Abstract
Vitamin B1 (thiamin) is an essential compound in all organisms acting as a cofactor in key metabolic reactions and has furthermore been implicated in responses to DNA damage and pathogen attack in plants. Despite the fact that it was discovered almost a century ago and deficiency is a widespread health problem, much remains to be deciphered about its biosynthesis. The vitamin is composed of a thiazole and pyrimidine heterocycle, which can be synthesized by prokaryotes, fungi, and plants. Plants are the major source of the vitamin in the human diet, yet little is known about the biosynthesis of the compound therein. In particular, it has never been verified whether the pyrimidine heterocycle is derived from purine biosynthesis through the action of the THIC protein as in bacteria, rather than vitamin B6 and histidine as demonstrated for fungi. Here, we identify a homolog of THIC in Arabidopsis and demonstrate its essentiality not only for vitamin B1 biosynthesis, but also plant viability. This step takes place in the chloroplast and appears to be regulated at several levels, including through the presence of a riboswitch in the 3′-untranslated region of THIC. Strong evidence is provided for the involvement of an iron–sulfur cluster in the remarkable chemical rearrangement reaction catalyzed by the THIC protein for which there is no chemical precedent. The results suggest that vitamin B1 biosynthesis in plants is in fact more similar to prokaryotic counterparts and that the THIC protein is likely to be the key regulatory protein in the pathway.
Keywords: metabolites, plant viability, riboswitch, thiamin, Arabidopsis
Vitamin B1, in the form of thiamin diphosphate (TPP), acts as a cofactor for several enzymes in key cellular metabolic pathways such as glycolysis, the pentose phosphate pathway and the citric acid cycle (TCA), in addition to amino acid and nonmevalonate isoprenoid biosynthesis, respectively (1). More recently, it has been implicated in tolerance to DNA damage (2) and as an activator of disease resistance in plants (3, 4). It is an essential compound in all-living systems, but de novo biosynthesis is only found in prokaryotes, fungi, and plants; therefore, animals must acquire it from dietary sources. Deficiency of the vitamin is a widespread health problem particularly in countries where rice is a major constituent of the diet, because grain polishing removes most of the thiamin in the bran. Therefore, it is of major interest to define the pathways of biosynthesis, which may assist in the overproduction of the vitamin for beneficial purposes.
Thiamin biosynthesis occurs through the separate formation of the pyrimidine and thiazole heterocycle, which are then coupled to form the cofactor [supporting information (SI) Scheme 1]. Although the kinases involved in the phosphorylation of the precursor molecules have been identified in many organisms, the de novo formation of the heterocycles is poorly understood. In bacteria, the mechanism of thiazole formation is reasonably well documented and involves at least five enzymes that transform 1-deoxy-d-xylulose-5-phosphate, cysteine and either glycine or tyrosine (Bacillus subtilis or Escherichia coli, respectively), in a complex oxidative condensation (5, 6). Much less is known about the formation of the pyrimidine moiety, which is thought to be derived from 5-aminoimidazole ribonucleotide (AIR), an intermediate in the purine biosynthetic pathway (SI Scheme 1) (7, 8). AIR is then converted into 4-amino-2-methyl-5-hydroxymethylpyrimidine phosphate (HMP-P), or its alcohol, through the action of THIC. The mechanism by which THIC performs this remarkable rearrangement reaction is of particular interest, as there appears to be no chemical precedent, but little is known of its requirements (9).
In eukaryotes, neither the biosynthesis of the thiazole nor of the pyrimidine moiety has been fully elucidated. A study in spinach has implicated deoxyxylulose-5-phosphate, cysteine and tyrosine as precursors for thiazole synthesis (10). Yet, to date, only a single thiazole biosynthetic enzyme has been identified, called THI4 in Saccharomyces cerevisiae (11) and THI1 in Arabidopsis (2), but shows no homology to the bacterial enzymes. Although the involvement of this protein in thiazole biosynthesis has been deduced from genetic studies, its biochemical function has not been clarified. Interestingly, recent studies based on structural and mechanistic data of the S. cerevisiae protein have led to the intriguing implication of the redox cofactor NADH as a precursor in this organism (12). In contrast to bacteria, in eukaryotes the pyrimidine moiety appears to be derived from vitamin B6 and histidine (13–16). Again, only a single protein involved in this transformation is known, THI5 or NMT1 for “no message in thiamin1” in S. cerevisiae and Schizosaccharomyces pombe, respectively (17, 18). Thus, although much remains to be deciphered, it is assumed that the pathways of thiamin biosynthesis are distinct from each other in prokaryotes and eukaryotes (19).
Here, we have focused on the synthesis of the pyrimidine moiety in plants employing Arabidopsis. It could be envisaged that the pyrimidine heterocycle is derived either from AIR as in bacteria, or vitamin B6 and histidine as in fungi, or both (SI Scheme 1). In this study, we demonstrate that the predominant and perhaps sole source of the pyrimidine heterocycle in Arabidopsis is via a bacterial type pathway. A single THIC homolog is present and is essential for plant viability. However, the thiC knockdown mutant can be rescued with thiamin supplementation. Metabolic profiling allows us to demonstrate the essentiality of the vitamin for primary metabolic function in plants. Localization studies show the protein to be partitioned to chloroplasts, and an expression analysis reveals positive regulation of the expression of the gene by light and severe negative regulation by thiamin itself. We provide strong evidence that the THIC protein contains an iron–sulfur (Fe-S) cluster that is required for functionality, and we suggest a further level of regulation of the protein by the chloroplast thioredoxin–ferredoxin system.
Results and Discussion
De Novo Biosynthesis of the Pyrimidine Moiety of Vitamin B1 in Planta via THIC.
