Abstract
The ERK pathway responds to extracellular stimuli and oncogenes by modulating cellular processes, including transcription, adhesion, survival, and proliferation. ERK has diverse substrates that carry out these functions. The processes that are modulated are determined in part by the substrates that ERK phosphorylates. We demonstrate that PEA-15 (phosphoprotein enriched in astrocytes, 15 kDa) targets ERK to RSK2 and thereby enhances RSK2 activation. PEA-15 independently bound ERK and RSK2 and increased ERK association with RSK2 in a concentration-dependent manner. PEA-15 increased RSK2 activity and CREB-mediated transcription, and this process was regulated by phosphorylation of PEA-15. Finally, phorbol ester stimulation of PEA-15-null lymphocytes resulted in impaired RSK2 activation that was rescued by exogenous PEA-15 expression. Therefore, PEA-15 functions as a scaffold to enhance ERK activation of RSK2, and this activity is regulated by phosphorylation. Thus, PEA-15 can integrate signal transduction to provide a specific physiological outcome from activation of the multipotent ERK MAP kinase pathway.
Keywords: signal transduction, scaffold, transcription, lymphocytes
The ERK MAPK signaling cascade has been implicated in diverse physiological processes, including differentiation, proliferation, migration, and adhesion (1). This ability to impact several processes is partly because ERK recognizes an array of substrates both cytoplasmic and nuclear to elicit specific responses. However, how such a multipotent pathway achieves specificity of response is an ongoing problem in understanding ERK function. ERK interaction with scaffolding proteins, regulation of the duration of the ERK signal, and restriction of ERK to specialized subcellular compartments have all been suggested to contribute to this specificity (2–4). However, thus far, the mammalian scaffolding proteins for ERK such as KSR are known to act only upstream of ERK to enhance ERK activation without directing ERK to a particular substrate (5, 6). PEA-15 (phosphoprotein enriched in astrocytes, 15 kDa) is a small death effector domain protein that binds ERK and RSK2 and sequesters them in the cytoplasm (7, 8). RSK2 is a substrate of ERK and a kinase that can activate transcription, regulate apoptosis, and control cancer cell proliferation (9–11). Therefore, we investigated whether PEA-15 influences ERK activation of RSK2. We show that increasing expression of PEA-15 enhances ERK binding to RSK2, ERK phosphorylation of RSK2, and RSK2 activity. Moreover, genetic deletion of PEA-15 substantially abrogates ERK activation of RSK2 in astrocytes and lymphocytes. RSK2 activation can be rescued by ectopic expression of PEA-15 in these cells. Thus, PEA-15 functions as a scaffold to enhance ERK binding and phosphorylation of RSK2. It does not affect the phosphorylation of other ERK substrates, such as stathmin and Mnk1. This work provides a model to understand how scaffold proteins can integrate signal transduction and provide specificity in MAP kinase signaling.
Results
PEA-15 Forms a Complex with ERK and RSK2 and Enhances ERK Binding to RSK2.
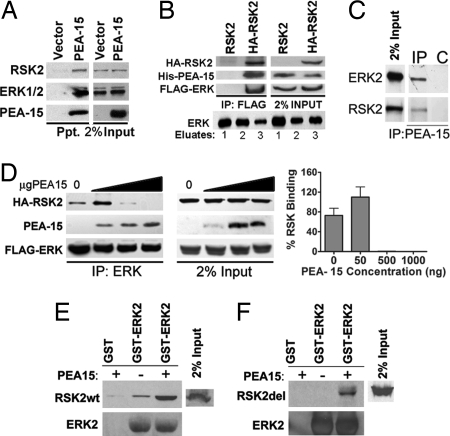
PEA-15 binds to ERK1/2 and RSK2 (7, 8, 12, 13). RSK2 is a substrate of ERK1/2 and, therefore, we investigated whether PEA-15 is present in a complex with both ERK and RSK2. We first tested whether PEA-15 coprecipitates ERK1/2 and RSK2 from cells. Indeed PEA-15 coprecipitated both ERK1/2 and RSK2 (Fig. 1A). We then determined whether these components existed in a single complex within the cell. We coexpressed all three proteins with fusion tags to allow for immunoprecipitation (FLAG-ERK2, HA-RSK2, and His-PEA-15). We first immunoprecipitated FLAG-ERK2 and then eluted the immune complexes by competing with a FLAG peptide. RSK2 bound to the eluted ERK was then immunoprecipitated. The resultant RSK2/ERK complex was examined for the presence of PEA-15. PEA-15 was present in the RSK2/ERK complex (Fig. 1B). Finally, we determined whether endogenous PEA-15 would coprecipitate ERK2 and RSK2. Immunoprecipitation of PEA-15 from astrocytes coprecipitated both RSK2 and ERK2 (Fig. 1C). Thus, PEA-15, ERK, and RSK2 can exist in a complex in the cell.
