Abstract
Elevated expression of members of the BCL-2 pro-survival family of proteins can confer resistance to apoptosis in cancer cells. Small molecule obatoclax (GX15-070), which is predicted to occupy a hydrophobic pocket within the BH3 binding groove of BCL-2, antagonizes these members and induces apoptosis, dependent on BAX and BAK. Reconstitution in yeast confirmed that obatoclax acts on the pathway and overcomes BCL-2-, BCL-XL-, BCL-w-, and MCL-1-mediated resistance to BAX or BAK. The compound potently interfered with the direct interaction between MCL-1 and BAK in intact mitochondrial outer membrane and inhibited the association between MCL-1 and BAK in intact cells. MCL-1 has been shown to confer resistance to the BCL-2/BCL-XL/BCL-w-selective antagonist ABT-737 and to the proteasome inhibitor bortezomib. In both cases, this resistance was overcome by obatoclax. These findings support a rational clinical development opportunity for the compound in cancer indications or treatments where MCL-1 contributes to resistance to cell killing.
Keywords: Bcl-2, melanoma, Bax, Bak, caspase
In addition to uncontrolled cell proliferation, transformation to the malignant cell phenotype is associated with abrogation of apoptosis, a finding that derived initially from studies of the BCL-2 oncogene (1, 2). BCL-2, however, has emerged as a family of both anti- and proapoptotic proteins that function in part to regulate and execute the core mitochondrial pathway of apoptosis (3). Moreover, the cell proliferative and cell death potential of cancer cells seem to be intimately linked because oncogenes like c-myc are potent activators of the apoptotic machinery and rely on pro-survival regulators like BCL-2 to blunt this apoptotic response to drive tumorigenesis (4–7). Together with the fact that BCL-2 can inhibit the induction of apoptosis by many anti-cancer agents, this finding has led to the conclusion that therapeutic modulation of the BCL-2 pathway may represent a new treatment option in a variety of cancer settings (8, 9).
A key element of the BCL-2 mechanism is to control the permeability of the mitochondrial outer membrane, permitting release of regulatory factors that are located in the intermembrane space, including cytochrome c. Upon release, they activate caspases and the core apoptotic pathway (10). Such control by the BCL-2 family is achieved by the binary protein–protein interactions that can occur between pro- and antiapoptotic members, an interaction that is mediated by the insertion of the amphiphilic BH3 domain of proapoptotic members into an extended groove formed by the BH1, BH2, and BH3 domains at the surface of pro-survival members (11). How the pro-survival members BCL-2, BCL-XL, MCL-1, BCL-w, or A1 antagonize membrane permeabilization, however, is the subject of considerable debate (12). Nevertheless, whether directly or indirectly via the BH3-only members, the pro-survival members must ultimately prevent the activation of the multi-BH domain proapoptotic members BAX and BAK, which are the effectors of membrane permeabilization. This antagonism occurs within the outer membrane and, in the case of inhibition of BAX by BCL-2, may involve substantial conformational changes and further penetration into the membrane lipid bilayer of both proteins in response to stimuli, to drive interactions between the two proteins (13, 14).
Despite the complexities of the protein–protein interactions and inhibition of apoptosis by pro-survival members, there is now strong support for antagonism of the pro-survival members by BH3 mimetics as a potential for therapeutic intervention (15, 16). There may be different ways, however, to achieve this outcome. For example, the nonequivalent functional redundancies among pro-survival family members, the different conformational states that can exist for an individual member in response to death stimuli, and the degree of penetration of these proteins into their target membrane, may mean that therapeutic antagonists of BCL-2 proteins could be developed with different mechanisms of action and selectivity. One example is illustrated by the unique properties of MCL-1. It has a structure that exhibits specific differences compared with that of BCL2/BCL-XL (17). Coupled with other properties of the protein, it may explain the fact that ABT-737, a recently described small molecule inhibitor of BCL2/BCL-XL, does not exhibit potency against MCL-1 (16). Moreover, current evidence suggests that MCL-1 regulates BAK by constitutively interacting with BAK within the mitochondrial outer membrane (18, 19). Inhibition of MCL-1, therefore, may require disruption of a preformed MCL-1/BAK membrane complex. Finally, MCL-1 is distinct among pro-survival BCL-2 members in that it rapidly turns over at steady state, because of degradation by the 26S proteasome (18, 20). As a result, inhibition of the proteasome causes accumulation of MCL-1 in a number of systems. Given the importance of MCL-1 in conferring resistance to apoptosis in the cancer setting, there is a clear need to overcome this resistance therapeutically.
Here, we describe a small molecule inhibitor of pro-survival BCL-2 members, including MCL-1, and illustrate how this compound, named obatoclax (GX15-070), can overcome the resistance to apoptosis conferred by MCL-1. Obatoclax is a hydrophobic molecule that was developed as a BCL-2 family antagonist. It is currently undergoing evaluation in multiple single agent and combination phase Ib and phase II clinical trials directed at a variety of leukemia, lymphoma, and solid tumor malignancies. The ability of obatoclax to overcome resistance to apoptosis mediated specifically by MCL-1 provides the basis for rationally combining obatoclax with other targeted anti-cancer agents whose activity is restrained by MCL-1.
