Abstract
In the elongation cycle of translation, translocation is the process that advances the mRNA–tRNA moiety on the ribosome, to allow the next codon to move into the decoding center. New results obtained by cryoelectron microscopy, interpreted in the light of x-ray structures and kinetic data, allow us to develop a model of the molecular events during translocation.
Keywords: protein synthesis, ribosome, translation, EF-G, cryo-EM
Synthesis of proteins from their building blocks, the amino acids, is a fundamental process in the cells of all living organisms, be it animal, plant, or bacteria. The discovery that the macromolecular assembly that facilitates this process, the ribosome, is highly conserved in all essential parts has lent additional credence to the idea of the unity of all life at the molecular level.
The ribosome is a very large (2.4 MDa in eubacteria) ribonucleic-protein complex composed of two distinct subunits, the small subunit (30S) charged with the task of decoding the genetic message carried by the messenger RNA (mRNA), the large subunit (50S) to the catalysis of peptide bond formation. Instrumental for these fundamental processes is the interaction of the ribosome with transfer RNA (tRNA), a small L-shaped molecule that embodies in its various forms the association of each amino acid with a three-base “word” of the genetic code, the codon. Translation is based on the mutual recognition, by partial Watson–Crick pairing, between the codon on the mRNA and the anticodon of the tRNA carrying the corresponding amino acid. In facilitating tRNA selection, decoding, and the stepwise formation of the polypeptide, ribosomal RNA (rRNA) acts as both a structural framework and a catalyst.
Despite the success in the elucidation of ribosomal structure by x-ray crystallography, the detailed mechanism by which translation of mRNA code into peptide proceeds is still only scantly understood. One of the obstacles we face is that although the process is complex and dynamic, x-ray crystallography represents the molecule in a static form—packed in a crystal, moreover, whose very stability depends on intermolecular contacts that are largely nonphysiological. Of crucial importance for the understanding of the multistep translation process is the knowledge of how the ribosome interacts with its ligands, notably (apart from the most crucial ligands mRNA and tRNAs) the various factors catalyzing initiation, elongation, termination, and recycling. To date, with the exception of ribosomal complexes containing eubacterial release (RF1 or RF2) (1) or recycling (RRF) (2, 3) factors, there exists no x-ray structure of a factor–ribosome complex. The crystal structures of individual subunits complexed with initiation (4–6) and recycling (7) factors have also been solved; however, crystallographic data of elongation factors bound to the ribosome are currently still not available. Moreover, to date, despite many efforts, no atomic structure is available for a eukaryotic ribosome.
Increasingly, within the past decade, cryo-electron microscopy (cryo-EM) has filled this gap; in fact, the very first three-dimensional images of the ribosome (8, 9) were obtained by this technique well before the first x-ray structure was solved. In the mean time, cryo-EM has furnished quite detailed information on the interaction of the Escherichia coli ribosome with initiation factors (10), elongation factors (11–14), release factors (15–18), and ribosome recycling factor (19, 20). Some of the corresponding complexes have been visualized for the 80S eukaryotic ribosome (21–23).
As the initially low-resolution maps gave way to density maps in the subnanometer range, evidence of conformational changes both in the ribosome and its ligands was uncovered, and qualitative descriptions were increasingly replaced by quantitative measurements based on fitting and docking of x-ray structures. As a result, we can begin to piece together mechanistic models that explain kinetic, genetic, and other data collected over the five decades of ribosome research in structural terms.
The Elongation Cycle, and Translocation
In the course of protein synthesis, tRNA occupies successively the universally conserved A (aminoacyl), P (peptidyl), and E (exit) sites of the mRNA-programmed ribosome. The elongation cycle of translation is a repeating, three-step process catalyzed by the ribosome and two GTPases, EF-Tu and EF-G (eEF1A and eEF2 in eukaryotes). First, in decoding, EF-Tu delivers an aminoacylated-tRNA (aa-tRNA) molecule as part of the ternary complex of aa-tRNA·EF-Tu·GTP to the A/T site. The orientation with which tRNA enters the ribosome is not favorable for codon–anticodon interaction, and the anticodon stem-loop of the tRNA needs to be strongly kinked at the A/T site to allow the anticodon to interact with mRNA (24). If the aa-tRNA is cognate—i.e., when its three-base anticodon forms complementary base pairs with the codon in the messenger RNA—GTP is hydrolyzed, and the aa-tRNA, upon dissociation of EF-Tu·GDP, is accommodated in the ribosomal A site. The second step, the peptidyl-transferase reaction, is catalyzed by the rRNA of the large subunit and occurs immediately following the accommodation of the aa-tRNA. The transfer of the nascent peptide chain, from peptidyl-tRNA in the ribosomal P site to the accommodated aa-tRNA in the A site, results in the “pretranslocational” (PRE) state of the translating ribosome, with the deacylated tRNA in the P site and a peptidyl-tRNA in the A site.
The final step of the elongation cycle is translocation, which brings the ribosome into the “posttranslocational” (POST) state, with an empty A site, peptidyl-tRNA in the P site, and deacylated tRNA in the E site. Translocation is catalyzed by the binding of EF-G and subsequent GTP hydrolysis. The movement of the tRNA molecules is concomitant with the movement of the bound mRNA chain by the length of three bases, allowing the next codon of mRNA in the ribosomal A site to be presented for decoding.
Each step of the elongation cycle of translation, namely decoding, peptidyl-transfer, and translocation, presents an enigma on its own. Whereas the mechanism of peptidyl-transfer has received particular attention (see ref. 25), and at least part of the tRNA selection step of decoding has been unraveled in molecular detail (6), the molecular processes of EF-G-mediated translocation have been least understood until recently. Key findings include the discovery of the tRNA hybrid states (26) and the results of kinetic experiments that GTP hydrolysis precedes translocation (27).
Important cryo-EM findings relevant to translocation are the visualization of EF-G binding to the ribosome (11, 12, 14); the visualization of tRNA in the P/E hybrid state (13, 18, 23, 28); the discovery of the ratchet motion, a rotation of the small vs. large subunit upon binding of EF-G (29) or eEF2 (22) to the ribosome; and, most recently, the elucidation of the role of GTP hydrolysis in eEF2-induced translocation of the eukaryotic ribosome (23).
Hybrid-State tRNAs and Intersubunit “Ratchet” Motion
It was originally proposed in the 1960s that translocation of tRNAs might occur independently on the two ribosomal subunits during elongation (30, 31). Later, biochemical data such as the results from direct chemical footprinting led to the hybrid-state tRNA model, which describes an intermediate step in translocation (26). Here, the acceptor ends of the A- and P-site tRNAs move into the P and E sites of the large subunit, while the anticodon stem-loops of the tRNAs remain in the A and P sites of the small subunit. This configuration results in occupation by the tRNAs of the A/P and P/E hybrid states. Cryo-EM reconstructions indeed revealed structural evidence for a position of tRNA in the P/E hybrid state (11, 13, 18, 23); however, the A/P hybrid state has thus far eluded structural analysis using cryo-EM or x-ray crystallography.