Fig. 1.
PEA-15 enhances ERK1/2 association with RSK2. (A) HeLa cells were transfected with His6-tagged PEA-15 or empty vector. Cell lysates were passed over a Probond Ni2+ column. Protein precipitates were immunoblotted to detect endogenous RSK2, ERK1/2, and His-PEA-15. (B) (Upper) Cos-7 cells were transfected with FLAG-ERK2, His-PEA-15, and HA-RSK2 or untagged RSK2 control. (Lower) Lysates were pooled and precipitated with FLAG-ERK2. FLAG-ERK was eluted off the column with FLAG peptide. HA-RSK2 was immunoprecipitated from the eluate. The immune complexes were immunoblotted for PEA-15, FLAG-ERK2, and HA-RSK2. (C) Endogenous PEA-15 was immunoprecipitated (IP) from primary astrocytes. Beads alone (C) were used as control. The precipitate was immunoblotted for ERK and RSK2. (D) Cos-7 cells were transfected with equal amounts of HA-RSK2 (1 μg) and FLAG-ERK2 (1 μg) and increasing amounts of His-PEA-15 plasmid (0, 50, 500, or 1,000 ng). FLAG-ERK2 was immunoprecipitated, and the precipitated complexes were immunoblotted to identify RSK2, ERK1/2, and PEA-15. Control immunoblots indicate 2% of the input protein levels. The graph of the percentage of RSK2 precipitate was derived from spot densitometry of three independent experiments. (E and F) Cos-7 cells were transfected with RSK2 (RSK2wt) or a deletion mutant of RSK2 (RSK2del) that does not bind ERK. Lysates of these cells were run over GST-ERK in the presence or absence of 300 μg of PEA-15 protein. Bound RSK2 was determined by immunoblot with anti-HA antibody. (E) Western blots showing full-length RSK2 bound to ERK with and without PEA-15. (F) Western blot showing the amount of RSK2 deletion mutant (Rsk2del) bound to GST-ERK2.
Because PEA-15 exists in a complex with both ERK and RSK2, we hypothesized that it may alter ERK activation of RSK2 by acting as a scaffold. Scaffolds enhance signaling by bringing together signaling components. Because a scaffold's effects can depend on the amount of protein expressed in relation to the kinase components (14), we used a range of expression levels to determine the effect of PEA-15 on ERK binding to RSK2. We first determined whether PEA-15 increased the interaction between ERK and RSK2. We immunoprecipitated FLAG-ERK2 from cells expressing HA-RSK2, FLAG-ERK2, and increasing amounts of His-PEA-15. In the absence of PEA-15, we observed an interaction between ERK and RSK2 supporting previous evidence that ERK and RSK2 exist in a complex within the cell (Fig. 1D). However, the presence of 50 ng of PEA-15 significantly enhanced the interaction between ERK and RSK2 (Fig. 1D). At higher concentrations of PEA-15, we observed a drop in the interaction, suggesting that the formation of the complex is inhibited by overexpression of PEA-15 (Fig. 1D). Thus, PEA-15 present in the complex in optimal amounts can increase the interaction between ERK and RSK2.
PEA-15 Binds to RSK2 Lacking the ERK-Binding Site and Forms a Complex with This Mutant and ERK.