Results and Discussion
Obatoclax [GX15-070; supporting information (SI) Fig. 6] stemmed from a discovery and development program aimed at therapeutically modulating the pro-survival members of the BCL-2 family. A recent study comparing the ability of a number of such small molecules to inhibit binding of a BH3 peptide to recombinant fragments of the cytosolic domains of BCL-2, BCL-XL, MCL-1, BCL-w, A1, and BCL-b in a fluorescence polarization assay confirmed that obatoclax inhibited this interaction for all 6 members (Ki values ≈ 1–7 μM), with no effect on a control protein–protein interaction (21). Under the conditions of this and other aqueous-based assays used to measure BCL-2 binding activity, however, hydrophobic obatoclax is largely insoluble (calculated logP > 4.0); experimental logD measurements revealed that the compound partitioned exclusively into the octanol phase at pH 7.4. Because of this insolubility, its activity in these assays is either under-estimated or cannot be determined. This problem extends both to inhibition of protein-peptide interactions and to NMR.
Although the compound would be expected to readily partition into lipophilic environments such as membranes where BCL-2 family members reside, we investigated the binding mode of obatoclax to the cytosolic fragment of a BCL-2 chimera in silico by taking advantage of the NMR structure of the recombinant fragment bound to a small molecule BH3 peptidomimetic (1YSW in SI Fig. 6; ref. 16). An in silico docking algorithm was developed (22) that accurately predicted the observed orientation of this peptidomimetic within the BCL-2 BH3 binding groove obtained by NMR as the highest scoring pose. Docking of obatoclax to BCL-2 using this algorithm (see SI Materials and Methods) suggested that the compound occupies a hydrophobic pocket at one end of the BH3 binding groove (Fig. 1A; SI Fig. 7), which is distal to that recently described for ABT-737 (16, 23). Because this hydrophobic pocket resides in close proximity to a lipid bilayer in the normal membrane-integrated form of full length BCL-2 in situ, the membrane could potentiate the accessibility of this pocket to a hydrophobic molecule like obatoclax.
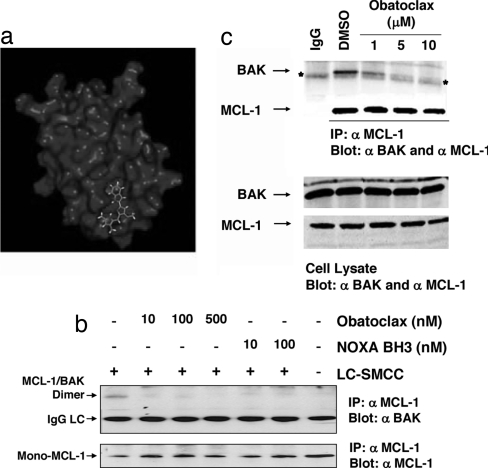
Fig. 1.
Obatoclax interferes with MCL-1/BAK interactions. (a) Predicted orientation of obatoclax within the BH3 binding groove of BCL-2 (see also SI Fig. 7 and SI Materials and Methods). (b) Disruption of constitutive MCL-1/BAK interactions in isolated mitochondria. Mitochondria were isolated from SK-Mel5 cells and incubated with the indicated concentrations of obatoclax or NOXA BH3 peptide for 30 min at 37°C and then subjected to chemical cross-linking with 0.1 mM LC-SMCC (+) or vehicle (DMSO) (−). After blocking excess LC-SMCC, the mitochondria were lysed with detergent and subjected to immunoprecipitation with anti MCL-1 antibody, and the precipitates were probed by immunoblot with anti-MCL-1 and anti BAK antibodies. (c) Obatoclax antagonizes MCL-1/BAK association in cells. SK-Mel5 cells were incubated with DMSO or obatoclax in DMSO for 5 h, after which time total cell lysate was subjected to immunoprecipitation with anti MCL-1 antibody. Immunoprecipitates and cell lysates were separated by SDS/PAGE and analyzed by Western blot with anti-MCL-1 and anti-BAK antibodies.
A predicted affinity constant for the binding of obatoclax to the BCL-2 cytosolic structure shown in Fig. 1a and SI Fig. 7 was calculated by using the SIE scoring function (ref. 22; SI Materials and Methods). As a control for the reliability of the calculation, binding affinities were calculated for a training set of 12 small molecule ligands with experimentally published binding affinities to BCL-2. The calculated and experimental binding affinities for this group were highly correlated (SI Table 1). The results suggested that obatoclax binds this cytosolic form of BCL-2 with a calculated Ki ≈ 220 nM. However, because the pro-survival BCL members function at or within cellular membranes, we focused our experimental analyses on the activity of the compound in intact cells or organelles. As expected, the subcellular distribution of obatoclax was seen to be regulated by the specific distribution of ectopic BCL-2 (SI Fig. 8).
Obatoclax Potently Inhibits Constitutive Interactions Between MCL-1 and BAK in Intact Mitochondria.
Because the hydrophobic character of obatoclax is compatible with membranes, the ability of the compound to interfere with BCL protein–protein interactions was determined in isolated, intact mitochondria. We took advantage of the fact that MCL-1 constitutively interacts with BAK in the mitochondrial outer membrane (18, 19). Chemical cross-linking was used to measure this interaction because it can be conducted while maintaining the structural integrity of the membrane. A screen of potential cross-linking agents was performed and revealed LC-SMCC [3-aminopropyltriethoxysilane (APTES), succinimidyl-4-(N-maleimidomethyl)cyclohexane-1-carboxy-(6-amidocaproate)] (a membrane-permeant hetero-bifunctional compound that covalently links a primary amine and a thiol) as the optimal agent. LC-SMCC is not reactive with obatoclax. Mitochondria from human SK-Mel5 melanoma cells were incubated with either vehicle (DMSO) or vehicle containing 0.1 mM LC-SMCC, and the mitochondria subsequently solubilized with detergent. MCL-1 was recovered by immunoprecipitation with anti-MCL-1 antibody, and the presence in the precipitate of MCL-1 cross-linked to BAK (MCL-1/BAK dimer), as well as monomeric MCL-1, determined by immunoblot (Fig. 1b). Pretreatment of the mitochondria with 10 nM of either obatoclax or a synthetic 25-aa peptide corresponding to the BH3 domain of human NOXA (which interacts selectively with MCL-1; ref. 24) before the addition of cross-linking agent strongly inhibited the formation of MCL-1/BAK dimers (Fig. 1b). Of note, however, addition of concentrations as high as 2.0 μM obatoclax to isolated mitochondria did not stimulate release of cytochrome c in vitro, whereas a corresponding concentration of BID BH3 peptide, which can activate BAX and BAK (25), depleted the mitochondrial pool of cytochrome c (SI Fig. 9).