The initial report of hybrid-state tRNAs by Moazed and Noller (26) suggested that tRNAs occupy the hybrid state immediately after peptide bond formation, but structures representing the PRE ribosome determined by cryo-EM (13) and x-ray crystallography (32–35) showed that in the absence of elongation factors, the tRNAs occupy the classic A/A, P/P, and E/E sites exclusively. Potential explanations for this discrepancy are that the tRNAs are sampling conformations between classic and hybrid states in the PRE ribosome (36), or that the buffer used in the various studies stabilizes one state of binding over the other (i.e., classic over hybrid) (28).
The P/E hybrid state tRNA in the cryo-EM reconstructions was visualized only when EF-G was bound to the ribosome. In addition to bringing the tRNA into the P/E hybrid state, the binding of the factor also induces a new conformation of the ribosome, where the small subunit is in a different orientation with respect to the large subunit, related to the normal conformation by a counterclockwise rotation (“ratchet motion”) (29, 37) [supporting information (SI) Fig. 6 and SI Movie 1]. Evidence for the existence of an intersubunit motion first came from small-angle neutron scatting experiments, but the precise form of this motion could not be deduced (38). It is likely that the ratchet motion of the ribosome even occurs in the absence of ribosomal factors and that it is coincident with the movement of tRNAs from the classic to the hybrid states, which facilitate translocation (39, 40). Hence, the view is emerging that, as suggested by the cryo-EM data, the binding of EF-G/eEF2 in the GTP form stabilizes the ratcheted conformation, and with it, the hybrid-state tRNAs (40, 41). The ratcheted ribosome with hybrid-state tRNAs, therefore, represents a distinct structural intermediate between the PRE and POST ribosomes. In solution, it is likely that the PRE ribosome oscillates between the ratcheted and nonratcheted states, with EF-G catalyzing translocation to the POST ribosome. The POST ribosome, with peptidyl-tRNA in the P-site, is locked in the nonratcheted state (13).
A convincing proof that the ratchet motion is instrumental for translocation is contained in a series of three recent studies. In one, the motion was inhibited by the placement of a crosslink between proteins of the two subunits facing each other in the normal conformation of the ribosome, and complete suppression of translocation was observed (42). Furthermore, two FRET studies, one bulk (40) and the other single-molecule,¶ have provided conclusive evidence that the motion inferred from the comparison of cryo-EM maps indeed occurs in a translating ribosome.
It is noteworthy, in this context, that the kinetic parameters of tRNA movement have been investigated in detail with the use of fluorescent labels (43, 44). In addition to supporting the existence of the A/P and P/E hybrid states, these data identify distinct hybrid-state intermediates. The movements of the two tRNAs bound to the A and P sites of the small subunit occur separately, such that A/A tRNA and P/E-hybrid tRNA may exist simultaneously in an intermediate ribosomal state. In the absence of elongation factors, single-molecule FRET measurements suggest that the tRNAs transition between the classic, A/P-P/E hybrid, and A/A-P/E intermediate states but spend the majority of time (≈60%) in the classic states (43).
Factor-Free Translocation
Some of the observations discussed above clearly show that the transition between two dramatically different conformations of the ribosome and, along with it, the transition between canonical and hybrid states of the tRNA occur spontaneously, in the absence of EF-G/eEF2. It is compelling to link the structural and dynamic evidence to the long-known observations of factor-free translocation.
Reports in the very early years of ribosome studies (45–47), when very little was known about its biochemistry and structure, indicated that the ribosome is capable of performing the synthesis of oligo-phenylalanine from a poly(U) template even in the absence of elongation factors and GTP. Subsequent work by Spirin's group (48–50) further indicated that the treatment of the ribosome with pCMB, or the removal of ribosome protein S12 (51), stimulates the rate of factor-free translocation significantly. Recent work from Rachel Green's laboratory (52) identified three target cysteine residues, in protein S12 (C104 and C27) and S13 (C87), whose modification by pCMB results in an elevated factor-free translocation rate, albeit with decreased accuracy (see Fig. 1).
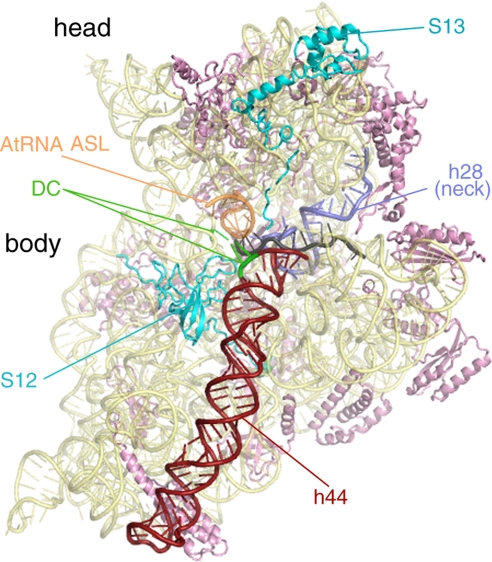
Fig. 1.
Atomic model of the 30S subunit of the pretranslocational ribosome from Thermus thermophilus (ref. 34; Protein Data Bank ID code 2J00). This intersubunit view of the 30S highlights several important features, which are color-coded and labeled. Ribosomal proteins S12 and S13 interact directly with the 50S subunit and affect ratcheting efficiency of the ribosome. The mRNA channel passes between the head and the body of the SSU (mRNA is colored gray). Upon EF-G binding, the top of helix 44 (h44) bends from the A site toward the P site. Because helix 44 is directly connected to helix 28 (h28 or the “neck”), the movement in h44 could provide torsional force on h28. GTP hydrolysis by EF-G decouples the tether between the A-site tRNA bound to the head and the decoding center (DC) in the body of the SSU. This event would release the head to rotate about its neck and relieve the torsional force in h28. The head rotation is likely to be important for movement of the mRNA and tRNA anticodon stem-loops on the SSU.
Interestingly, ribosomal protein S12 is the very protein identified in cryo-EM maps as the pivot of the ratchet-like motion (13) and was shown to interact with EF-G both by cross-linking studies (53, 54) and cryo-EM (13). A number of subsequent cryo-EM studies found that other translation factors, including IF2 (10), RF3 (18, 55), and RRF (20), all shown to interact with S12, induce the same type of intersubunit movement as EF-G (Fig. 2 and SI Fig. 7). In fact, the above-mentioned C104 residue of S12 is located exactly at the interface between the 30S subunit and these factors. These cryo-EM observations thus provide a link between factor-free and factor-promoted translocation, where S12 plays an apparent regulatory role in controlling the intersubunit motion of the ribosome (see below for a similar role of S13).
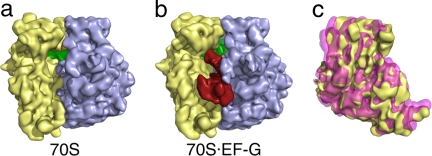
Fig. 2.
Binding of EF-G stabilizes the ratcheted conformation of the ribosome. (a) Cryo-EM reconstruction of the pretranslocational ribosome. The large subunit is colored blue, the small subunit is colored yellow, and the P-site tRNA is colored green. (b) Cryo-EM reconstruction of the EF-G·70S complex, showing EF-G in red. The large subunit of the two reconstructions (a and b) was aligned and the small subunit of each was superimposed (c). This view, from the solvent side of the small subunit, shows that binding of EF-G induces a counterclockwise rotation of the small subunit (pink) when compared with the small subunit of the pretranslocational ribosome (yellow).