If PEA-15 acts as a scaffold, it should bind ERK and RSK2 at sites distinct from those that mediate ERK binding to RSK2. Therefore, we examined the ability of purified PEA-15 to influence the binding of ERK to mutant RSK2 lacking the ERK-binding site (RSK2del), compared with binding to control WT RSK2. As expected, GST-ERK2 was able to bind to WT RSK2, and this binding was enhanced by PEA-15 (Fig. 1E). We also found that deletion of the ERK-binding domain of RSK2 abrogated GST-ERK2 binding (Fig. 1F) as previously reported (15). GST-ERK2 was able to pull down this RSK2-deletion mutant when purified PEA-15 was added (Fig. 1F). This finding indicates that PEA-15 binds to ERK and RSK2 through different sites concomitantly and can mediate the formation of an ERK/RSK2/PEA-15 complex in the absence of direct binding of ERK to RSK2.
PEA-15 Enhances ERK-Dependent RSK2 Phosphorylation and Kinase Activity.
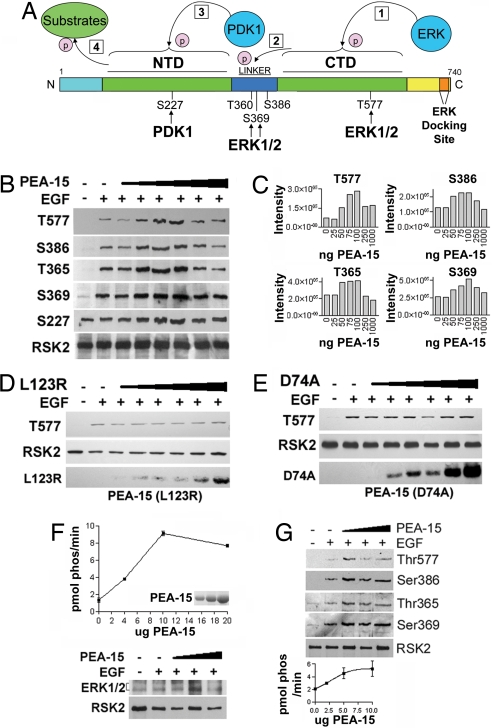
ERK phosphorylates RSK2 at multiple sites (Fig. 2A) (9). To determine whether the enhanced association of ERK with RSK2 affected phosphorylation of RSK2, we cotransfected increasing amounts of PEA-15 with ERK and RSK2. We observed increasing phosphorylation of RSK2 to a maximum point of 100 ng of PEA-15, at which the levels began to decrease (Fig. 2B). Quantitation of phosphorylation at ERK-dependent sites showed an increase in phosphorylation at all sites when an optimal amount of PEA-15 was expressed (Fig. 2C). However, as the amount of PEA-15 increased, phosphorylation returned to basal levels at some sites (particularly T365, S369, S386, and T577). Moreover, PEA-15 mutants that do not bind ERK (L123R and D74A) (16) do not enhance ERK phosphorylation of RSK2 at threonine 577 (Fig. 2 D and E).
Fig. 2.