Thus, obatoclax seems to disrupt MCL-1/BAK interactions in intact mitochondria but BAK at this site is either not optimal for activation in isolated mitochondria under the conditions of this assay or requires a second stimulus to become activated (e.g., provided by tonic signaling pathways in cancer cells), causing release of cytochrome c. In lymphoma cells expressing MCL-1 and treated with obatoclax as a single agent, activation of BAX and BAK and release of cytochrome c from mitochondria was observed, upstream of caspase activation (26). However, to confirm that obatoclax can inhibit endogenous MCL-1/BAK interactions within the cell, SK-Mel5 cells were treated with compound, the cells recovered and washed, the cell pellet dissolved in excess of solution containing the detergent Nonidet P-40, and MCL-1 recovered by overnight immunoprecipitation. The amount of MCL-1 and BAK in the immunoprecipitate was determined by immunoblotting. Despite significant dilution of the cell content of obatoclax during immunoprecipitation, exposure of cells to 1 μM obatoclax for 5 h caused a significant reduction in the amount of BAK recovered with MCL-1, with near complete inhibition recorded at 5 μM (Fig. 1c). Coimmunoprecipitations have also revealed disruption of BCL-2 interactions with BIM (data not shown).
Obatoclax Antagonizes the BCL Pathway, Dependent on BAX or BAK.
Examination of the ability of obatoclax to directly antagonize MCL-1 in mammalian cells is complicated by the fact that multiple indirect pathways, for example proapoptotic stress pathways, can also influence the activity of this BCL member. The yeast, Saccharomyces cerevisiae, however, does not contain any known BCL-2 homolog, but it has been noted that overexpression of either BAX or BAK inhibits its cell growth (27). This inhibition can be countered by expression of MCL-1 (and BCL-2, BCL-XL, or BCL-w) (SI Fig. 10a). At 50 μM, obatoclax did not affect the growth of control yeast cells; however, it significantly overcame the ability of the pro-survival members to antagonize BAX- or BAK-mediated inhibition of cell growth. Although 50 μM is a relatively high external concentration of the compound, the proteins were expressed by using high-copy number plasmids; moreover, uptake and retention of drugs by yeast differs from that of mammalian cells and is consistent with the finding that yeast cells typically require at least 100- to 1,000-fold higher concentrations of anti-cancer compounds to inhibit cell growth compared with the concentrations that are effective in mammalian cells (28). Examples pertaining to camptothecin and doxorubicin relative to several human cell lines are provided in SI Fig. 10b. Importantly, expression of each of the pro-survival BCL members in the absence of BAX or BAK did not confer sensitivity of the yeast cells to obatoclax, indicating that the effect of the compound on yeast cell growth depended on the presence of BAX or BAK.
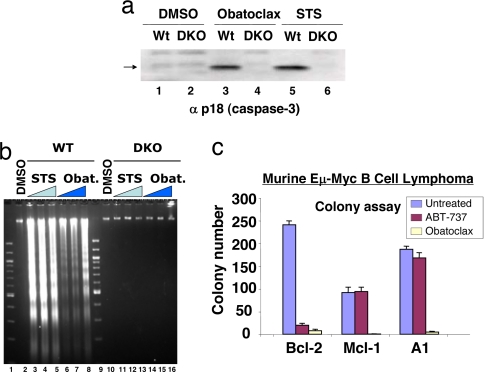
The findings in yeast cells suggest that mammalian cells lacking the expression of BAX and BAK should exhibit resistance to apoptosis in response to obatoclax. This prediction was confirmed in E1A/p53DN-transformed kidney epithelial cells derived from the Bax,Bak double knock out mouse (29). Compared with transformed WT cells, the DKO cells resisted the activation of caspase-3 (Fig. 2a) and the cleavage of DNA into oligo-nucleosomal fragments in response to obatoclax (Fig. 2b), both of which are hallmarks of apoptosis. Moreover, the ability of obatoclax to induce apoptosis in the Bax,Bak-expressing WT cells also extended to its ability to inhibit colony formation of malignant cancer cells. Such activity was examined in the murine Eμ-Myc lymphoma cell model (30, 31), in which protection against apoptotic stress mediated by Myc expression was provided by overexpression of MCL-1, BCL-2, or A1 (Fig. 2c). In all three cell lines, colony formation and survival was strongly inhibited by obatoclax.
Fig. 2.