As either the bacterial or fungal route of pyrimidine heterocycle biosynthesis, or both, could be in operation in planta, we first searched for homologs of known genes of either route in Arabidopsis, i.e., THIC or NMT1, respectively. Whereas one homolog of THIC was identified (At2g29630), no homolog of NMT1 could be found. The THIC gene contains three introns and four exons (Fig. 1A). The predicted protein sequence is 644 amino acids with a calculated molecular mass of 71,994 Da. Homologs of THIC were identified in prokaryotes and plants, but not in fungi or apicomplexa. An amino acid sequence alignment (Fig. 1B) demonstrates that the Arabidopsis protein is highly homologous to THIC from B. subtilis and E. coli (58% and 53% identity, respectively) and is ≈80% identical to the two homologs identified in rice.
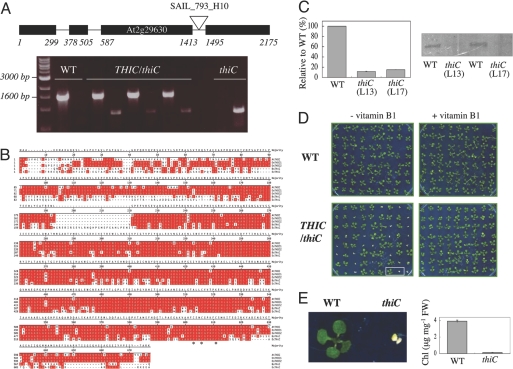
Fig. 1.
Arabidopsis THIC characteristics. (A) (Upper) Exon–intron structure of Arabidopsis THIC indicating the location of the SAIL_793_H10 insertion. (Lower) PCR analysis indicating plant genotypes employing primer pairs specific for either WT or the T-DNA insertion in thiC, respectively. From left to right: molecular marker, WT, three lines heterozygous for thiC, and a homozygous line (L13). (B) Amino acid sequence alignment of THIC homologs identified in Arabidopsis thaliana (At), Oryza sativa (Os), Bacillus subtilis (Bs), and Escherichia coli (Ec). Amino acids identical in at least two of the sequences are shaded in red. The asterisk denotes the identified C(X)2C(X)4C motif. (C) (Left) Quantitative RT-PCR of THIC transcript in WT or thiC plants. The values indicated are the percentage mRNA levels relative to WT (value = 100). Error bars indicate the standard deviation of the experiment performed in triplicate. (Right) Western blot analysis of the same samples probed with a THIC antibody. L13 and L17 refer to lines 13 and 17, respectively. (D) thiC mutant phenotype. (Left) WT and progeny of selfed THIC/thiC plants grown in sterile culture lacking vitamin B1. Approximately one quarter of the population shows a pale green phenotype and does not develop beyond the cotyledon stage. (Right) Supplementation with vitamin B1 (0.5 μM) restores growth of thiC to that of WT. Ten-day-old seedlings grown under long day conditions. (E) (Left) Enlarged picture of two seedlings from D. (Right) Total chlorophyll content (Chl) of WT and thiC (L13) under the same conditions. Error bars indicate the standard deviation of three independent experiments.
To address the question of whether THIC is involved in the biosynthesis of the pyrimidine moiety of thiamin, loss of function studies were performed employing a putative insertion mutant of THIC identified in the SAIL collection (SAIL_793_H10) (20). SAIL_793_H10 is a T-DNA insertion line that carries the BAR gene conferring resistance to phosphinothricin (Basta). PCR analysis with primers designed to hybridize close to either end of the inserted element and the flanking gene sequence revealed bands of the expected size, thus confirming the respective insertion(s) in THIC (Fig. 1A). Sequencing of the amplified fragments revealed that the T-DNA insertion is located at base pair 1464 relative to the start codon, i.e., within the third intron of THIC (Fig. 1A). A segregation analysis among the progeny of selfed THIC/thiC plants revealed that Basta-resistant and -sensitive seedlings, respectively, segregated in a ratio of 3:1 (SI Table 3), indicating that the insertion is at a single locus. A quantitative RT-PCR analysis revealed that the THIC transcript is severely reduced (≈10-fold) in the thiC mutant plants (Fig. 1C). Furthermore, Western blot analysis using a THIC antibody detected the ascribed protein band at substantially lower abundance in the mutant as compared with WT (Fig. 1C). Thus, we conclude that this mutant represents a strong knockdown version of THIC in Arabidopsis.
To assess for a thiC phenotype, seeds heterozygous for the T-DNA insertion were germinated and seedlings were grown on thiamin-deficient medium. One quarter of the progeny exhibited a chlorotic phenotype and did not develop beyond the cotyledon stage (Fig. 1 D and E). PCR analysis confirmed that the segregating phenotype were thiC seedlings (data not shown). This phenotype was rescued by supplementation with thiamin (Fig. 1D). Moreover, thiC lines grown in vitamin B1-free sterile culture for 10 days and then transferred to soil could be maintained by continuously supplying them with a thiamin solution (Fig. 2A). If the thiC plants were not supplemented with thiamin, they eventually died. A concentration of thiamin as low as 1.5 μM was sufficient to allow growth of the seedlings, but these were chlorotic; at 100 μM thiamin, seedlings had normal coloration, but were smaller than control WT seedlings (Fig. 2A Left). To further corroborate that the seedling lethal phenotype of thiC is due to an impairment in thiamin biosynthesis, a quantitative microbiological assay employing a strain of S. cerevisiae auxotrophic for thiamin was established. Ten-day-old thiC seedlings grown in the absence of thiamin had only 30% total vitamin B1 content compared with WT seedlings (Table 1).