One apparent benefit of introducing protein factors into the elongation cycle, in the course of evolution, is that it helps lower the energetic barriers of the various transition states, because factor-free translation occurs at a much lower rate (45, 48). Translocation itself has been shown to be an exergonic reaction (56, 57), but with a high free energy of activation [ΔG≠ ≈ 96 kJ/mol for factor-free translocation (58, 59)]. Kinetic studies indicated that the free energy of activation is lowered by ≈18 kJ/mol in the presence of EF-G, and by another 10 kJ/mol with the hydrolysis of GTP, which correspond to an ≈1,000-fold increase in translocation rate and another 50-fold increase, respectively (27, 60).
Architecture of the Ribosome: A Molecule Made to Rock
X-ray studies show that the ribosome possesses a unique architecture with built-in instabilities, among them architectural features conducive to the ratchet motion. As we have seen, these properties are apparently required for a whole range of translational steps, not just for translocation. Among the architectural features giving rise to instability are the following.
The ribosome is composed of two relatively loosely coupled subunits, a fact that prompted Spirin (31) quite early on to postulate the existence of intersubunit motion as a necessary part of the translational process.
The coupling is such that rotational movement around a central core of connections accompanied by torsional stress of these connections is prevalent among the modes shown by normal mode analysis (61, 62). Specifically, the central bridge B2a is formed by RNA–RNA minor groove interactions (32, 63, 64). According to Valle and coworkers (13), the rotational pivot axis passes through a point between bridges B3 and B5a on h44, just above h27 of 16S rRNA (see also ref. 61).
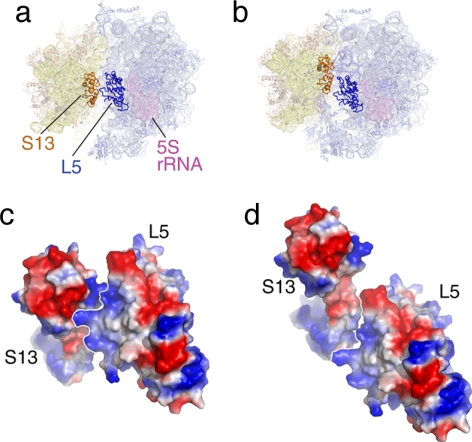
The peripheral bridges (i.e., bridges located far away from the rotational pivot axis) are quite flexible and involve at least one protein [e.g., B4, involving S15 and the 715 stem loop of 23S rRNA (65); for definition of bridge nomenclature, see refs. 9 and 19]. The bridge having to absorb the largest movement, B1b, is formed by two proteins, S13 and L5, which disengage and reengage in different constellations (13). It is interesting to look at this bridge in the two constellations, using the quasi-atomic models obtained by real-space refinement (Fig. 3): in the normal configuration of the ribosome (macrostate I), the electrostatic charges of regions of S13 and L5 facing each other are of opposite polarity, lending stability to the ribosome. In the ratcheted configuration (macrostate II), in contrast, the electrostatic charges of regions facing each other are of equal polarity, conveying instability to the proteins and a propensity for gliding along their interface. By extending the motion further, we would reach a constellation where opposite polarities would again face each other, creating another stabilizing interaction. The distance traveled between the “normal” position (macrostate I) and the postulated second antipolar juxtaposition would be 30 Å, which might act as a yardstick defining the maximum amplitude of “allowed” ratchet motion.
There are a large number of tertiary contacts, identified by Noller (66), that are slightly outside of the range of canonical A-minor interactions (67) and hence would allow the gliding of adjacent strands of rRNA without locking them into a defined position.
The rRNA possesses a number of kink-turns, a new element discovered in the x-ray structure of the 50S subunit of Haloarcula marismortui (68). Kink-turns are elements with built-in instability, allowing extensive hinge motions of some ribosomal components, among these the L1 stalk, the A-site finger (69), and the GTPase-associated center (GAC). These elements and their biological significance are detailed in the following.
The L1 stalk, which is implicated in the transport of the tRNA, pivots around a hinge region in h76 of 23S rRNA and has been observed in different positions both by cryo-EM (13, 21, 28) and x-ray crystallography (64, 70). The position of this stalk in an “inward” position, found in macrostate II, likely blocks the exit path of tRNA. Conversely, an “outward” position, found in macrostate I, leaves the exit path open. Movement from the “inward” to the “outward” positions of the L1 stalk likely coordinates translocation of tRNAs by allowing ejection of E-site tRNA from the ribosome (see ref. 13).
Fig. 3.
The bridge B1b, formed by S13 and L5, in the two ratchet-related conformations of the ribosome. (a and b) Positions of proteins S13 and L5 in the two states. (c and d) Surface charge representation of S13-L5 in the two states. In c (normal conformation), the charges at the interface are of opposite polarity, leading to stabilization. In d (ratcheted conformation), the charges are of equal polarity, leading to instability.
The A-site finger is a long rRNA helix (h38) reaching from the 23S rRNA into the intersubunit region where it forms a contact (termed B1a) with S13 of the small subunit. In Thermus thermophilus, h38 possesses a kink-turn (Kt-38). In E. coli, it has a similar region of instability. Because the tip of the A-site finger must follow the ratchet motion of the small subunit at a peripheral location, its built-in instability may serve to reduce the activation energy for the ratcheting. Shortening of the helix, presumably leading to a disruption of B1a, results in an acceleration of translocation activity, pointing to a role of the A-site finger as a control element maintaining the reading frame (71).
Finally, the GAC, formed by 23S rRNA h43 and h44, is flexibly linked to the main body of the large subunit by h42, which contains kink-turn 42. According to cryo-EM studies, the mobility of the GAC is essential for factor binding, forming part of an “induced fit” mechanism during the binding of EF-Tu (24, 72), EF-G (11), RF3 (18), and IF2 (10).
Hence, in conclusion, the ribosome's architecture is intrinsically unstable, conducive to making the ribosome alternate between two conformations. In the context of translocation, the first of these conformations (macrostate I) is required for initial factor binding, to initiate the first step of the translocation process, whereas the second (macrostate II) is required for staging the hybrid states, triggering GTP hydrolysis, and disengaging the firm grip that holds the mRNA–tRNA moiety in the decoding center, thereby initiating the second step of the translocation process.
Observed Conformations of EF-G
EF-G-catalyzed translocation requires EF-G to assume at least four conformations: (i) the GTP-bound, ribosome-free form; (ii) the GTP-bound conformation of EF-G on the ribosome immediately before GTP hydrolysis (as binding to the ribosome induces the GTPase of EF-G); (iii) the GDP-bound conformation, in which EF-G remains bound to the ribosome; and (iv) the GDP-bound conformation after dissociation of the factor from the ribosome. All four conformations have been observed structurally—by cryo-EM for the ribosome-bound conformations and by x-ray crystallography for the unbound conformations (SI Fig. 8). Such conformational alterations, specifically a hinge-like motion between the N-terminal domains (I, II, and G′) and C-terminal domains (III–V) of EF-G, undoubtedly play a role in the physiological function of the factor.