Induction of apoptosis and inhibition of colony formation by obatoclax. (a) Deletion of Bax and Bak causes resistance to obatoclax. Baby kidney epithelial cell lines derived from the WT and Bax,Bak double knockout (DKO) mouse were treated with vehicle (DMSO) or vehicle containing 2.0 μM obatoclax or 0.5 μM staurosporin (STS), and 20 h later the processed large subunit of caspase-3 (arrow) was detected by SDS/PAGE and immunoblot analysis. (b) As in a, except that oligonucleosomal fragments were detected by agarose gel electrophoresis and staining with ethidium bromide after treatments with 0.05 μM (lanes 3 and 10), 0.2 μM (lanes 4 and 11) and 1 μM (lanes 5 and 12) STS for 16 h; obatoclax treatment was 0.5 μM (lanes 6 and 14), 1 μM (lanes 7 and 15) and 2 μM (lanes 8 and 16) for 24 h. (c) Sensitivity of murine Eμ Myc B cell lymphoma cell lines overexpressing Bcl-2, Mcl-1, or A1 to obatoclax. Cells were exposed to 1 μM obatoclax or ABT737 for 24 h, the media (containing compound) diluted 30-fold, and colony formation on soft agar recorded after 7 days, as described in ref. 45.
Obatoclax Overcomes Resistance to Apoptosis Conferred by MCL-1.
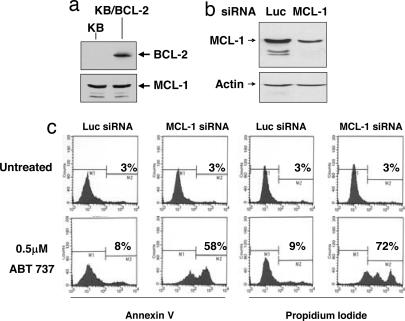
In contrast to obatoclax, the BH3 mimetic ABT-737 inhibited colony formation of Eμ-Myc lymphoma cells that overexpress BCL-2, but not those overexpressing MCL-1 or A1 (Fig. 2c). This result is consistent with both the finding that ABT-737 does not antagonize A1 or MCL-1 (16, 21) and the observation that MCL-1 can confer cellular resistance to ABT-737-induced apoptosis (32, 33). Such a resistance mechanism to ABT-737 can thus be exploited to determine whether obatoclax can overcome MCL-1-mediated resistance to apoptosis. To that end, a KB oral carcinoma cell line was generated that overexpresses ectopic BCL-2 (to restrict cell death) but also expresses sufficient endogenous MCL-1 to restrict cytotoxicity conferred by ABT-737 (Fig. 3 a–c). The KB/BCL-2 model for MCL-1-mediated resistance to ABT-737 was validated by siRNA-mediated knockdown of MCL-1 (Fig. 3 b and c). As judged by annexin V reactivity and uptake of propidium iodide, neither knock down of MCL-1 alone nor treatment of the cells with 0.5 μM ABT-737 alone induced cell death; however, combining the two treatments was strongly cytotoxic (Fig. 3c).
Fig. 3.
BCL-2-overexpressing KB cells whose endogenous MCL-1 confers resistance to ABT-737. (a) Stable overexpression of BCL-2 in KB cells. (b) Reduction of MCL-1 expression in KB/BCL-2 cells by MCL-1 siRNA. (c) MCL-1 knockdown confers sensitivity to ABT-737 in KB/BCL-2 cells. KB/BCL-2 cells were transfected with human MCL-1 siRNA or luciferase (Luc) siRNA and plated into media containing 0.5 μM ABT-737. After 72, the percentage of cells incorporating annexin V FITC and propidium iodide was determined by flow cytometry.
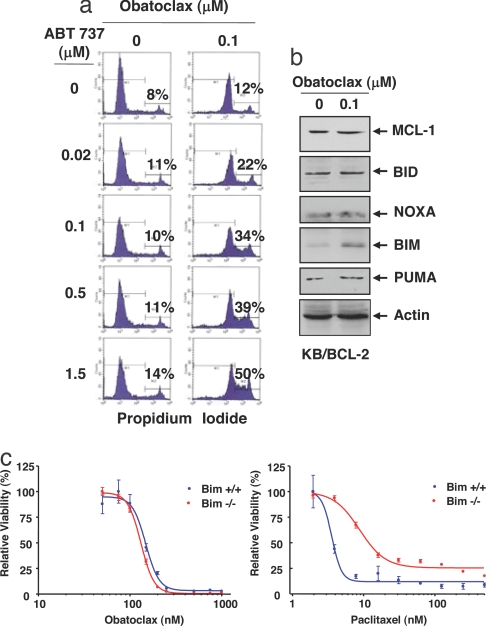
The KB/BCL-2 cells were also resistant to 0.1 μM obatoclax. However, like MCL-1 siRNA, 0.1 μM obatoclax overcame MCL-1-mediated resistance to ABT-737, leading to cell death in combination (Fig. 4a). The treatment with obatoclax had no effect on MCL-1 levels but among those BH3-only members that have been reported to antagonize MCL-1 (BID, NOXA, BIM, and PUMA; ref. 24), a modest increase in BIM was noted (Fig. 4b). However, E1A/p53DN transformed kidney epithelial cells derived from the Bim−/− mouse were as sensitive to obatoclax-mediated killing as were WT cells (Fig. 4c), whereas Bim deletion conferred partial resistance to paclitaxel, as described in ref. 34. Although BIM is not required for obatoclax-mediated cytotoxicity, its increase in response to obatoclax could potentiate the inhibition of MCL-1 by the drug.
Fig. 4.
Obatoclax overcomes the resistance of KB/BCL-2 cells to ABT-737. (a) KB/BCL-2 cells were incubated with the indicated concentrations of ABT-737 in the absence or presence of 0.1 μM obatoclax. The percentage of cells that took up propidium iodide was determined by flow cytometry. (b) Influence of obatoclax on expression of MCL-1 and BH3-only antagonists of MCL-1 in KB/BCL-2 cells. After treatment with or without 0.1 μM obatoclax for 72 h, 25 μg of total cell lysate was analyzed by SDS/PAGE and immunoblot for the indicated proteins. (c) Bim deletion does not interfere with obatoclax cytotoxicity. Bim−/− or Bim+/+ transformed baby mouse kidney cells were treated with obatoclax or paclitaxel at the indicated concentrations for 72 h and cell viability determined. Standard error is shown (bars).