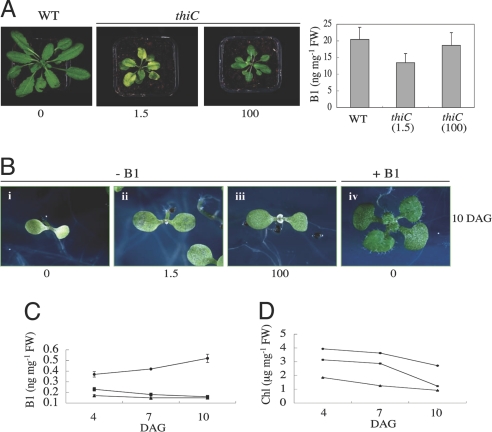
Fig. 2.
Rescue of Arabidopsis thiC mutants. (A) (Left) Rescue of thiC by supplementation with thiamin after transfer to soil. Ten-day-old segregating thiC seedlings grown on medium lacking vitamin B1 (see Fig. 1D) were transferred to soil and watered with the indicated concentrations of thiamin (μM). Plants are 30 days after transfer to soil. (Right) Total vitamin B1 content of seeds of rescued plants. (B) Progeny of rescued thiC lines as described in A compared with segregating nonsupplemented thiC from a heterozygous population. The numbers indicate the concentration of thiamin (μM) supplied to the mother plants. Seedlings (10 days old) were grown in the absence of vitamin B1. As a control, a segregating thiC seedling from a heterozygous mother grown in the presence of vitamin B1 is shown. (C) Total vitamin B1 content of WT (circles), thiC progeny rescued with 100 μM (squares) and 1.5 μM (triangles) thiamin, respectively, grown in the absence of thiamin over the time period indicated. (D) Total chlorophyll content (Chl) of the same seedlings as in C. Error bars (some of which are sufficiently small that they fall within the symbols) indicate the standard deviation of three independent experiments. DAG, days after germination.
Table 1.
Vitamin B1 content of WT and thiC mutant plants
| Sample | Vitamin B1, ng mg−1 FW | % of WT |
|---|---|---|
| WT | 0.47 ± 0.04 | 100 |
| thiC | 0.14 ± 0.05 | 30 |
Total vitamin B1 content was quantified in whole seedlings employing a microbiological assay. Seedlings were grown in sterile culture in medium lacking vitamin B1 and analyzed 10 days after germination. The results shown are the average of four independent experiments. In the case of the thiC mutant, segregating homozygous seedlings from a heterozygous population of line 17 were used.
Interestingly, we observed that the total vitamin B1 level in seeds of rescued thiC is 66% and 91% of WT when supplemented with 1.5 and 100 μM thiamin, respectively (Fig. 2A Right). This was reflected in a noticeable developmental advance of the progeny even when grown in the absence of vitamin B1, compared with segregating thiC seedlings from a nonsupplemented heterozygous population (Fig. 2B). The level of vitamin B1 in the thiC progeny was seen to decrease over time, whereas that of the WT increased over the same period (Fig. 2C). Most likely, stored vitamin B1 gets used up in the thiC mutant, whereas thiamin biosynthesis is induced in WT plants when the compound becomes limiting (see expression analysis below). Indeed, the increase in thiamin levels observed over time implies that a certain threshold level is required for plant viability at this stage. Even when thiC seedlings have an enhanced thiamin content, they are not able to proceed to primary leaf development in the absence of supplementation, thus emphasizing the need for a certain level of the vitamin for survival. Furthermore, we observed that seedlings diminished in thiamin content were paler due to a decrease in total chlorophyll (compare Fig. 2 C and D) and were approaching albino in appearance 12 days after germination. This can be explained by the fact that the biosynthesis of the phytol moiety of chlorophyll depends on the action of 1-deoxy-d-xylulose 5-phosphate synthase, a key enzyme of the nonmevalonate pathway of isoprenoid biosynthesis, which in turn is critically dependent on TPP as a cofactor (21). As 1-deoxy-d-xylulose 5-phosphate synthase is the initial enzyme of this pathway, it is likely that all isoprenoids biosynthesized via this route are reduced.
Metabolite Profiling Reveals the Importance of THIC in Primary Metabolism.
As vitamin B1 in the form of TPP is a necessary cofactor for key enzymes of primary metabolism, we compared the relative level of certain metabolites in thiC and WT. Whole seedlings were analyzed 11 days after germination, reflecting the state of more severe depletion in total thiamin content (Fig. 2). The most notable alteration in metabolites of thiC plants, when they were grown in the absence of thiamin supplementation, appears in the dramatic relative increase in the level of certain amino acids, in particular alanine, asparagine, methionine, phenylalanine, and tryptophan (Table 2). In addition, certain constituents of the TCA cycle were seen to increase, i.e., malate, whereas there was a significant decrease in the level of fumarate. It is possible to explain these changes based on the requirement of certain primary metabolic enzymes on TPP as a cofactor. Notably, impairment of pyruvate dehydrogenase would lead to a breakdown of the TCA cycle. In this context, fumarate is thought to represent a major fraction of fixed carbon in Arabidopsis (22), thus the thiC seedlings can maintain the cycle by using these reserves, which ultimately leads to the observed depletion of this metabolite at day 11. An impairment of pyruvate dehydrogenase could also lead to an increase in glycolytic intermediates, e.g., pyruvate or phosphoenolpyruvate (PEP), which in turn may explain the increases in alanine, phenylalanine, and tryptophan. An increase in the accumulation of either pyruvate or PEP could also elevate the oxaloacetate pool through the action of either pyruvate carboxykinase or PEP-carboxylase. This then may explain not only the observed increase in the pool sizes of methionine and asparagine, but also of malate through the reverse reaction of enzymes of the TCA cycle. It should also be pointed out that transketolase, a participant in both the Calvin and nonoxidative pentose phosphate cycle, whose products and substrates in turn are starting points of several other chloroplast pathways, is a TPP-dependent enzyme. A decrease in transketolase activity is expected to be reflected not only in the Calvin cycle, where the enzyme seems to be rate limiting, but also in carbohydrate, amino acid and natural product metabolism (23). Although a partial reversion of thiC metabolites to WT levels was observed when grown in the presence of thiamin, notably some metabolites remained at elevated levels, in particular certain amino acids (Table 2). The reason for this is unknown at present.