PEA-15 enhances ERK phosphorylation of RSK2 and subsequent RSK2 kinase activity. (A) Activated ERK phosphorylates RSK2 on residues in the linker region and the C-terminal domain (CTD; step 1). The CTD-kinase domain autophosphorylates RSK2 at S386 (step 2). This step results in the recruitment of PDK1, which phosphorylates RSK2 at S227 in the N-terminal domain (NTD; step 3). The NTD kinase domain phosphorylates substrates (step 4). (B) Cos-7 cells were cotransfected with 250 ng of RSK2; 250 ng of ERK2; and 0, 25, 50, 75, 100, 250, or 500 ng of PEA-15. Cells were stimulated with 10 ng/ml EGF or were left unstimulated. Cell lysates were analyzed by Western blotting to detect RSK2 phosphorylation as indicated. (C) Phosphorylation was quantitated at four ERK-dependent sites, and net intensity was normalized for protein loading and is shown as relative to no EGF control. (D and E) Cos-7 cells were cotransfected with 250 ng of RSK2; 250 ng of ERK2; and 0, 25, 50, 75, 100, 250, or 500 ng of the indicated mutant PEA-15. PEA15(D74A) does not bind ERK, and PEA15(L123R) does not bind ERK or RSK2. Cells were stimulated with 10 ng/ml EGF or were left unstimulated. Cell lysates were analyzed by Western blotting to detect RSK2 phosphorylation. (F) Cos-7 cells were transfected with HA-RSK2. Cells were serum-starved and stimulated with EGF for 10 min. HA-RSK2 was immunoprecipitated, and the immune complexes were split into two aliquots. One aliquot was used in a kinase assay with 0, 4, 10, and 20 μg of GST-PEA-15 protein. The other aliquot was used to detect precipitation of RSK2 and the presence of ERK1/2 in the complexes. (Inset) Coomassie staining of PEA-15 was carried out to indicate protein loading. The values were normalized to background counts from no EGF control. The kinase assay is representative of two independent experiments done in triplicate. (G) Cos-7 cells were transfected and stimulated as described earlier. HA-RSK2-immunoprecipitated complexes were used to detect phosphorylation at ERK-dependent sites (T365, S369, S386, and T577) in the presence of 0, 2, 5, or 10 μg of PEA-15 protein. Total RSK2 levels are indicated. An aliquot was used to verify the kinase activity of RSK2 as described earlier.
Increased ERK-dependent RSK2 phosphorylation may result in increased RSK2 activity. To determine the effects of PEA-15 on kinase activity of RSK2, we carried out in vitro RSK2 kinase assays with increasing amounts of PEA-15. Purified, recombinant PEA-15 was added to the RSK2 kinase assay. The addition of 4 μg of PEA-15 protein to the reaction increased the kinase activity of the RSK2 (Fig. 2F). As the concentration of PEA-15 increased, the kinase activity of the enzyme also increased. Comparable amounts of ERK1/2 and RSK2 were present in the precipitates (Fig. 2F). We next determined whether ERK phosphorylation of RSK2 was enhanced in the kinase assay samples. Immunoprecipitated HA-RSK2 from EGF-stimulated or unstimulated samples was split into equal aliquots and subjected to the kinase assay as before. The reaction was either immunoblotted to detect ERK-dependent RSK2 phosphorylation or counted to determine S6 substrate phosphorylation. We found that the addition of PEA-15 enhanced RSK2 phosphorylation at the major ERK phosphorylation sites (T365, S369, S386, and T577) (Fig. 2G). Thus, PEA-15 may act as a scaffold for ERK and RSK2, thereby increasing ERK activation of RSK2.
PEA-15 Enhances RSK2-Dependent Transcription.
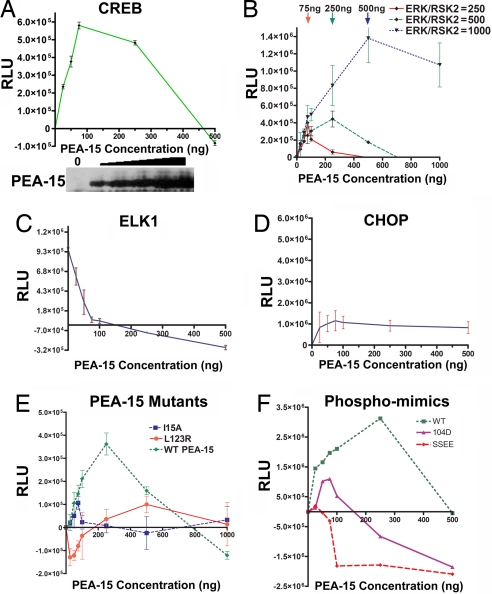
To determine whether there is a functional result of PEA-15 targeting of ERK to RSK2, we examined the effects of increasing amounts of PEA-15 on RSK2-dependent CREB transcription. We cotransfected constant amounts of ERK and RSK2 with increasing amounts of PEA-15 to study the effect on CREB-mediated transcription. As intracellular PEA-15 concentrations were increased, CREB-mediated signaling increased (Fig. 3A). However, at higher levels of PEA-15, CREB-mediated transcription was inhibited (Fig. 3A). Therefore, at specific cellular levels, PEA-15 enhances the interaction between ERK and RSK2 and facilitates the downstream activation of RSK2 by ERK.