Obatoclax Overcomes MCL-1-Mediated Resistance to the Proteasome Inhibitor Bortezomib in Melanoma.
A unique property of MCL-1, relative to other pro-survival members of the BCL-2 family, is its rapid steady-state turnover by the 26S proteasome complex (18, 20), resulting from constitutive ubiquitination of MCL-1 by the E3 ligase MULE/LASU1 (35, 36). Interference with either the E3 ligase or the activity of the proteasome itself can result in significant elevation in the level of the MCL-1 protein. Moreover, such elevation of MCL-1 can interfere with the therapeutic response to the proteasome inhibitor bortezomib (Velcade) in cells derived from malignancies for which the drug is currently approved for treatment, multiple myeloma (37) and mantle cell lymphoma (26). In the case of both multiple myeloma (38) and mantle cell lymphoma (39), combining bortezomib with obatoclax partially overcame resistance to bortezomib and resulted in an enhanced cytotoxic response. The ability of obatoclax to overcome MCL-1-mediated resistance to bortezomib, therefore, could ameliorate the response barrier to the use of bortezomib as a single agent in other indications.
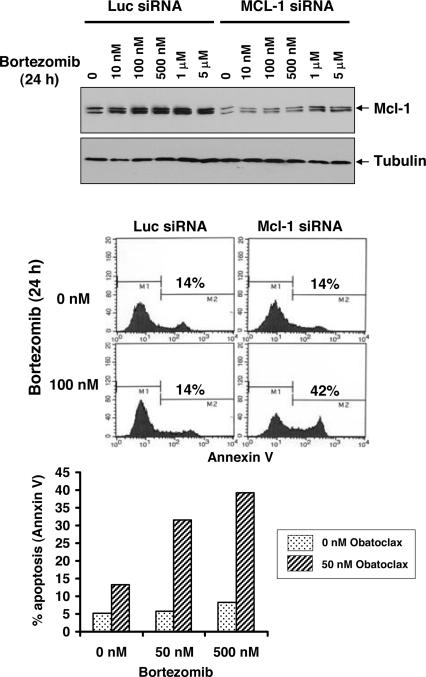
An example is illustrated in the melanoma cell line B16-F1 (Fig. 5). This cell line was selected because bortezomib was found not to influence Noxa (data not shown), which, like MCL-1, can also be elevated in response to proteasome inhibition (39, 40). This cell line was particularly resistant to bortezomib even at relatively high concentrations of the drug. Bortezomib caused elevation of MCL-1 to near maximum levels at 100 nM of drug (Fig. 5 Top) and the cells resisted treatment with the drug at concentrations as high as 500 nM for 24 h (Fig. 5 Middle and Bottom). MCL-1 siRNA prevented the induction of MCL-1 protein levels over a wide range of bortezomib concentrations (Fig. 5 Top) and sensitized the cells to induction of apoptosis by 100 nM bortezomib, as measured by annexin V staining, while having no effect on its own (Fig. 5 Middle). Because obatoclax can antagonize MCL-1, it would be expected to have an effect similar to that of MCL-1 siRNA. A concentration of obatoclax was selected that alone had only modest effects on annexin V staining (Fig. 5 Bottom, left side of histogram), but it was found to potentiate either 50 or 500 nM bortezomib. Thus, melanoma as well as potentially other cancer indications could benefit from the combination of a proteasome inhibitor and an MCL-1 antagonist.
Fig. 5.
Obatoclax overcomes MCL-1-mediated resistance to bortezomib in melanoma cells. (Top) Mouse melanoma B16-F1 cells were transfected with luciferase siRNA or mouse Mcl-1 siRNA for 24 h followed by a 24-h treatment with bortezomib at the indicated concentrations. Total cell lysates were analyzed by immunoblot for Mcl-1 expression. (Middle) As in Top, except that the percentage of cells that incorporated annexin V was determined by flow cytometry. (Bottom) B16-F1 cells were incubated with bortezomib alone or in combination with 50 nM obatoclax for 24 h and apoptosis (annexin V staining) determined as in Middle.
Obatoclax Exhibits Single-Agent Antitumor Activity.
Multiple cancer cell lines exhibit cytotoxicity in response to treatment with obatoclax. When formulated for i.v. administration, obatoclax was found to have single-agent antitumor effects in several standard mouse tumor models. Examples are provided in SI Fig. 11 a and b. Antitumor activity without animal weight loss was observed in mice bearing solid tumors, including those derived from mouse mammary and human colon, prostate and cervical carcinoma cell lines.
Concluding Remarks.
The hydrophobic properties of obatoclax seem to be compatible with the mitochondrial outer membrane milieu in which the BCL-2 pro-survival proteins operate. Indeed, we were able to demonstrate potent inhibition of the constitutive interactions between MCL-1 and BAK within the intact mitochondrial outer membrane. As a consequence, we conducted most of our analyses in systems where the integrity of cellular membranes was maintained, including intact mitochondria and cells. Together with the findings in yeast cells reconstituted with the BCL/BAX or BCL/BAK pathways and in murine epithelial cells deleted of Bax and Bak, the results collectively indicated that obatoclax can inhibit the ability of pro-survival members of the BCL-2 family to antagonize BAX or BAK, causing cytotoxicity. Such cytotoxicity also extended to inhibition of colony formation in the murine Eμ-Myc model of B cell lymphoma and to antitumor activity in murine xenograft models of human carcinomas.