In the 1990s, the crystal structures of GDP-bound (73) and nucleotide-free (74) EF-G were solved. Both were shown to have similar conformations, with disordered regions in domain 3 and the switch 1 loop. The ribosome-free structure of EF-G·GTP has been solved in two conformations. First, the structure of a mutant EF-G·GDPNP revealed only slight differences when compared with the GDP-bound form (75). Noteworthy differences between these structures are seen in the P-loop, the visible regions of the switch 1 loop, the switch 2 loop, and a slight shift at the tip of domain IV, where a conserved histidine resides. Whereas the γ-phosphate of GDPNP is ordered in the structure, the switch 1 loop is not. This could be due to an artifact from crystal packing or because the crystals were seeded from mutant EF-G·GDP crystals. Recently, the x-ray structure of an EF-G homolog, EF-G-2, complexed with GTP was reported (76). This structure possesses a fully ordered switch 1 loop and a conformation that is distinct from the GDP-bound EF-G structure; specifically, a reorganization of domains III–V with respect to domains I, II, and G′. This rearrangement results in a 25-Å shift at the tip of domain IV when compared with the GDP-bound EF-G, a conformation of EF-G undoubtedly favorable for binding to the ribosome as evidenced by the increased affinity for EF-G·GTP over its GDP-bound counterpart (see ref. 77 and references therein).
Attempts to find a system in the bewildering variety of observed structures for EF-G (see, for instance, ref. 78) need to account for the fact that the factor is in different environments, facing different conformational constraints when placed in a crystal context versus bound to the ribosome. Conformational differences relevant for the functioning of the factor while performing its work on the ribosome are best inferred from structures of EF-G bound to the ribosome in an authentic state. Because of the lack of x-ray structures for such a complex, the existing quasi-atomic models of EF-G computed from cryo-EM maps are currently the only source of information. A comparison of the cryo-EM reconstructions of EF-G (11–14) bound to the ribosome with the crystal structures of the factors (assuming that these are close to the solution forms) revealed that EF-G undergoes gross conformational changes when it binds to the ribosome. These changes are again described as a hinge-like joint motion of domains III, IV, and V with respect to domains I, II, and G′ (11, 12) and are coincident with the ratchet motion (29). The hinge-like conformational change in EF-G induced by ribosome binding is in the same direction as that of the GDP- to GTP-bound conformations but with even greater magnitude (≈27 Å) (SI Fig. 8).
The conformations of ribosome-bound EF-G before and after GTP hydrolysis are quite likely similar to those recently elucidated for eEF2 by cryo-EM reconstructions of 80S–eEF2 complexes (23). The cryo-EM maps of modified eEF2 bound to the 80S ribosome in the GTP-state (using GDPNP) and GDP-state (GDP plus sordarin) revealed additional, smaller conformational changes in eEF2 upon GTP hydrolysis, which include a 6-Å shift of domain IV of eEF2 toward the decoding center (23). This conformational change is likely responsible for decoupling the decoding center from the mRNA–tRNA duplex in the A site, a prerequisite for translocation.
The GTP Hydrolysis Mechanism in EF-G-Catalyzed Translocation
GTPases, including EF-Tu and EF-G and their eukaryotic counterparts, are a family of proteins characterized by a GTP-binding domain, which is responsible for the binding and hydrolysis of GTP. GTPases function as molecular switches in several processes of cellular regulation by cycling between an active, GTP-bound state and an inactive, GDP-bound state (see ref. 79). The core, or G domain, is composed of three conserved features in the nucleotide-binding pocket: the phosphate-binding loop (P-loop) and the switch 1 and switch 2 motifs. The P-loop binds the nucleotide via the α- and β-phosphates, while the switch 1 and 2 loops coordinate the γ-phosphate of GTP.
Based on the similarities in structure, particularly of the switch 1 and 2 loops and their conserved ribosomal binding site near the GAC, it is likely that the mechanism of GTP hydrolysis is conserved among the elongation factors. In fact, this mechanism may be extended to all ribosomal binding GTPases involved in initiation, elongation, and termination because of the high conservation in factor structures and the universal GAC binding site on the ribosome. However, despite the availability of several crystal structures of these factors bound with various nucleotides and antibiotics, the mechanism of GTP hydrolysis of the factors is not well understood. This is due mainly to a lack of atomic models of the elongation factors bound to the ribosome. Such a model is obviously important for the understanding of the mechanism because GTP hydrolysis by elongation factors is accelerated on the ribosome by more than seven orders of magnitude (80).
GTPase activation of the smaller G proteins is catalyzed by the binding of a GTPase-activating protein (GAP), which induces conformational changes in, and thus alters the activity of, the G proteins (79). Because there is no GAP per se for the elongation factors, the GAC of the ribosome apparently assumes this role. Recent cryo-EM reconstructions of EF-G·70S (76) and eEF2·80S (23) complexes revealed that the switch 1 loop of EF-G/eEF2 is in fact ordered in the GTP-bound state. After GTP hydrolysis, the switch 1 loop of the ribosome-bound eEF2 becomes disordered, similar to that of the EF-Tu·GDP·aurodox crystal structure (81) and the structure of 70S·EF-Tu·Phe-tRNAPhe·kirromycin complex studied by cryo-EM, which also represents a post-GTPase state (24). This observation indicates a critical role of the switch 1 loop, as well as the existence of a similar mechanism of GTP hydrolysis in EF-Tu and EF-G/eEF2.
The sarcin–ricin loop (SRL) of the ribosome is a conserved tetraloop in the rRNA of the large subunit, also implicated in the activation of the GTPase activity. Cleavage of the SRL by the ribotoxin α-sarcin or ricin inhibits binding of elongation factors (82, 83). When free in solution, the SRL binds EF-G (84). Cryo-EM showed that the SRL is in close proximity of the switch 1 loop of GTP-bound EF-G and eEF2 (23, 76). Chemical protection assays revealed that the SRL is protected by EF-G in either the GTP-bound (GDPNP) or the GDP-bound (stabilized by fusidic acid) conformation (85). A conserved feature in the cryo-EM reconstructions of the ribosome-bound GTPases, IF2 (86) and eEF2 (23), is a shift in the GTP-binding domain of the factors toward the GAC of the ribosome in the transition from the GTP-bound states to GDP-bound states. This transition may alter the interactions between the SRL and the switch regions of the factors, leading to GTP hydrolysis and the disordering of the switch 1 loop.
Head Rotation of the Small Subunit: A Pivotal Step in Translocation
While the movement of the tRNAs on the large subunit (i.e., the formation of the hybrid-state tRNA) is facilitated by the binding of EF-G·GTP and the concomitant ratcheting of the ribosome, the second step of translocation, the movement of the tRNAs along with the mRNA on the small subunit by one codon, is more difficult to explain. Evidence has accumulated that supports the notion that GTP hydrolysis precedes translocation on the small subunit (44, 87). Toe-printing experiments of EF-G·70S complexes in the presence of either GTP or GDPNP demonstrated that mRNA movement relative to the small subunit occurs after GTP hydrolysis (88). Kinetic data suggest that GTP hydrolysis by EF-G leads to some kind of “unlocking” event that induces a conformational change of the ribosome, on such a scale that it limits the rate of translocation (89).
An explanation of this “unlocking” event was recently proposed on the basis of cryo-EM reconstructions of modified eEF2 bound to the 80S ribosome (23). Using the ADP moiety as a density marker, these authors showed that ADP-ribosylated eEF2 (ADPR-eEF2) undergoes conformational changes that are a direct result of GTP hydrolysis. These changes result in a 6-Å shift in the tip of domain IV of ADPR-eEF2, where a diphthamide residue resides in eEF2 (or a conserved histidine, in EF-G). Judged from the location of the tip, this movement likely severs the connection between the ribosomal decoding center in the body of the small subunit and the mRNA–tRNA duplex in the A site, bound to the head of the small subunit. Once this link is severed, the head is free to rotate around the neck of the small subunit, simultaneously translocating the mRNA–tRNA complex.