Table 2.
Relative metabolite levels between WT and thiC
| WT − B1 | thiC − B1 | thiC + B1 | |
|---|---|---|---|
| Amino acids | |||
| Alanine | 1.000 ± 0.053 | 9.659 ± 0.913 | 1.448 ± 0.053 |
| Arginine | 1.000 ± 0.080 | 1.283 ± 0.110 | 2.458 ± 0.144 |
| Asparagine | 1.000 ± 0.070 | 6.316 ± 0.534 | 5.037 ± 0.248 |
| Aspartate | 1.000 ± 0.042 | 0.964 ± 0.025 | 1.098 ± 0.006 |
| β-alanine | 1.000 ± 0.027 | 3.545 ± 0.155 | 1.396 ± 0.019 |
| Cysteine | 1.000 ± 0.043 | 3.513 ± 0.123 | 4.329 ± 0.135 |
| Glutamate | 1.000 ± 0.050 | 3.018 ± 0.128 | 1.416 ± 0.054 |
| Glycine | 1.000 ± 0.255 | 0.131 ± 0.009 | 0.051 ± 0.003 |
| Lysine | 1.000 ± 0.013 | 4.563 ± 0.243 | 4.392 ± 0.101 |
| Methionine | 1.000 ± 0.027 | 8.292 ± 0.598 | 8.103 ± 0.290 |
| Phenylalanine | 1.000 ± 0.071 | 23.397 ± 1.129 | 16.529 ± 0.514 |
| Proline | 1.000 ± 0.046 | 1.216 ± 0.029 | 2.119 ± 0.088 |
| Serine | 1.000 ± 0.022 | 1.285 ± 0.014 | 0.539 ± 0.005 |
| Threonine | 1.000 ± 0.008 | 1.290 ± 0.023 | 0.575 ± 0.005 |
| Tryptophan | 1.000 ± 0.142 | 44.130 ± 4.096 | 42.691 ± 1.915 |
| Tyramine | 1.000 ± 0.038 | 1.129 ± 0.023 | 1.061 ± 0.028 |
| Valine | 1.000 ± 0.048 | 2.840 ± 0.159 | 1.980 ± 0.040 |
| Organic acids | |||
| Butyrate | 1.000 ± 0.057 | 6.111 ± 0.347 | 2.479 ± 0.045 |
| Citrate | 1.000 ± 0.144 | 1.211 ± 0.172 | 0.835 ± 0.122 |
| Fumarate | 1.000 ± 0.054 | 0.231 ± 0.016 | 0.220 ± 0.005 |
| Glycerate | 1.000 ± 0.107 | 1.309 ± 0.094 | 0.526 ± 0.033 |
| Glycerol | 1.000 ± 0.064 | 0.693 ± 0.031 | 0.690 ± 0.089 |
| Isocitrate | 1.000 ± 0.078 | 1.769 ± 0.134 | 1.704 ± 0.040 |
| Malate | 1.000 ± 0.058 | 2.509 ± 0.271 | 0.938 ± 0.033 |
| Sugars | |||
| Fructose | 1.000 ± 0.078 | 1.443 ± 0.066 | 1.412 ± 0.121 |
| Gentiobiose | 1.000 ± 0.085 | 0.196 ± 0.031 | 0.435 ± 0.006 |
| Glucose | 1.000 ± 0.019 | 1.681 ± 0.128 | 1.801 ± 0.201 |
| Raffinose | 1.000 ± 0.154 | 0.453 ± 0.041 | 0.382 ± 0.016 |
| Ribose | 1.000 ± 0.022 | 0.293 ± 0.012 | 0.373 ± 0.017 |
| Sucrose | 1.000 ± 0.023 | 0.789 ± 0.036 | 0.782 ± 0.046 |
| Trehalose | 1.000 ± 0.023 | 1.031 ± 0.047 | 0.718 ± 0.011 |
Samples were grown in the absence or presence of thiamin (B1) (0.5 μM) as indicated and cotyledons were harvested 11 days after germination. Substantial increases are shown in bold (≥2-fold), whereas decreases (≤0.5-fold) are italicized. Standard errors are the average of six experimental repetitions. Quantitative data are provided in SI Table 4.
Subcellular Localization of THIC.