An important opportunity arising from the development of targeted therapies for the treatment of cancer is the ability to rationally combine these agents to achieve drug synergies. To illustrate such a potential for obatoclax, we explored its combination with the 26S proteasome inhibitor bortezomib (Velcade) and the BCL-2/BCL-XL,BCL-w-selective inhibitor ABT-737. In both cases, resistance to these agents was conferred by MCL-1: in the case of bortezomib, because this drug interferes with the rapid steady-state turnover of MCL-1, resulting in MCL-1 elevation; and in the case of ABT-737, because this BCL-2 antagonist does not significantly inhibit MCL-1. Cell models were selected for which siRNA analyses confirmed that the endogenous levels of MCL-1 conferred resistance to these agents. As expected, obatoclax was found to overcome the MCL-1-mediated resistance to bortezomib or to ABT-737, and in both cases strong synergistic cytotoxicities with obatoclax were recorded. In the case of a potent cytotoxic like bortezomib with a restrictive side-effect profile (41), the combination with obatoclax may permit a reduction in the dose of bortezomib required to achieve a therapeutic benefit and promote its therapeutic potential in cancer indications where it does not have utility as a single agent. Moreover, evidence of induction of MCL-1 by bortezomib, with or without concomitant induction of NOXA, could be exploited as a biomarker to predict synergy with an MCL-1 antagonist such as obatoclax.
Finally, the findings reported here are consistent with the principle that inhibition of MCL-1 below a critical threshold level, as opposed to complete inhibition, is sufficient to prevent MCL-1 from resisting signals that result in cell death. This point is relevant because MCL-1 is an essential protein in development, and complete inhibition may be toxic to certain normal tissues. Germ line ablation of the mouse Mcl-1 gene, for example, resulted in peri-implantation embryonic lethality (42), and conditional gene deletion indicated an essential role for MCL-1 in the development and maintenance of B and T lymphocytes (43). Importantly, however, it required only partial knock down of endogenous MCL-1 to sensitize a cell line overexpressing BCL-2 to cytotoxicity induced by ABT-737. The ability of a small molecule like obatoclax to lower the functional threshold of MCL-1 below a critical level, therefore, may provide a therapeutic window for an effective and safe treatment in an appropriate cancer setting.
Materials and Methods
Analyses with Isolated Mitochondria.
Mitochondria were isolated from SK-Mel5 as described in ref. 44. For MCL-1/BAK interaction, 40 μg of isolated mitochondria was incubated with obatoclax or NOXA BH3 peptide in 50 μl of 200 mM mannitol, 70 mM sucrose, 10 mM Hepes (pH 7.5), and 1 mM EDTA for 30 min at 37°C. The mitochondria were reisolated and subjected to cross-linking with 0.1 mM LC-SMCC in the same buffer for 30 min at 25°C. Cross-linked mitochondria were reisolated, lysed in 50 mM Hepes (pH 7.7), 150 mM NaCl, 0.1% Triton X-100, 0.5% Nonidet P-40, and 1 mM EDTA and subjected to immunoprecipitation with anti-MCL-1 antibody (Stressgen). The precipitates were analyzed by SDS/PAGE followed by immunoblotting with anti-MCL-1 and anti-BAK (Upstate) antibodies. For analyses of cytochrome c, 25 μg of isolated mitochondria from KB cells were incubated with obatoclax or BID BH3 peptide as described above. Mitochondrial pellets and supernatants were separated and analyzed by SDS/PAGE and immunoblot with anti-cytochrome c antibody (BD Biosciences).
Coimmunoprecipitation from Cells.
SK-Mel5 cells were incubated at 37°C for 5 h with obatoclax or DMSO. Cell pellets were resuspended in ice-cold 10 mM Tris (pH 8.0), 60 mM KCl, 1 mM EDTA, 1 mM DTT, 0.5% Nonidet P-40 (IGEPAL), 10 mM sodium vanadate, 50 mM NaF, 0.1 mM PEFABLOC, 1 μg/ml pepstatin A and leupeptin, and 10 μg/ml aprotinin and gently pipetted up and down 10–15 times. The samples were incubated for 10 min on ice and then centrifuged at 10,000 × g for 15 min at 4°C. Cell extracts (250 μg of protein) were brought to a volume of 1 ml with immunoprecipitation (IP) buffer (50 mM Hepes, pH 7.7, 150 mM NaCl, 0.1% Nonidet P-40 (IGEPAL), 5 mM EDTA) and incubated with 2 μg of rabbit anti-MCL-1 antibody (SC-819; Santa Cruz Biotechnology) overnight at 4°C. Immune complexes were precipitated by incubation with protein A-coated Sepharose beads previously equilibrated with IP buffer, and washed five times with 1 ml of IP buffer. Immunoprecipitated proteins were separated on a 12.5% SDS/PAGE gel, and analyzed by Western blot using the following antibodies: mouse anti-MCL-1 antibody (PharMingen) (1/1,000 dilution); rabbit anti-BAK (Upstate) antibody (1/1,000 dilution); and HRP-conjugated goat anti-rabbit or anti-mouse antibody (1/5,000 dilution). Proteins were detected by using the ECL plus Western blot detection kit (Amersham).
siRNA Transfection.
Pools of four siRNAs against human MCL-1 or mouse Mcl-1 mRNAs were purchased from Dharmacon. KB/BCL-2 cells were transfected twice with the pool of human MCL-1 siRNA or luciferase siRNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Twenty-four hours after the second transfection, cells were plated into media containing the compounds. Seventy-two hours later, cells were analyzed by flow cytometry. For mouse B16-F1 cells, pools of mouse Mcl-1 siRNAs or luciferase siRNAs were transfected once by using Lipofectamine 2000. Twenty-four hours after transfection, cells were plated into medium containing the compounds; cells were collected the following day and analyzed by flow cytometry.
