1128/mcb.20.15.5454-5468.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bruick RK. Proc Natl Acad Sci USA. 2000;97:9082–9087. doi: 10.1073/pnas.97.16.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sowter HM, Ratcliffe PJ, Watson P, Greenberg AH, Harris AL. Cancer Res. 2001;61:6669–6673. [PubMed] [Google Scholar]
- 17.Tracy K, Dibling BC, Spike BT, Knabb JR, Schumacker P, Macleod KF. Mol Cell Biol. 2007;27:6229–6242. doi: 10.1128/MCB.02246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galvez AS, Brunskill EW, Marreez Y, Benner BJ, Regula KM, Kirschenbaum LA, Dorn GW. J Biol Chem. 2006;281:1442–1448. doi: 10.1074/jbc.M509056200. [DOI] [PubMed] [Google Scholar]
- 19.Fei P, Wang W, Kim SH, Wang S, Burns TF, Sax JK, Buzzai M, Dicker DT, McKenna WG, Bernhard EJ, et al. Cancer Cell. 2004;6:597–609. doi: 10.1016/j.ccr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 20.Aerbajinai W, Giattina M, Lee YT, Raffeld M, Miller JL. Blood. 2003;102:712–717. doi: 10.1182/blood-2002-11-3324. [DOI] [PubMed] [Google Scholar]
- 21.Yussman MG, Toyokawa T, Odley A, Lynch RA, Wu G, Colbert MC, Aronow BJ, Lorenz JN, Dorn GW. Nat Med. 2002;8:725–730. doi: 10.1038/nm719. [DOI] [PubMed] [Google Scholar]
- 22.Diwan A, Koesters AG, Odley AM, Pushkaran S, Baines CP, Spike BT, Daria D, Jegga AG, Geiger H, Aronow BJ, et al. Proc Natl Acad Sci USA. 2007;104:6794–6799. doi: 10.1073/pnas.0610666104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nobes PR, Carter AB. J Clin Pathol. 1990;43:675–678. doi: 10.1136/jcp.43.8.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Leyen K, Duvoisin RM, Engelhardt H, Wiedmann M. Nature. 1998;395:392–395. doi: 10.1038/26500. [DOI] [PubMed] [Google Scholar]
- 25.Lindsten T, Ross AJ, King A, Zong WX, Rathmell JC, Shiels HA, Ulrich E, Waymire KG, Mahar P, Frauwirth K, et al. Mol Cell. 2000;6:1389–1399. doi: 10.1016/s1097-2765(00)00136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takeuchi O, Fisher J, Suh H, Harada H, Malynn BA, Korsmeyer SJ. Proc Natl Acad Sci USA. 2005;102:11272–11277. doi: 10.1073/pnas.0504783102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Letai A, Bassik MC, Walensky LD, Sorcinelli MD, Weiler S, Korsmeyer SJ. Cancer Cell. 2002;2:183–192. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 28.Wagner KU, Claudio E, Rucker EB, III, Riedlinger G, Broussard C, Schwartzberg PL, Siebenlist U, Hennighausen L. Development (Cambridge, UK) 2000;127:4949–4958. doi: 10.1242/dev.127.22.4949. [DOI] [PubMed] [Google Scholar]
- 29.Rhodes MM, Kopsombut P, Bondurant MC, Price JO, Koury MJ. Blood. 2005;106:1857–1863. doi: 10.1182/blood-2004-11-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 31.Maiuri MC, Le TG, Criollo A, Rain JC, Gautier F, Juin P, Tasdemir E, Pierron G, Troulinaki K, Tavernarakis N, et al. EMBO J. 2007;26:2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yorimitsu T, Klionsky DJ. Cell Death Differ. 2005;12(Suppl 2):1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. J Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 35.Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 36.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsui M, Yamamoto A, Kuma A, Ohsumi Y, Mizushima N. Biochem Biophys Res Commun. 2006;339:485–489. doi: 10.1016/j.bbrc.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 38.Kissova I, Salin B, Schaeffer J, Bhatia S, Manon S, Camougrand N. Autophagy. 2007;3:329–336. doi: 10.4161/auto.4034. [DOI] [PubMed] [Google Scholar]
- 39.Tal R, Winter G, Ecker N, Klionsky DJ, Abeliovich H. J Biol Chem. 2007;282:5617–5624. doi: 10.1074/jbc.M605940200. [DOI] [PubMed] [Google Scholar]
- 40.de Botton S, Sabri S, Daugas E, Zermati Y, Guidotti JE, Hermine O, Kroemer G, Vainchenker W, Debili N. Blood. 2002;100:1310–1317. doi: 10.1182/blood-2002-03-0686. [DOI] [PubMed] [Google Scholar]
- 41.Arama E, Agapite J, Steller H. Dev Cell. 2003;4:687–697. doi: 10.1016/s1534-5807(03)00120-5. [DOI] [PubMed] [Google Scholar]
- 42.Wagner KU, Wall RJ, St-Onge L, Gruss P, Wynshaw-Boris A, Garrett L, Li M, Furth PA, Hennighausen L. Nucleic Acids Res. 1997;25:4323–4330. doi: 10.1093/nar/25.21.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J, MacLean KH, Han J, Chittenden T, Ihle JN, et al. Cancer Cell. 2003;4:321–328. doi: 10.1016/s1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- 44.Koury MJ, Sawyer ST, Bondurant MC. J Cell Physiol. 1984;121:526–532. doi: 10.1002/jcp.1041210311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.