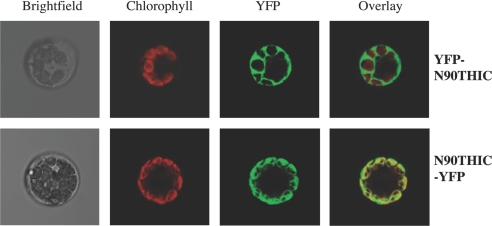
So far, proteins involved in thiamin biosynthesis in plants have been reported to have differential subcellular localizations. For example, THI1 involved in the synthesis of the thiazole moiety, is targeted to both mitochondria and plastids (24), whereas the recently described thiamin pyrophosphokinase is reported to be cytosolic (25). To assemble the site(s) of synthesis of the thiamin molecule, it is thus of interest to know the subcellular localization of THIC. An in silico analysis indicates that the THIC protein should be localized to the chloroplast, but an amino acid sequence alignment does not reveal an obvious N-terminal presequence in the Arabidopsis protein in comparison with bacterial counterparts (Fig. 1B). Moreover, the algorithms differ with respect to the length of the expected target sequence, e.g., TargetP (26) and PSORT (27) predict 37 and 48 amino acids, respectively. We investigated the subcellular location of THIC by fusing the N-terminal 90 amino acids to YFP at either the N or C terminus (N90THIC-YFP and YFP-N90THIC, respectively). The constructs were transformed into Arabidopsis mesophyll protoplasts and the intracellular compartmentation was observed by confocal microscopy (Fig. 3). In the case of the N90THIC-YFP fusion protein, the overlay with chlorophyll autofluorescence clearly demonstrates that the protein is exclusively found in the chloroplasts. Moreover, a stromal localization within the chloroplast is indicated by the fluorescence pattern. This statement is corroborated by a recent study of the stromal proteome of Arabidopsis, in which THIC was identified as a component (28). The YFP-N90THIC fusion protein, on the other hand, is observed throughout the cytosol, as is the case for YFP alone (data not shown). This demonstrates that THIC is localized to the plastid and that moreover the targeting sequence must be contained within the N-terminal 90 amino acids.
Fig. 3.
Subcellular localization of THIC in Arabidopsis. The N-terminal 90 amino acids of THIC were fused to either the N or C terminus of YFP to give N90THIC-YFP and YFP-N90THIC, respectively, and transiently expressed in isolated A. thaliana mesophyll protoplasts. Confocal laser scanning microscopy was used to monitor fluorescence.
Regulatory Mechanisms Revealed by Expression Analysis.
To further characterize THIC, its transcript abundance was analyzed by using RT-PCR. Consistent with a chloroplastic localization, a tissue expression analysis revealed that the transcript was predominantly found in green tissue, with barely detectable expression in roots or stems (Fig. 4A). This is also consistent with the involvement of thiamin diphosphate primarily in photosynthesis and carbohydrate metabolism. As the thiC mutant has a seedling lethal phenotype, we also analyzed the expression of THIC during early developmental stages. The THIC transcript was not detectable two days after germination, but was readily detectable at day five (Fig. 4B). This suggests that there may be no net thiamin biosynthesis in Arabidopsis during the first days of germination, which agrees with an earlier report on maize (29). Therefore, we assume that thiamin stored in the seed (most likely translocated from other parts of the plant) is used during the first stages of seedling development, after which expression of THIC is induced and homeostasis is restored. Indeed, the variable thiamin levels described above in the differentially supplemented thiC progeny corroborate this statement.
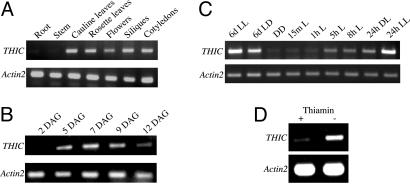
Fig. 4.
Expression analysis of THIC as assessed by RT-PCR. Transcript abundance in the indicated tissues (A) and days after germination (DAG) in the absence of thiamin (B) grown under long day conditions (LD, 100 μmol photons m−2 s−1 white light for 16-h/8-h dark). (C) Samples of whole seedlings were analyzed 6 days after growing in either continuous light (LL) or LD, or continuous dark (DD). In addition, samples of 6-day-old etiolated seedlings were analyzed at the indicated times after transfer to light, as well as after a 24-h LD cycle. (D) Seedlings grown in the presence and absence of 0.5 μM thiamin under LD. In all cases, Actin2 mRNA serves as the control.
As THIC is localized to the chloroplast, regulation of expression by light was examined. THIC transcript was detected in both dark and light grown seedlings (Fig. 4C). After transfer of etiolated seedlings to light, the level of the THIC transcript increases gradually over a 24-h period. The transcript level in seedlings grown in 24 h of light is substantially higher than that in seedlings exposed to a single long day cycle (16-h light/8-h dark). This is even more pronounced in 6-day-old seedlings subjected to the same conditions (Fig. 4C). The THIC protein could also be detected by Western blot analysis in extracts from the seedlings grown for 24 h or 6 days in the light (data not shown), i.e., when transcript abundance was high, indicating that protein and transcript levels may correlate.
In an attempt to further investigate the regulation of THIC, we probed a rather exciting phenomenon recently discovered: the concept of gene regulation by conserved regions of mRNA that bind specific metabolites, known as riboswitches (30, 31). Discovered in 2002, riboswitches were originally thought to occur only in the 5′ UTR of genes and moreover to be restricted to prokaryotes. This notion is no longer tenable with the very recent discovery of a thiamin pyrophosphate binding riboswitch in the 3′ UTR of the THIC gene of Arabidopsis (32, 33). As the gene had not been studied in any detail, it was not known at the time if it is actually involved in thiamin biosynthesis in planta. Clearly, with this study, this latter question has now been answered. Moreover, we observed that expression of the THIC gene is negatively regulated by thiamin itself (Fig. 4D). It is thought that this occurs due to instability of the mRNA conferred by thiamin diphosphate binding. As the thiamin riboswitch only responds to thiamin diphosphate but not thiamin (33), the down-regulation of THIC mRNA observed here implies that the externally provided thiamin is converted to thiamin diphosphate inside the cell leading to the conformational change inducing mRNA instability (33).
THIC is a Fe-S Cluster Protein.