Such a head rotation of the subunit, which is independent of ratcheting, has been inferred from the difference in head positions observed both in cryo-EM reconstructions of 80S ribosomes (22, 23) and in x-ray structures of the 70S ribosome (64). Even earlier small-angle neutron scattering data predicted a head rotation of the small subunit to provide the mechanical act of translocation (90). Genetic mutations also suggest a necessary interaction between the head of the small subunit and the tRNAs during translocation, because mutations to a conserved adenine in the head-region of 16S rRNA (A1339 in E. coli) confer a >18-fold decrease in translation activity (91). Not surprisingly, A1339 interacts intimately with P-site tRNA in the PRE ribosome (32).
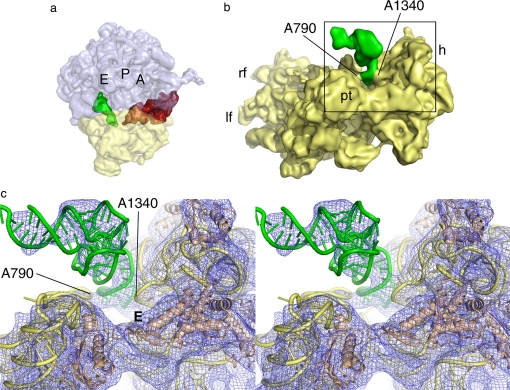
Analysis of the small subunit from the x-ray structures of vacant 70S ribosomes from E. coli revealed that residues G1338 to U1341 (E. coli numbering) in the head and residue A790 in the shoulder of the small subunit form a block between the P and E sites that opens or closes as a function of head movement (64). In the x-ray structure of the 70S ribosome in the PRE state (32) and the cryo-EM structure of the 80S ribosome in that same state (92), the distance between A1340 and A790 α-carbons is ≈18 Å and thus would impede movement of the tRNA stem from the P to E site. Binding of eEF2 opens this distance to >26 Å in the 80S ribosome (22, 23), which would accommodate the passing of the anticodon stem-loop of tRNA on the small subunit from the P to the E site (64). In the cryo-EM maps, however, eEF2 is bound and this block is “open,” but the P/E-site tRNA remains in the hybrid state, interacting strongly with the G1338–U1341 region of the head (Fig. 4). The strong interaction seen between the P/E-site tRNA and the head of the small subunit contrasts with tRNAs in the classic states, where the main interactions are with the body of the small subunit (32–34). This result suggests that in a translating ribosome, the head movement physically drags the anticodon stem-loop of P/E site tRNA toward the E site through a connection with the G1338–U1341 region in the head of the small subunit after ratcheting and GTP hydrolysis. At this point, the P/E hybrid tRNA is likely to be committed to the E site through a strong interaction formed with the L1 stalk of the large subunit after ratcheting of the ribosome (13, 21, 22), an interaction that may assist the transition phase. Disruptions of the interaction between the head region of the small subunit and the tRNAs must be induced by a combination of back-ratcheting of the small subunit and back-rotation of the head, both of which occur upon dissociation of eEF2/EF-G·GDP. We speculate that it is this event that allows P/E tRNA to finally pass through the gate between the head (G1338–U1341) and body (A790) of the small subunit to fully occupy the classic E/E state.
Fig. 4.
The P/E-site tRNA visualized in cryo-EM reconstructions. (a) A side view of the 80S·ADPR-eEF2·GDPNP complex (23), showing the CCA end of the tRNA occupying the E site of the large subunit, while the anticodon stem-loop occupies the P site of the small subunit. The A, P, and E sites of the ribosome are individually labeled. (b) The anticodon stem-loop of the P/E-site tRNA forms a strong interaction with the small subunit head, near the G1338-U1341 ridge, and a weaker interaction with the small subunit body, via the A790 loop of the 18S rRNA. The gap between A790 and A1340 separates the ribosomal P and E sites of the small subunit. (c) Stereo view of the interactions between the small subunit and P/E-site tRNA described in b. The head of the small subunit in this complex is rotated, increasing the distance between A790 in the body and A1339 in the head of the small subunit from ≈18 Å to ≈26 Å. This distance would allow passage of the anticodon stem-loop from the P to the E site; however, the tRNA remains bound in the hybrid state. Passage of the tRNA anticodon stem-loop from the P to the E site of the small subunit therefore must occur during back-ratcheting and back-rotation of the head of the small subunit, once EF-G/eEF2 dissociates from the ribosome. E denotes the E site of the small subunit.
Finally, it is likely that the binding of EF-G indirectly provides the driving force for head rotation in the small subunit: The binding of EF-G·GTP and ribosomal ratcheting are coupled events that are accompanied by a bending of h44 at a point 2/3 along its length, producing a shift of the decoding center by ≈8 Å toward the P site (93). This shift is required to fully accommodate domain IV of EF-G in the ribosomal A site. Directly connected with h44 is h28 in 16S rRNA, which makes up the neck of the small subunit (Fig. 1). It is plausible that EF-G-induced movement at the top of h44 will cause torsional stress in h28. Thus, the head of the small subunit would be primed to rotate as soon as the rigid link between the mRNA–tRNA duplex, still bound to the small subunit's head in the A site, and the decoding center in the body of the small subunit is removed upon GTP hydrolysis (see Fig. 1).
Synthesis of a Model for mRNA–tRNA Translocation
Recent evidence from structural, kinetic, and biochemical data, some of which has been reviewed above, point toward a two-step mechanism of translocation. The binding of EF-G·GTP (or EF-G·GDPNP) induces a first phase of translocation by stabilizing the tRNA molecules in the hybrid state. This occurs because the deacylation of the tRNA at the P site liberates the tRNA CCA, such that it can proceed to the E site on the 50S subunit, and allows the small subunit to ratchet forward while the mRNA–tRNA remains locked with the small subunit at its P site. At the same time, the vacating of the P site on the large subunit by the P/E-tRNA creates the precondition for the formation of the A/P hybrid state (43). Once this state is attained and stabilized by EF-G·GTP, the second step of translocation will start. This step is catalyzed by GTP hydrolysis on EF-G and entails the coupled movement of the mRNA and the anticodon loops of the tRNAs with respect to the small subunit.
The recent crystal structure of EF-G-2·GTP presents a conformation of EF-G that is ideally tailored to interacting with the PRE ribosome (76). Modeling of EF-G in this conformation with the PRE ribosome suggests that the binding of domains I, II, and G′ to the large subunit of the ribosome would position the tip of domain IV precisely at the codon–anticodon binding region of A-site tRNA (SI Fig. 8). Domain IV of EF-G, specifically the conserved histidine residue at its tip, comes into intimate contact with the codon–anticodon moiety formed by the mRNA and A-site tRNA. This interaction could stabilize the base pairing of the codon–anticodon of A-site tRNA during ratcheting of the ribosome, as suggested earlier (22). This reasoning would in fact explain the adverse effects on translocation when the conserved histidine in domain IV of EF-G is mutated (94). (For the eukaryotic case, the same scenario will hold, with EF-G replaced by eEF2 and the critical histidine residue by diphthamide.)