Fig. 3.
Optimal levels of PEA-15 increase RSK2-dependent CREB transcription. (A) Cos-7 cells were transfected with 100 ng of ERK2; 100 ng of RSK2; and 0, 25, 50, 75, 250, or 500 ng of PEA-15 plasmid, along with the CREB luciferase reporter. Cells were stimulated with serum for 45 min, and luciferase activity was determined. (B) Cos-7 cells were transfected with 0, 25, 50, 75, 250, 500, or 1,000 ng of PEA-15 plasmid and 250, 500, and 1,000 ng of ERK2 and RSK2; along with the CREB luciferase reporter. Cells were stimulated with EGF (10 ng/ml) for 60 min, and luciferase activity was determined. (C) Cos-7 cells were transfected with PEA-15 as described earlier, along with 250 ng of ERK2 and RSK2 and the ELK-1 luciferase reporter. Cells were stimulated with EGF for 60 min, and luciferase activity was determined. (D) Cos-7 cells were transfected with PEA-15 as described earlier, along with 250 ng of ERK2 and RSK2 and the CHOP luciferase reporter. Cells were stimulated with EGF for 60 min, and luciferase activity was determined. (E) Cos-7 cells were transfected with WT PEA-15, the ERK/RSK2-binding defective mutant (L123R), or the nuclear-localized mutant (I15A) in increasing amounts as described earlier, along with 500 ng of ERK2 and RSK2 and the CREB luciferase reporter. Cells were stimulated with EGF for 60 min, and luciferase activity was determined. (F) Cos-7 cells were transfected with WT PEA-15, a phospho-Ser-104 mimic (S104D), or a mutant that mimics phosphorylation at both Ser-104 and Ser-116 (SSEE) in increasing amounts as in B, along with 500 ng of ERK2 and RSK2 and the CREB luciferase reporter. Cells were stimulated with EGF for 60 min, and luciferase activity was determined. In all experiments, equal amounts of protein were used to obtain absolute readings. These values were normalized to that of 0 PEA-15. Each experiment was done in triplicate and is representative of at least three independent experiments. Error bars represent the SEM of triplicate values.
If PEA-15 works as a scaffold, the amount of PEA-15 required for peak enhancement of ERK activation of RSK2 will increase as the levels of ERK and RSK2 increase in a cell (14). Moreover, the point at which PEA-15 begins to act as a dominant negative (when it is presumed to bind only ERK or RSK2 individually and, thus, inhibit ERK access to RSK2) (Fig. 1D) also would increase. We found that as we express more ERK and RSK2 in a cell, more PEA-15 is required to reach maximum enhancement of ERK activation of RSK2 (Fig. 3B). The amount of PEA-15 required to act as a dominant negative also is greater, and the level of CREB-mediated transcription is higher. In contrast, PEA-15 blocked ERK activation of ELK1 (Fig. 3C), whereas it had little effect on CHOP transcription, which is not directly targeted by ERK or RSK2 (Fig. 3D). The blockade of ERK activation of ELK1 likely results from PEA-15 retention of ERK in the cytoplasm, thereby preventing ERK access to ELK1 in the nucleus (7).
We also tested whether mutants of PEA-15 that were localized to the nucleus (PEA15-I15A) (7) or unable to bind ERK and RSK2 (PEA15-L123R) (8, 16) could enhance activation of CREB transcription. Neither ERK-binding-deficient PEA-15 affected RSK2 activation comparably to WT PEA-15, although they had moderate effects (Fig. 3E). Indeed, the nuclear-localized mutant may be enhancing ERK activation of RSK2 in the nucleus. This finding suggests that PEA-15 must be capable of binding ERK and have access to the cytoplasm (where ERK binds RSK2) to substantially enhance RSK2 activation of CREB. Therefore, PEA-15 behaves as predicted for a scaffold to direct ERK activation of RSK2.