THIC is thought to convert AIR into HMP-P (SI Scheme 1). This fascinating reaction has been described as the most complex unresolved rearrangement in primary metabolism (9) and has no chemical precedent. Remarkably, the reaction has been reconstituted in vitro employing the Escherichia coli protein (9). However, reconstitution necessitated the addition of a crude extract from the bacterium rendering it difficult to dissect factors necessary for functionality. We observed that all THIC homologs harbor the consensus motif C(X)2C(X)4C at the C terminus (Fig. 1B). However, the motif is not exclusive to THIC as illustrated by the fact that the same motif is displayed, in the E. coli genome, by at least 27 additional enzymes. This motif is similar to the C(X)3C(X)2C motif normally found at the N terminus of enzymes dependent on the 5′-deoxyadenosyl radical generated from S-adenosyl methionine (SAM) and a Fe-S cluster (34), albeit that of THIC is at the other end of the protein and has altered the order and number of residues between the cysteines. In this context, it is noteworthy that the capacity to reconstitute the THIC reaction as described by Lawhorn et al. (9) was reduced if SAM was omitted. As “radical SAM”-dependent enzymes require a Fe-S cluster for functionality, we were prompted to check for evidence of such a cofactor in THIC.
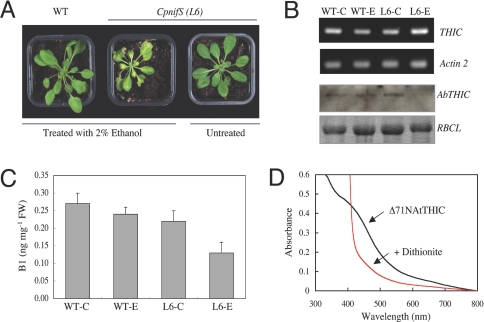
In plants, cysteine desulfurase (NifS) supplies the sulfur for Fe-S clusters. In Arabidopsis, two distinct NifS proteins exist, localized to the chloroplast and mitochondria (CpNifS and MtNifS, respectively). Recently, a study employing a CpnifS mutant has shown that the protein is essential for the maturation of plastidic Fe-S proteins (35). As THIC is localized to the chloroplast, we examined both THIC abundance and thiamin content in the CpnifS mutant. Silencing of the gene is achieved by using an ethanol-inducible RNA interference (RNAi) approach and results in a chlorotic phenotype (35) (Fig. 5A). Two independent transgenic lines (6 and 9, kindly provided by Elizabeth A. H. Pilon-Smits, Colorado State University, Ft. Collins) were used for analysis. Although line 6 (L6) showed a clear phenotype after ethanol treatment (Fig. 5A), induction of CpNifS silencing could not be consistently observed in line 9 in our hands; thus, only the results for L6 are shown. Concomitant with silencing of CpNifS, mutant plants exhibiting the phenotype have a clear reduction in THIC accumulation, whereas when silencing of CpNifS does not occur, the levels of THIC are similar to WT (Fig. 5B Lower). In contrast, a reduction of THIC mRNA was not observed in CpNifS silenced plants (Fig. 5B Upper), suggesting that the stability of the protein depends on the Fe-S cluster, and that apoTHIC does not accumulate. Moreover, the total thiamin content of CpnifS plants is reduced (Fig. 5C).
Fig. 5.
THIC is an iron–sulfur cluster protein. (A) RNA interference of CpnifS as evidenced by the chlorotic phenotype. Plants were grown on soil for 14 days, after which silencing was induced by spraying with 2% (vol/vol) ethanol every 4 days. The plants shown are 19 days after induction of CpnifS silencing. (B) (Upper) Transcript abundance of THIC in WT or CpnifS plants (as shown in A). WT-C, WT-E, L6-C, and L6-E refer to wild-type or CpnifS line 6 control or ethanol-treated plants, respectively. Actin2 mRNA serves as the control. (Lower) Western blot of the same samples probed with a THIC antibody in addition to the Ponceau-S stained membrane where RBCL indicates the large subunit of Rubisco and serves as a protein loading control. (C) Total thiamin content of the same samples described in B. (D) UV-visible spectrum of Arabidopsis THIC as isolated aerobically (black line), and 1 min after addition of 0.3 M sodium dithionite (red line).
To corroborate the above result, the Arabidopsis THIC protein was produced recombinantly in E. coli with a C-terminal hexa-histidine tag. Whereas the full-length protein was completely insoluble, a N-terminally truncated version lacking the first 71 amino acids was soluble (data not shown). Using Ni-NTA chromatography, the protein was purified to homogeneity in a very high yield (42 mg/liter of bacterial culture). Although the protein was isolated under aerobic conditions, it was intensely brown in color with an absorption maximum at 410 nm and a shoulder at 320 nm that strongly decreased after reduction with sodium dithionite (Fig. 5D). These features are clear indicators of a Fe-S cluster (36). A more detailed characterization of the Fe-S cluster will appear in a future report, and demonstration of THIC activity will await the availability of substrates.
Concluding Remarks.
Vitamin B1 was discovered in 1932, its structure was elucidated in 1936, and it was the first such compound to be recognized as an essential metabolic cofactor. Yet, surprisingly many facets of its metabolism still remain unresolved. Nowhere is this more apparent than in plants. In this study, we show that THIC plays an essential role in the synthesis of vitamin B1 and moreover is necessary for plant viability. The presence of a Fe-S cluster in THIC suggests that it must be coupled with a reductant to enable catalytic activity. In bacteria, this can be accomplished by the flavodoxin/flavodoxin reductase/NADPH system as for the non-mevalonate isoprenoid biosynthesis proteins, IspG and IspH (37). However, flavodoxin is absent from the plastids of phototrophic organisms, but reduction could be achieved by ferredoxin, which can supply the electrons either in the presence of light or in the dark via ferrredoxin-NADP+ reductase and NADPH (38). In plants, many chloroplastic enzymes are also activated and deactivated through oxidation/reduction reactions via the thioredoxin system. THIC has been found as a potential thioredoxin target protein in chloroplasts (39) and could influence the protein at several levels, including activity, oxidative regulation, and assembly or folding (40). Thus, the regulation of thiamin biosynthesis in plants may occur at several levels, i.e., riboswitch and redox. We attempted to complement an E. coli thiC mutant with the full-length Arabidopsis THIC and various N-terminally truncated versions; however, with the exception of the E. coli protein itself, none of these could restore thiamin prototrophy to the E. coli mutant, suggesting that a specific “plant/chloroplast factor” is necessary for functionality of the plant protein. Given the central role that both THIC and its riboswitch likely serve in modulating the concentration of thiamin, this protein or its mRNA may serve as a new target for drug discovery. Furthermore, previous efforts to engineer RNAs that perform as ligand-dependent molecular switches have proven that RNA has an enormous potential for molecular sensing.