The movement of the decoding center in the direction of mRNA–tRNA translocation also stabilizes the A-site tRNA in the A/P hybrid state. Importantly, placement of tRNAs in the hybrid state is more favorable for translocation to occur as compared with tRNAs in the classic states (39, 43). Therefore, by stabilizing tRNAs in the hybrid states, EF-G binding directs the translocation process to go forward, which explains the catalytic potential of EF-G even in the absence of GTP hydrolysis (27).
Binding of EF-G and ratcheting of the ribosome are followed by GTP hydrolysis, causing a shift of domain IV, which in turn detaches the mRNA–tRNA complex from the decoding center (23). The mRNA–tRNA complex is temporarily freed from the body of the small subunit but likely maintains the strong interactions with the subunit's head. A head rotation in the small subunit is likely responsible for translocation of the mRNA chain by one codon but, because the tRNAs maintain their contacts with the head throughout the rotation, they technically remain in their hybrid states. Disruptions of these interactions must be induced by a combination of reverse-ratcheting of the small subunit and back-rotation of the head that both occur upon release of eEF2/EF-G. The final result is the POST-state ribosome with tRNAs in the canonical P and E sites (Fig. 5).
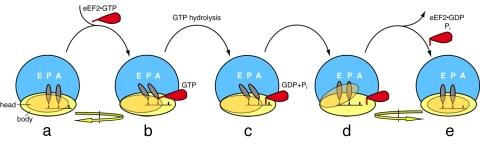
Fig. 5.
Proposed sequence of events during the translocation process. Schematic depicts the top view of the translating ribosome. EF-G interacts with the pretranslocational ribosome (a), stabilizing the A- and P-site tRNAs into A/P and P/E hybrid sites, respectively. Binding of EF-G is coincident with the ratcheting motion of the entire small subunit (i.e., head and body ratchet together) with respect to the small subunit (b). GTP hydrolysis causes the tip of domain IV of EF-G to sever the connection between the small subunit head–tRNA–mRNA complex and the small subunit body (c). Once the link to the body is severed, the head rotates, translocating the mRNA by one codon and the anticodon stem-loop ends of the hybrid site tRNAs to the P and E sites (d). Translocation is completed by back-rotation of the head and reverse-ratcheting of the entire small subunit as EF-G leaves the ribosome with the tRNAs in full P/P and E/E sites (e).
Conclusion
The ribosome, as we have seen, has intrinsic properties facilitating translocation. The most important one is an architecture that lends a specific kind of instability to the molecule and allows the ribosome to alternate between two distinct conformations separated by a small activation barrier. The change from one conformation to the other is along a pathway predicted as one of the highest-ranking modes by normal mode analysis of the x-ray structure (61, 62). This means, in other words, that the architecture has developed such that a minimum of energy is needed to go from the “normal” conformation to the “ratcheted” conformation, which is productive for the ultimate events of translocation.
This kind of parsimonious design is ubiquitous in all life forms. Human locomotion is an example on the macroscopic scale: during the act of walking, the legs are used as pendula, swinging with close to their innate resonance frequency and thus requiring a minimum of energy for sustenance of the movement. In the same way, energy supplied by thermal bombardment is channeled, by virtue of the ribosome's unique architecture, into a motion that is most productive in terms of translation.
Cryo-EM and the methods of flexible fitting by real-space refinement have allowed us to produce quasi-atomic models of the ribosome in both states (SI Fig. 6). An analysis of these models—particularly by watching an animation in which the two models alternate while the ribosome rotates (SI Movie 1)—makes it clear that the motion involves two interrelated events: (i) a conformational change in the intersubunit bridges and (ii) a change in the entire rRNA framework of each of the subunits, which resembles an elastic deformation, and accommodates the changes in the bridges.
The principal role of EF-G/eEF2, then, is in decreasing the energy barrier between the two ribosomal conformations. Specifically, the role of these factors is twofold: in bringing the ribosome into a conformation that is consistent with the binding of the tRNAs at the hybrid sites, and in unlocking the mRNA–tRNA moiety from the decoding center, thereby allowing the small subunit head movement to occur that is required for completion of translocation.
An “ur-ribosome” with the same basic architecture, and the same properties of instability, in a world in which elongation factors had not yet made their appearance, would already have functioned, albeit with very much decreased efficiency and accuracy. The very small rate of protein synthesis in these initial stages means that the evolution of the complex G-proteins (EF-G/eEF2, EF-Tu/eEF1A, IF2/eIF5B, and RF3/eRF3) tailored precisely to the ribosome's architecture could only proceed at a snail's pace. A similar line of reasoning holds for the impact of the addition of ribosomal proteins such as S12 and S13, which greatly improved the accuracy of factor-free translocation. Once these helper proteins had developed, however, the resultant much higher efficiency and accuracy of protein synthesis would lead to today's overflowing richness of life forms.
Supplementary Material
Acknowledgments
We thank M. Watters for assistance in generating figures and Y. Chen for help in creating SI Movie 1. This work was supported by the Howard Hughes Medical Institute and National Institutes of Health Grants R37 GM29169, R01 GM55440, and P41 RR01219.
Footnotes
The authors declare no conflict of interest.
See accompanying Profile on page 19668.
This article contains supporting information online at www.pnas.org/cgi/content/full/0708517104/DC1.
Cornish, P. V., Ermolenko, D., Noller, H. F., Ha, T., Ribosomes Meeting, June 3–8, 2007, Cape Cod, MA, p. 69 (abstr.).