PEA-15 is phosphorylated at Ser-104 and Ser-116 by PKC and CamKII or AKT, respectively (17). Phosphorylation of these serines regulates PEA-15 binding to ERK and FADD (18). Therefore, we investigated whether phosphorylation of PEA-15 at these serines alters its ability to enhance ERK activation of RSK2. We expressed increasing amounts of PEA-15 mutants that contain aspartic acid or glutamic acid substitutions at these serines and examined CREB transcription. We found that PEA-15-containing mutations that mimic phosphorylation at either serine 104 or both serines (19) failed to enhance RSK2 activation of CREB (Fig. 3F). Indeed, these mutants were effective at blocking CREB activation even at low concentrations. This finding may result from sequestering ERK or RSK2 in unproductive complexes. Therefore, the scaffold function of PEA-15 may be regulated by phosphorylation.
PEA-15 Is Required for Normal RSK2 Activation in Vivo.
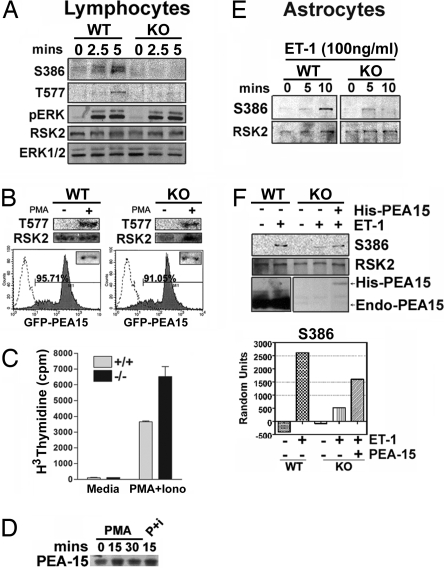
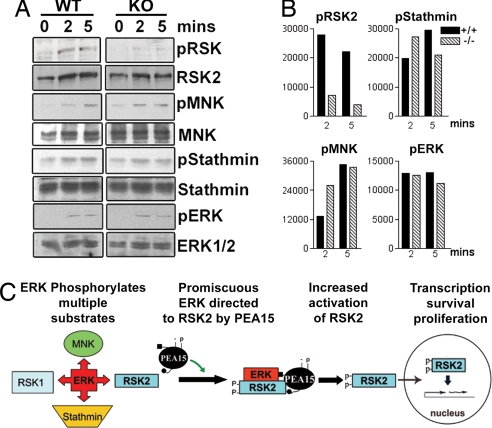
We next investigated whether genetic deletion of PEA-15 alters RSK2 activity in vivo by using primary cells from WT and PEA-15 KO mice. PEA-15 is expressed in lymphocytes (data not shown), and previous work has shown that PMA activates RSK2 in lymphocytes (20). We examined whether PEA-15-null lymphocytes had deficient RSK2 activation. Lymphocytes from WT and PEA-15 KO mice were stimulated with PMA. In WT lymphocytes, RSK2 activation reached a peak at 5 min (Fig. 4A, WT). In PEA-15 KO lymphocytes, RSK2 activation was diminished in response to PMA, although activation of ERK remained unaffected (Fig. 4A, KO). To determine whether the lack of PEA-15 was responsible for the decrease in RSK2 activation in these cells, we expressed exogenous PEA-15 in null lymphocytes by using a lentiviral vector-expressing GFP-tagged PEA-15. Reexpression of PEA-15 in the PEA-15 KO lymphocytes rescued the activation of RSK2 (Fig. 4B, KO). A primary result of the PMA activation of lymphocytes is proliferation. We examined whether proliferation was altered in the PEA-15 KO cells where RSK2 activity is diminished. We found that, indeed, PEA-15 KO cells proliferated significantly more than control WT littermate cells (Fig. 4C). Finally, PEA-15 expression levels in PMA stimulated lymphocytes were similar (Fig. 4D).
Fig. 4.