Materials and Methods
Plant Material and Growth Conditions.
Unless indicated otherwise, Arabidopsis [wild-type (ecotype Col-0), SAIL_793_H10 (20) and CpNifS RNAi-inducible lines (35)] was either grown in soil or sterile culture under a 16-h-day (100 μmol of photons m−2 s−1 of light intensity) and 8-h-night cycle at 22°C/19°C, respectively, and in the presence or absence of thiamin as indicated. In the case of the light induction analysis, plates were exposed to 100 μmol of photons m−2 s−1 for 1 h after vernalisation and then transferred to continuous dark at 20°C for 6 days. The etiolated seedlings were then transferred to light (as above) and whole plantlets were collected after various times of exposure to light or a 24-h light/dark cycle (i.e., 16-h light/8-h dark). In all cases, samples of plant material collected were immediately frozen in liquid nitrogen and maintained at −80°C until analysis.
Vitamin B1 Determination.
Total vitamin B1 was measured by a microbiological assay employing S. cerevisiae strain thi4 (41) auxotrophic for vitamin B1 grown in thiamin deficient medium (ForMedium). Vitamin B1 was extracted from plant tissue as described for vitamin B6 in (42), with the exception that the extract was treated with alkaline phosphatase before quantification. The vitamin B1 content of plant extracts (either mature dry seeds or seedlings as indicated) was extrapolated from a standard curve of growth of cultures supplemented with known amounts of thiamin (0–9 ng).
Subcellular Localization.
The N-terminal 90 amino acids of THIC were amplified from a full-length cDNA clone obtained from the Arabidopsis Biological Resource Center (stock U16941) and fused to both the N- and C-terminus of YFP. Transient expression was carried out in Arabidopsis protoplasts essentially as described in ref. 42.
CpNifS Silencing.
Silencing of the chloroplast localized cysteine desulfurase (CpNifS) was carried out essentially as described in ref. 35. Briefly, RNA interference was induced in 14-day-old CpnifS RNAi inducible plants grown in soil by spraying every 4 days with a 2% (vol/vol) ethanol solution. Samples were taken for analysis 19 days after induction.
Supplementary Material
Acknowledgments
We dedicate this study to Prof. Meinhart H. Zenk (Donald Danforth Plant Science Center, St. Louis, MO) on the occasion of his 75th birthday. We thank Prof. Elizabeth A. H. Pilon-Smits for her kind gift of the CpNifS RNAi lines, Fabian Ramseyer and Sandro Steiner (ETH, Zurich) for assistance in the preliminary stages of this project, Dr. Hironori Niki at the National Institute of Genetics in Japan for supplying the E. coli thiC knockout mutant and the pCA24NEcThiC construct that were used in the mentioned complementation experiments, Dr. Dieter Rubli for photographic work, and Prof. Julia Fritz-Steuber and Po-Chi Lin at the University of Zürich, and Prof. Sam Zeeman at the ETH Zürich, for useful discussions. This work was supported by Swiss National Science Foundation Grant 3100A0-107975/1 (to N.A. and T.B.F.). D.A. acknowledges support from Novartis International AG. The Arabidopsis Biological Resource Center is greatly acknowledged for supplying line 793_H10 and stock U16941 (THIC cDNA).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709597104/DC1.
References
- 1.Pohl M, Sprenger GA, Müller M. Curr Opin Biotechnol. 2004;15:335–342. doi: 10.1016/j.copbio.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Machado CR, de Oliveira RL, Boiteux S, Praekelt UM, Meacock PA, Menck CF. Plant Mol Biol. 1996;31:585–593. doi: 10.1007/BF00042231. [DOI] [PubMed] [Google Scholar]
- 3.Ahn I-P, Kim S, Lee Y-H. Plant Physiol. 2005;138:1505–1515. doi: 10.1104/pp.104.058693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahn I-P, Kim S, Lee Y-H, Suh SC. Plant Physiol. 2007;143:838–848. doi: 10.1104/pp.106.092627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Settembre E, Begley TP, Ealick SE. Curr Opin Struct Biol. 2003;13:739–747. doi: 10.1016/j.sbi.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Begley TP, Downs DM, Ealick SE, McLafferty FW, Van Loon APGM, Taylor S, Campobasso N, Chiu H-J, Kinsland C, Reddick JJ, Xi J. Arch Microbiol. 1999;171:292–300. doi: 10.1007/s002030050713. [DOI] [PubMed] [Google Scholar]
- 7.Himmeldirk K, Sayer B, Spenser ID. J Am Chem Soc. 1998;120:3581–3589. [Google Scholar]
- 8.Estramareix B, David S. J Am Chem Soc. 1984;106:3857–3860. [Google Scholar]
- 9.Lawhorn BG, Mehl RA, Begley TP. Org Biomol Chem. 2004;2:2538–2546. doi: 10.1039/B405429F. [DOI] [PubMed] [Google Scholar]