Flow Cytometry.
Treated cells were collected, washed with PBS, and resuspended in buffer [10 mM Hepes (pH 7.4), 140 mM NaCl, 2.5 mM CaCl2] containing annexin V (Biovision) or propidium iodide (Molecular Probes) as described by the manufacturer. Cells were incubated for 15 min at 25°C and analyzed by flow cytometry. For additional information, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Eileen White (Rutgers University, Piscataway, NJ) for kindly providing WT and Bax,Bak double knockout mouse epithelial cell lines. We thank the following for contributions: Salah Aouad, Angela Babineau, Sylvie Baillie, Mark Berger, Geneviève Brindle, Helen Chan, Gang Chen, Gaétan Gagnon, Michel-Olivier Gratton, Jeff Hincks, Anne Jang, Abdelkrim Khadir, Mark Mariani, Sandra Naranjo, Youssouf Soumounou, Brigitte St.-Denis, Nancy Steenaart, Émilie Turcotte, Zhiying Zhang, Giorgio Attardo, Anne Bélanger, Stéphane Branchaud, Sylvie Charron, Kenza Dairi, Lionel Dumas, Walid Elbast, Noura El-Djabri, Éric Fournier, Gerson Gonzalez, Andrea Hill, Jean-François Lavallée, Manon Lavoie, Daniel Rabouin, Élise Rioux, and Sasmita Tripathy. This work was supported in part by research grants from the Canadian Institutes of Health Research, the Leukemia Foundation of Australia, Cancer Council Victoria, the Canadian Cancer Society, and the National Health and Medical Research Council of Australia.
Footnotes
Conflict of interest statement: R.C.M., A.R., M.W., L.S., S.R.M.M., D.G., J.V., L.B., X.B., P.B., and G.C.S. are or were employees of Gemin X Biotechnologies, Inc.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709443104/DC1.
References
- 1.Cory S, Vaux DL, Strasser A, Harris AW, Adams JM. Cancer Res. 1999;59(Suppl 7):1685s–1692s. [PubMed] [Google Scholar]
- 2.Korsmeyer SJ. Cancer Res. 1999;59(Suppl 7):1693s–1700s. [PubMed] [Google Scholar]
- 3.Danial NN, Korsmeyer SJ. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 4.Vaux DL, Cory S, Adams JM. Nature. 1988;335:440–442. doi: 10.1038/335440a0. [DOI] [PubMed] [Google Scholar]
- 5.Reed JC, Cuddy M, Slabiak T, Croce CM, Nowell PC. Nature. 1988;336:259–261. doi: 10.1038/336259a0. [DOI] [PubMed] [Google Scholar]
- 6.McDonnell TJ, Deane N, Platt FM, Nunez G, Jaeger U, McKearn JP, Korsmeyer SJ. Cell. 1989;57:79–88. doi: 10.1016/0092-8674(89)90174-8. [DOI] [PubMed] [Google Scholar]
- 7.Pelengaris S, Khan M, Evan GI. Cell. 2002;109:321–334. doi: 10.1016/s0092-8674(02)00738-9. [DOI] [PubMed] [Google Scholar]
- 8.Reed JC. Cell Death Differ. 2006;13:1378–1386. doi: 10.1038/sj.cdd.4401975. [DOI] [PubMed] [Google Scholar]
- 9.Adams JM, Cory S. Oncogene. 2007;26:1324–1337. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang X, Wang X. Annu Rev Biochem. 2004;73:87–106. doi: 10.1146/annurev.biochem.73.011303.073706. [DOI] [PubMed] [Google Scholar]
- 11.Petros AM, Olejniczak ET, Fesik SW. Biochim Biophys Acta. 2004;1644:83–94. doi: 10.1016/j.bbamcr.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 12.Youle RJ. Science. 2007;315:776–777. doi: 10.1126/science.1138870. [DOI] [PubMed] [Google Scholar]
- 13.Annis MG, Soucie EL, Dlugosz PJ, Cruz-Aguado JA, Penn LZ, Leber B, Andrews DW. EMBO J. 2005;24:2096–2103. doi: 10.1038/sj.emboj.7600675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dlugosz PJ, Billen LP, Annis MG, Zhu W, Zhang Z, Lin J, Leber B, Andrews DW. EMBO J. 2006;25:2287–2296. doi: 10.1038/sj.emboj.7601126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD, Wagner G, Verdine GL, Korsmeyer SJ. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, et al. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 17.Day CL, Chen L, Richardson SJ, Harrison PJ, Huang DC, Hinds MG. J Biol Chem. 2005;280:4738–4744. doi: 10.1074/jbc.M411434200. [DOI] [PubMed] [Google Scholar]
- 18.Cuconati A, Mukherjee C, Perez D, White E. Genes Dev. 2003;17:2922–2932. doi: 10.1101/gad.1156903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willis SN, Chen L, Dewson G, Wei A, Naik E, Fletcher JI, Adams JM, Huang DC. Genes Dev. 2005;19:1294–1305. doi: 10.1101/gad.1304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nijhawan D, Fang M, Traer E, Zhong Q, Gao W, Du F, Wang X. Genes Dev. 2003;17:1475–1486. doi: 10.1101/gad.1093903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhai D, Jin C, Satterthwait AC, Reed JC. Cell Death Differ. 2006;13:1419–1421. doi: 10.1038/sj.cdd.4401937. [DOI] [PubMed] [Google Scholar]