RSK2 activation is impaired in PEA-15-null lymphocytes and astrocytes. (A) Equal numbers of lymphocytes were stimulated with 50 ng/ml PMA for 0, 2.5, or 5 min and lysed. Equal amounts of cellular protein were immunoblotted to detect pRSK2 (S386, T577), pERK1/2, total RSK2, and total ERK1/2. (B) (Upper) PEA-15 WT and KO lymphocytes were infected with lentiviral GFP-PEA-15. The cells were then treated with PMA for 5 min and blotted for phosphorylated RSK2 as before. (Lower) GFP-PEA15 and GFP expression were verified by flow cytometry. (C) Lymphocytes isolated from WT and KO mice were treated with PMA and ionomycin for 48 h, followed by incubation in [3H]thymidine for 16–20 h. Proliferation was assessed as thymidine incorporation. The experiments were performed in triplicate, and the error bars represent the SEM in one representative experiment. (D) Lymphocytes were treated for 0, 15, or 30 min with PMA or PMA plus ionomycin (P+I) as indicated. Cells were lysed and analyzed by immunoblot for PEA-15 expression levels. (E) Astrocytes cultured from WT or PEA-15-null mice were stimulated with 100 ng/ml endothelin-1 for the indicated times. Cells were lysed, and equal amounts of protein were analyzed for pRSK2 (S386) and total RSK2. (F) Cultured astrocytes were transfected with 750 ng of PEA-15. Cells were stimulated with ET-1 for 10 min. Cell lysates were immunoblotted to detect pRSK2 (S386), total RSK2, and PEA-15. Spot densitometry is shown and is corrected for the amount of protein on the blot.
To determine whether PEA-15 is required for full activation of RSK2 in other cells types, we examined RSK2 activation in astrocytes. Both RSK2 and PEA-15 are highly expressed in astrocytes, and RSK2 is activated in astrocytes by endothelin 1 (ET-1) (21). We found that RSK2 activation was reduced in PEA-15-null astrocytes (Fig. 4E) and that we could partially rescue this outcome by exogenously expressing PEA-15 (Fig. 4F). Although the rescue is not complete, this finding may be explained by the low expression level of exogenous PEA-15, compared with the endogenous PEA-15 levels in WT astrocytes (Fig. 4F). These results suggest that PEA-15 is a scaffold that enhances ERK activation of RSK2 in vivo.
PEA-15 Directs ERK Signaling Specifically to RSK2.
Finally, we investigated whether PEA-15 expression affected the activation of other ERK substrates. ERK phosphorylation of MNK and stathmin was determined by using phosphospecific antibodies in PMA-stimulated PEA-15 KO lymphocytes. Phosphorylation of MNK and stathmin was not reduced in the PEA-15-null cells (Fig. 5). Therefore, PEA-15 does not enhance ERK phosphorylation of its substrates in general, but rather appears to specifically enhance ERK activation of RSK2.
Fig. 5.
PEA-15 does not enhance ERK phosphorylation of all substrates. Equal numbers of lymphocytes from WT and PEA-15-null mice were stimulated with PMA for the indicated times. (A) Lysates were immunoblotted to detect phosphorylated RSK2 (S386), MNK, Stathmin, and ERK. Total RSK2, MNK1, Stathmin, and ERK1/2 levels were detected as controls for protein loading. (B) Densities of bands for phosphorylated RSK2, MNK, Stathmin, and ERK1/2 were quantitated, and the values are shown relative to controls without PMA. (C) PEA-15 is a regulated scaffold for ERK activation of RSK2. ERK recognizes many substrates. In cells expressing PEA-15, a fraction of cellular ERK is targeted specifically to RSK2. PEA-15 enhances the association of ERK with RSK2, resulting in increased phosphorylation and activation of RSK2. The scaffold complex is regulated by kinases such as PKC through phosphorylation of PEA-15.
Discussion
Astrocytes and lymphocytes express high levels of PEA-15 protein. We propose that, in these cells and others expressing PEA-15, ERK activation would lead to significant activation of RSK2 as a result of PEA-15 targeting a portion of the activated ERK to RSK2 (Fig. 5C). Furthermore, phosphorylation of these proteins might affect the formation of the PEA15/ERK/RSK2 complex. Here we found that PEA-15 mutants that mimic phosphorylation at serine 104 were unable to enhance RSK2 activity. PKC and AKT have been reported to phosphorylate PEA-15 at serine 104 (17, 22, 23). Thus, activation of these kinases may regulate PEA-15 binding to ERK and RSK2 and thereby promote or inhibit their productive interaction. In this way, PEA-15 may provide a point of integration of these pathways with the ERK MAP kinase pathway.