References
- 1.Petry S, Brodersen DE, Murphy FVIV, Dunham CM, Selmer M, Tarry MJ, Kelley AC, Ramakrishnan V. Cell. 2005;123:1255–1266. doi: 10.1016/j.cell.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 2.Borovinskaya MA, Pai RD, Zhang W, Schuwirth BS, Holton JM, Hirokawa G, Kaji H, Kaji A, Cate JH. Nat Struct Mol Biol. 2007;14:727–732. doi: 10.1038/nsmb1271. [DOI] [PubMed] [Google Scholar]
- 3.Weixlbaumer A, Petry S, Dunham CM, Selmer M, Kelley AC, Ramakrishnan V. Nat Struct Mol Biol. 2007;14:733–737. doi: 10.1038/nsmb1282. [DOI] [PubMed] [Google Scholar]
- 4.Carter AP, Clemons WM, Jr, Brodersen DE, Morgan-Warren RJ, Hartsch T, Wimberly BT, Ramakrishnan V. Science. 2001;291:498–501. doi: 10.1126/science.1057766. [DOI] [PubMed] [Google Scholar]
- 5.Pioletti M, Schlunzen F, Harms J, Zarivach R, Gluhmann M, Avila H, Bashan A, Bartels H, Auerbach T, Jacobi C, et al. EMBO J. 2001;20:1829–1839. doi: 10.1093/emboj/20.8.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogle JM, Brodersen DE, Clemons WM, Jr, Tarry MJ, Carter AP, Ramakrishnan V. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 7.Wilson DN, Schluenzen F, Harms JM, Yoshida T, Ohkubo T, Albrecht R, Buerger J, Kobayashi Y, Fucini P. EMBO J. 2005;24:251–260. doi: 10.1038/sj.emboj.7600525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frank J, Penczek P, Grassucci R, Srivastava S. J Cell Biol. 1991;115:597–605. doi: 10.1083/jcb.115.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frank J, Zhu J, Penczek P, Li Y, Srivastava S, Verschoor A, Radermacher M, Grassucci R, Lata RK, Agrawal RK. Nature. 1995;376:441–444. doi: 10.1038/376441a0. [DOI] [PubMed] [Google Scholar]
- 10.Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J. Cell. 2005;121:703–712. doi: 10.1016/j.cell.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 11.Agrawal RK, Penczek P, Grassucci RA, Frank J. Proc Natl Acad Sci USA. 1998;95:6134–6138. doi: 10.1073/pnas.95.11.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agrawal RK, Heagle AB, Penczek P, Grassucci RA, Frank J. Nat Struct Biol. 1999;6:643–647. doi: 10.1038/10695. [DOI] [PubMed] [Google Scholar]
- 13.Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 14.Stark H, Rodnina MV, Wieden HJ, van Heel M, Wintermeyer W. Cell. 2000;100:301–309. doi: 10.1016/s0092-8674(00)80666-2. [DOI] [PubMed] [Google Scholar]
- 15.Rawat U, Gao H, Zavialov A, Gursky R, Ehrenberg M, Frank J. J Mol Biol. 2006;357:1144–1153. doi: 10.1016/j.jmb.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 16.Rawat UB, Zavialov AV, Sengupta J, Valle M, Grassucci RA, Linde J, Vestergaard B, Ehrenberg M, Frank J. Nature. 2003;421:87–90. doi: 10.1038/nature01224. [DOI] [PubMed] [Google Scholar]
- 17.Klaholz BP, Pape T, Zavialov AV, Myasnikov AG, Orlova EV, Vestergaard B, Ehrenberg M, van Heel M. Nature. 2003;421:90–94. doi: 10.1038/nature01225. [DOI] [PubMed] [Google Scholar]
- 18.Gao H, Zhou Z, Rawat U, Huang C, Bouakaz L, Wang C, Cheng Z, Liu Y, Zavialov A, Gursky R, et al. Cell. 2007;129:929–941. doi: 10.1016/j.cell.2007.03.050. [DOI] [PubMed] [Google Scholar]
- 19.Agrawal RK, Sharma MR, Kiel MC, Hirokawa G, Booth TM, Spahn CM, Grassucci RA, Kaji A, Frank J. Proc Natl Acad Sci USA. 2004;101:8900–8905. doi: 10.1073/pnas.0401904101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao N, Zavialov AV, Li W, Sengupta J, Valle M, Gursky RP, Ehrenberg M, Frank J. Mol Cell. 2005;18:663–674. doi: 10.1016/j.molcel.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 21.Gomez-Lorenzo MG, Spahn CM, Agrawal RK, Grassucci RA, Penczek P, Chakraburtty K, Ballesta JP, Lavandera JL, Garcia-Bustos JF, Frank J. EMBO J. 2000;19:2710–2718. doi: 10.1093/emboj/19.11.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jorgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J. EMBO J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor DJ, Nilsson J, Merrill AR, Andersen GR, Nissen P, Frank J. EMBO J. 2007;26:2421–2431. doi: 10.1038/sj.emboj.7601677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M, Frank J. Nat Struct Biol. 2003;10:899–906. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- 25.Barta A, Dorner S, Polacek N. Science. 2001;291:203. doi: 10.1126/science.291.5502.203a. [DOI] [PubMed] [Google Scholar]
- 26.Moazed D, Noller HF. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 27.Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 28.Agrawal RK, Penczek P, Grassucci RA, Burkhardt N, Nierhaus KH, Frank J. J Biol Chem. 1999;274:8723–8729. doi: 10.1074/jbc.274.13.8723. [DOI] [PubMed] [Google Scholar]
- 29.Frank J, Agrawal RK. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 30.Bretscher MS. Nature. 1968;218:675–677. doi: 10.1038/218675a0. [DOI] [PubMed] [Google Scholar]
- 31.Spirin AS. Curr Mod Biol. 1968;2:115–127. doi: 10.1016/0303-2647(68)90017-8. [DOI] [PubMed] [Google Scholar]
- 32.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 33.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 34.Selmer M, Dunham CM, Murphy FV, IV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 35.Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. Nature. 2006;444:391–394. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]
- 36.Sharma D, Southworth DR, Green R. RNA. 2004;10:102–113. doi: 10.1261/rna.5148704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frank J, Agrawal RK. Cold Spring Harbor Symp Quant Biol. 2001;66:67–75. doi: 10.1101/sqb.2001.66.67. [DOI] [PubMed] [Google Scholar]
- 38.Serdyuk IN, Spirin AS. In: Structure, Function, and Genetics of Ribosomes. Hardesty B, Kramer G, editors. New York: Springer; 1986. pp. 425–437. [Google Scholar]
- 39.Dorner S, Brunelle JL, Sharma D, Green R. Nat Struct Mol Biol. 2006;13:234–241. doi: 10.1038/nsmb1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ermolenko DN, Majumdar ZK, Hickerson RP, Spiegel PC, Clegg RM, Noller HF. J Mol Biol. 2007;370:530–540. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 41.Spiegel PC, Ermolenko DN, Noller HF. RNA. 2007;13:1473–1482. doi: 10.1261/rna.601507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horan LH, Noller HF. Proc Natl Acad Sci USA. 2007;104:4881–4885. doi: 10.1073/pnas.0700762104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munro JB, Altman RB, O'Connor N, Blanchard SC. Mol Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan D, Kirillov SV, Cooperman BS. Mol Cell. 2007;25:519–529. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pestka S. J Biol Chem. 1968;243:2810–2820. [PubMed] [Google Scholar]
- 46.Pestka S. J Biol Chem. 1969;244:1533–1539. [PubMed] [Google Scholar]