- 22.Naim M, Bhat S, Rankin KN, Denni S, Chowdhury SF, Siddiqi I, Drabik P, Sulea T, Bayly CI, Jakalian A, Purisima EO. J Chem Inf Model. 2007;47:122–133. doi: 10.1021/ci600406v. [DOI] [PubMed] [Google Scholar]
- 23.Bruncko M, Oost TK, Belli BA, Ding H, Joseph MK, Kunzer A, Martineau D, McClellan WJ, Mitten M, Ng SC, et al. J Med Chem. 2007;50:641–662. doi: 10.1021/jm061152t. [DOI] [PubMed] [Google Scholar]
- 24.Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, Colman PM, Day CL, Adams JM, Huang DC. Mol Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 25.Letai A, Bassik MC, Walensky LD, Sorcinelli MD, Weiler S, Korsmeyer SJ. Cancer Cell. 2002;2:183–192. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 26.Perez-Galan P, Roue G, Villamor N, Montserrat E, Campo E, Colomer D. Blood. 2006;107:257–264. doi: 10.1182/blood-2005-05-2091. [DOI] [PubMed] [Google Scholar]
- 27.Zha H, Fisk HA, Yaffe MP, Mahajan N, Herman B, Reed JC. Mol Cell Biol. 1996;16:6494–6508. doi: 10.1128/mcb.16.11.6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simon JA, Szankasi P, Nguyen DK, Ludlow C, Dunstan HM, Roberts CJ, Jensen EL, Hartwell LH, Friend SH. Cancer Res. 2000;60(2):328–333. [PubMed] [Google Scholar]
- 29.Degenhardt K, Chen G, Lindsten T, White E. Cancer Cell. 2002;2:193–203. doi: 10.1016/s1535-6108(02)00126-5. [DOI] [PubMed] [Google Scholar]
- 30.Schmitt CA, Fridman JS, Yang M, Baranov E, Hoffman RM, Lowe SW. Cancer Cell. 2002;3:289–298. doi: 10.1016/s1535-6108(02)00047-8. [DOI] [PubMed] [Google Scholar]
- 31.Lindemann RK, Newbold A, Whitecross KF, Cluse LA, Frew AJ, Ellis L, Williams S, Wiegmans AP, Dear AE, Scott CL, et al. Proc Natl Acad Sci USA. 2007;104:8071–8076. doi: 10.1073/pnas.0702294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konopleva M, Contractor R, Tsao T, Samudi I, Ruvolo PP, Kitada S, Deng X, Zhai D, Shi YX, Sneed T, et al. Cancer Cell. 2006;10:375–388. doi: 10.1016/j.ccr.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 33.van Delft MF, Wei AH, Mason KD, Vandenberg CJ, Chen L, Czabotar PE, Willis SN, Scott CL, Day CL, Cory S, et al. Cancer Cell. 2006;10:389–399. doi: 10.1016/j.ccr.2006.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan TT, Degenhardt K, Nelson DA, Beaudoin B, Nieves-Neira W, Bouillet P, Villunger A, Adams JM, White E. Cancer Cell. 2005;7:227–238. doi: 10.1016/j.ccr.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 35.Zhong Q, Gao W, Du F, Wang X. Cell. 2005;121:1085–1095. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 36.Warr MR, Acoca S, Liu Z, Germain M, Watson M, Blanchette M, Wing SS, Shore GC. FEBS Lett. 2005;579:5603–5608. doi: 10.1016/j.febslet.2005.09.028. [DOI] [PubMed] [Google Scholar]
- 37.Gomez-Bougie P, Wuilleme-Toumi S, Menoret E, Trichet V, Robillard N, Philippe M, Bataille R, Amiot M. Cancer Res. 2007;67:5418–5424. doi: 10.1158/0008-5472.CAN-06-4322. [DOI] [PubMed] [Google Scholar]
- 38.Trudel S, Li ZH, Rauw J, Tiedemann RE, Wen XY, Stewart AK. Blood. 2007;109:5430–5438. doi: 10.1182/blood-2006-10-047951. [DOI] [PubMed] [Google Scholar]
- 39.Perez-Galan P, Roue G, Villamor N, Campo E, Colomer D. Blood. 2007;109:4441–4449. doi: 10.1182/blood-2006-07-034173. [DOI] [PubMed] [Google Scholar]
- 40.Qin JZ, Ziffra J, Stennett L, Bodner B, Bonish BK, Chaturvedi V, Bennett F, Pollock PM, Trent JM, Hendrix MJ, et al. Cancer Res. 2005;65:6282–6293. doi: 10.1158/0008-5472.CAN-05-0676. [DOI] [PubMed] [Google Scholar]
- 41.Caravita T, de Fabritiis P, Palumbo A, Amadori S, Boccadoro M. Nat Clin Pract Oncol. 2006;3:374–387. doi: 10.1038/ncponc0555. [DOI] [PubMed] [Google Scholar]
- 42.Rinkenberger JL, Horning S, Klocke B, Roth K, Korsmeyer SJ. Genes Dev. 2000;14:23–27. [PMC free article] [PubMed] [Google Scholar]
- 43.Opferman JT, Letai A, Beard C, Sorcinelli MD, Ong CC, Korsmeyer SJ. Nature. 2003;426:671–676. doi: 10.1038/nature02067. [DOI] [PubMed] [Google Scholar]
- 44.Ruffolo SC, Shore GC. J Biol Chem. 2003;278:25039–25045. doi: 10.1074/jbc.M302930200. [DOI] [PubMed] [Google Scholar]
- 45.Johnstone RW, Cretney E, Smyth MJ. Blood. 1999;93:1075–1085. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.