- 10.Juillard J-H, Douce R. Proc Natl Acad Sci USA. 1991;88:2042–2045. doi: 10.1073/pnas.88.6.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Praekelt UM, Byrne KL, Meacock PA. Yeast. 1994;10:481–490. doi: 10.1002/yea.320100407. [DOI] [PubMed] [Google Scholar]
- 12.Chatterjee A, Jurgenson CT, Schroeder FC, Ealick SE, Begley TP. J Am Chem Soc. 2007;129:2914–2922. doi: 10.1021/ja067606t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tazuya K, Yamada K, Kumaoka H. Biochem Mol Biol Int. 1993;30:893–899. [PubMed] [Google Scholar]
- 14.Tazuya K, Azumi C, Yamada K, Kumaoka H. Biochem Mol Biol Int. 1994;33:769–774. [PubMed] [Google Scholar]
- 15.Tazuya K, Adachi Y, Masuda K, Yamada K, Kumaoka H. Biochim Biophys Acta. 1995;1244:113–116. doi: 10.1016/0304-4165(94)00205-c. [DOI] [PubMed] [Google Scholar]
- 16.Zeidler J, Sayer BG, Spenser ID. J Am Chem Soc. 2003;125:13094–13105. doi: 10.1021/ja030261j. [DOI] [PubMed] [Google Scholar]
- 17.Maundrell K. J Biol Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- 18.Schweingruber AM, Dlugonski J, Edenharter E, Schweingruber ME. Curr Genet. 1991;19:249–254. doi: 10.1007/BF00355050. [DOI] [PubMed] [Google Scholar]
- 19.Hohmann S, Meacock PA. Biochim Biophys Acta. 1998;1385:201–219. doi: 10.1016/s0167-4838(98)00069-7. [DOI] [PubMed] [Google Scholar]
- 20.Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al. Plant Cell. 2002;14:2985–2994. doi: 10.1105/tpc.004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sprenger GA, Schorken U, Wiegert T, Grolle S, de Graaf AA, Taylor SV, Begley TP, Bringer-Meyer S, Sahm H. Proc Natl Acad Sci USA. 1997;94:12857–12862. doi: 10.1073/pnas.94.24.12857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chia DW, Yoder TJ, Reiter W-D, Gibson SI. Planta. 2000;211:743–751. doi: 10.1007/s004250000345. [DOI] [PubMed] [Google Scholar]
- 23.Henkes S, Sonnewald U, Badur R, Flachmann R, Stitt M. Plant Cell. 2001;13:535–551. doi: 10.1105/tpc.13.3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chabregas SM, Luche DD, Farias LP, Ribeiro AF, van Sluys M-A, Menck CFM, Silva-filho MC. Plant Mol Biol. 2001;46:639–650. doi: 10.1023/a:1011628510711. [DOI] [PubMed] [Google Scholar]
- 25.Ajjawi I, Rodriguez Milla MA, Cushman J, Shintani DK. Plant Mol Biol. 2007;65:151–162. doi: 10.1007/s11103-007-9205-4. [DOI] [PubMed] [Google Scholar]
- 26.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 27.Bannai H, Tamada Y, Maruyama O, Nakai K, Miyano S. Bioinformatics. 2002;18:298–305. doi: 10.1093/bioinformatics/18.2.298. [DOI] [PubMed] [Google Scholar]
- 28.Peltier J.-B., Cai Y, Sun Q, Zabrouskov V, Giacomelli L, Rudella A, Ytterberg J, Rutschow H, van Wijk KJ. Mol Cell Proteomics. 2006;1:114–133. doi: 10.1074/mcp.M500180-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Belanger FC, Leustek T, Chu B, Kriz AL. Plant Mol Biol. 1995;29:809–821. doi: 10.1007/BF00041170. [DOI] [PubMed] [Google Scholar]
- 30.Winkler W, Nahvi A, Breaker RR. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 31.Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- 32.Sudarsan N, Barrick JE, Breaker RR. RNA. 2003;9:644–647. doi: 10.1261/rna.5090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thore S, Leibundgut M, Ban N. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- 34.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Hoewyk D, Abdel-Ghany SE, Cohu CM, Herbert SK, Kugrens P, Pilon M, Pilon-Smits EAH. Proc Natl Acad Sci USA. 2007;104:5686–5691. doi: 10.1073/pnas.0700774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Külzer R, Pils T, Kappl R, Hüttermann J, Knappe J. J Biol Chem. 1998;273:4897–4903. doi: 10.1074/jbc.273.9.4897. [DOI] [PubMed] [Google Scholar]
- 37.Rohdich F, Zepeck F, Adam P, Hecht S, Kaiser J, Laupitz R, Grawert T, Amslinger S, Eisenreich W, Bacher A, Arigoni D. Proc Natl Acad Sci USA. 2003;100:1586–1591. doi: 10.1073/pnas.0337742100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seemann M, Tse Sum Bui B, Wolff M, Miginiac-Maslow M, Rohmer M. FEBS lett. 2006;580:1547–1552. doi: 10.1016/j.febslet.2006.01.082. [DOI] [PubMed] [Google Scholar]
- 39.Balmer Y, Koller A, del Val G, Manieri W, Schürmann P, Buchanan BB. Proc Natl Acad Sci USA. 2003;100:370–375. doi: 10.1073/pnas.232703799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buchanan BB, Balmer Y. Annu Rev Plant Biol. 2005;56:187–220. doi: 10.1146/annurev.arplant.56.032604.144246. [DOI] [PubMed] [Google Scholar]
- 41.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 42.Tambasco-Studart M, Titiz O, Raschle T, Forster G, Amrhein N, Fitzpatrick TB. Proc Natl Acad Sci USA. 2005;102:13687–13692. doi: 10.1073/pnas.0506228102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.