- 47.Gavrilova LP, Smolyaninov VV. Mol Biol. 1971;5:710–717. [PubMed] [Google Scholar]
- 48.Gavrilova LP, Spirin AS. FEBS Lett. 1971;17:324–326. doi: 10.1016/0014-5793(71)80177-1. [DOI] [PubMed] [Google Scholar]
- 49.Gavrilova LP, Spirin AS. FEBS Lett. 1974;39:13–16. doi: 10.1016/0014-5793(74)80005-0. [DOI] [PubMed] [Google Scholar]
- 50.Gavrilova LP, Kostiashkina OE, Koteliansky VE, Rutkevitch NM, Spirin AS. J Mol Biol. 1976;101:537–552. doi: 10.1016/0022-2836(76)90243-6. [DOI] [PubMed] [Google Scholar]
- 51.Gavrilova LP, Koteliansky VE, Spirin AS. FEBS Lett. 1974;45:324–328. doi: 10.1016/0014-5793(74)80872-0. [DOI] [PubMed] [Google Scholar]
- 52.Cukras AR, Southworth DR, Brunelle JL, Culver GM, Green R. Mol Cell. 2003;12:321–328. doi: 10.1016/s1097-2765(03)00275-2. [DOI] [PubMed] [Google Scholar]
- 53.Girshovich AS, Bochkareva ES, Ovchinnikov YA. J Mol Biol. 1981;151:229–243. doi: 10.1016/0022-2836(81)90513-1. [DOI] [PubMed] [Google Scholar]
- 54.Nechifor R, Wilson KS. J Mol Biol. 2007;368:1412–1425. doi: 10.1016/j.jmb.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 55.Klaholz BP, Myasnikov AG, Van Heel M. Nature. 2004;427:862–865. doi: 10.1038/nature02332. [DOI] [PubMed] [Google Scholar]
- 56.Makarov EM, Katunin VI, Odintsov VB, Semenkov Iu P, Kirillov SV. Mol Biol (Moscow) 1984;18:1342–1347. [PubMed] [Google Scholar]
- 57.Schilling-Bartetzko S, Bartetzko A, Nierhaus KH. J Biol Chem. 1992;267:4703–4712. [PubMed] [Google Scholar]
- 58.Wintermeyer W, Savelsbergh A, Semenkov YP, Katunin VI, Rodnina MV. Cold Spring Harbor Symp Quant Biol. 2001;66:449–458. doi: 10.1101/sqb.2001.66.449. [DOI] [PubMed] [Google Scholar]
- 59.Semenkov YP, Shapkina TG, Kirillov SV. Biochimie. 1992;74:411–417. doi: 10.1016/0300-9084(92)90080-x. [DOI] [PubMed] [Google Scholar]
- 60.Katunin VI, Savelsbergh A, Rodnina MV, Wintermeyer W. Biochemistry. 2002;41:12806–12812. doi: 10.1021/bi0264871. [DOI] [PubMed] [Google Scholar]
- 61.Tama F, Valle M, Frank J, Brooks CL., III Proc Natl Acad Sci USA. 2003;100:9319–9323. doi: 10.1073/pnas.1632476100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Y, Rader AJ, Bahar I, Jernigan RL. J Struct Biol. 2004;147:302–314. doi: 10.1016/j.jsb.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 63.Cate JH, Yusupov MM, Yusupova GZ, Earnest TN, Noller HF. Science. 1999;285:2095–2104. doi: 10.1126/science.285.5436.2095. [DOI] [PubMed] [Google Scholar]
- 64.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 65.Culver GM, Cate JH, Yusupova GZ, Yusupov MM, Noller HF. Science. 1999;285:2133–2136. doi: 10.1126/science.285.5436.2133. [DOI] [PubMed] [Google Scholar]
- 66.Noller HF. Science. 2005;309:1508–1514. doi: 10.1126/science.1111771. [DOI] [PubMed] [Google Scholar]
- 67.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. Proc Natl Acad Sci USA. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Klein DJ, Schmeing TM, Moore PB, Steitz TA. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Osswald M, Doring T, Brimacombe R. Nucleic Acids Res. 1995;23:4635–4641. doi: 10.1093/nar/23.22.4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 71.Komoda T, Sato NS, Phelps SS, Namba N, Joseph S, Suzuki T. J Biol Chem. 2006;281:32303–32309. doi: 10.1074/jbc.M607058200. [DOI] [PubMed] [Google Scholar]
- 72.Frank J, Sengupta J, Gao H, Li W, Valle M, Zavialov A, Ehrenberg M. FEBS Lett. 2005;579:959–962. doi: 10.1016/j.febslet.2004.10.105. [DOI] [PubMed] [Google Scholar]
- 73.Czworkowski J, Wang J, Steitz TA, Moore PB. EMBO J. 1994;13:3661–3668. doi: 10.1002/j.1460-2075.1994.tb06675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Aevarsson A, Brazhnikov E, Garber M, Zheltonosova J, Chirgadze Y, al-Karadaghi S, Svensson LA, Liljas A. EMBO J. 1994;13:3669–3677. doi: 10.1002/j.1460-2075.1994.tb06676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hansson S, Singh R, Gudkov AT, Liljas A, Logan DT. FEBS Lett. 2005;579:4492–4497. doi: 10.1016/j.febslet.2005.07.016. [DOI] [PubMed] [Google Scholar]
- 76.Connell SR, Takemoto C, Wilson DN, Wang H, Murayama K, Terada T, Shirouzu M, Rost M, Schuler M, Giesebrecht J, et al. Mol Cell. 2007;25:751–764. doi: 10.1016/j.molcel.2007.01.027. [DOI] [PubMed] [Google Scholar]
- 77.Czworkowski J, Moore PB. Biochemistry. 1997;36:10327–10334. doi: 10.1021/bi970610k. [DOI] [PubMed] [Google Scholar]
- 78.Liljas A. Structural Aspects of Protein Synthesis. Singapore: World Scientific; 2004. [Google Scholar]
- 79.Vetter IR, Wittinghofer A. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 80.Mohr D, Wintermeyer W, Rodnina MV. Biochemistry. 2002;41:12520–12528. doi: 10.1021/bi026301y. [DOI] [PubMed] [Google Scholar]
- 81.Vogeley L, Palm GJ, Mesters JR, Hilgenfeld R. J Biol Chem. 2001;276:17149–17155. doi: 10.1074/jbc.M100017200. [DOI] [PubMed] [Google Scholar]
- 82.Hausner TP, Atmadja J, Nierhaus KH. Biochimie. 1987;69:911–923. doi: 10.1016/0300-9084(87)90225-2. [DOI] [PubMed] [Google Scholar]
- 83.Wool IG, Gluck A, Endo Y. Trends Biochem Sci. 1992;17:266–269. doi: 10.1016/0968-0004(92)90407-z. [DOI] [PubMed] [Google Scholar]
- 84.Munishkin A, Wool IG. Proc Natl Acad Sci USA. 1997;94:12280–12284. doi: 10.1073/pnas.94.23.12280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Moazed D, Robertson JM, Noller HF. Nature. 1988;334:362–364. doi: 10.1038/334362a0. [DOI] [PubMed] [Google Scholar]
- 86.Myasnikov AG, Marzi S, Simonetti A, Giuliodori AM, Gualerzi CO, Yusupova G, Yusupov M, Klaholz BP. Nat Struct Mol Biol. 2005;12:1145–1149. doi: 10.1038/nsmb1012. [DOI] [PubMed] [Google Scholar]
- 87.Wilden B, Savelsbergh A, Rodnina MV, Wintermeyer W. Proc Natl Acad Sci USA. 2006;103:13670–13675. doi: 10.1073/pnas.0606099103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wilson KS, Nechifor R. J Mol Biol. 2004;337:15–30. doi: 10.1016/j.jmb.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 89.Savelsbergh A, Katunin VI, Mohr D, Peske F, Rodnina MV, Wintermeyer W. Mol Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- 90.Serdyuk I, Baranov V, Tsalkova T, Gulyamova D, Pavlov M, Spirin A, May R. Biochimie. 1992;74:299–306. doi: 10.1016/0300-9084(92)90107-p. [DOI] [PubMed] [Google Scholar]
- 91.Abdi NM, Fredrick K. RNA. 2005;11:1624–1632. doi: 10.1261/rna.2118105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- 93.VanLoock MS, Agrawal RK, Gabashvili IS, Qi L, Frank J, Harvey SC. J Mol Biol. 2000;304:507–515. doi: 10.1006/jmbi.2000.4213. [DOI] [PubMed] [Google Scholar]
- 94.Savelsbergh A, Matassova NB, Rodnina MV, Wintermeyer W. J Mol Biol. 2000;300:951–961. doi: 10.1006/jmbi.2000.3886. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.