We previously reported that PEA-15 binds RSK2 and that overexpression of PEA-15 blocks RSK2 activation and localizes RSK2 to the cytoplasm (8). We now demonstrate that, at lower PEA-15 expression levels, there is increased RSK2 activity by all measures and PEA-15 acts as a scaffold. Overexpression of the MEK/ERK scaffold, KSR, similarly impairs MEK activation of ERK (6, 24–26). KSR does so by combinatorial inhibition, where at high protein concentrations it binds MEK and ERK independently and thus separates them, whereas at low concentrations it brings them together (14, 27). PEA-15 may act similarly for ERK and RSK2. In agreement with this hypothesis, high expression levels of PEA-15 inhibit ERK binding to RSK2 (Fig. 1D) and subsequent RSK2 activation of CREB (Fig. 3). However, to fully test this finding requires further determination of the protein levels and binding constants involved. Alternatively, the effects of high PEA-15 levels on transcription may be the result of PEA-15 retention of active RSK2 in the cytoplasm (8). However, this finding does not account for the inhibition of ERK binding to RSK2 at high PEA-15 levels. Further work is needed to determine whether PEA-15 can influence endogenous ERK signaling by combinatorial inhibition.
The involvement of MAPKs in many physiological processes makes it important to understand how extracellular signals are interpreted by these kinases to generate a unique response. Signal duration, signal strength, and substrate expression have all been proposed as methods by which ERK activity could be biased to one physiological outcome versus another. Our work suggests an additional mechanism by which ERK signaling is targeted to a subset of substrates and thereby leads to a specific physiological outcome. The presence of scaffold molecules for other substrates of ERK would enable the cell to obtain a specific physiological response from the activation of a multipotent pathway. Therefore, this work may have broad implications for determining the targeting of MAP kinases in general and the ERK MAP kinase in particular.
Materials and Methods
Reagents and Cell Culture.
Cos-7 and HeLa cells were cultured in DMEM containing 10% FBS and antibiotics. All transfections were carried out by using Lipofectamine Plus (for Cos-7 cells and astrocytes; Invitrogen) or Polyfect (for HeLa cells; Qiagen) according to the manufacturers' protocols. Transfection efficiency for Cos-7 and HeLa cells was >80%, and increasing plasmid concentration led to increased protein expression in individual cells as determined by flow cytometry on GFP-transfected cells. FLAG-ERK2 was a gift from M. Weber (University of Virginia, Charlottesville, VA). pMT2-HA RSK2 was a gift from C. Bjorbaek (Harvard Medical School, Boston, MA). Generation of pcDNA3His-PEA-15 was described previously (8). Antibodies to PEA-15, phosphostathmin, and stathmin were a gift from H. Chneiweiss (College de France, Paris, France). Antibodies to RSK2, ERK1/2, MNK, and phospho-ERK were from Santa Cruz Biotechnology. Antibodies to phospho-RSK (for Ser-386 and Thr-577), phospho-MNK, and the HA tag were from Cell Signaling Technologies, and phospho-RSK Thr-365 and Ser-369 were from Upstate Biotechnology. Anti-mouse Lamin B2 antibodies were from Zymed Laboratories. Antibody to tubulin was a gift from S.-C. Hsu (Rutgers University).
Primary Cell Culture and Treatments.
The PEA-15-null mouse was obtained from H. Chneiweiss (28). Total lymphocytes were isolated from WT and PEA-15-null mice by crushing the thymus in serum-free RPMI medium (Invitrogen). For RSK2 activation, 5 × 106 cells were stimulated with 50 ng/ml PMA (Sigma–Aldrich) for the indicated times. The reaction was arrested on ice, and the cells were pelleted at 1,190 × g. The cell pellets were washed with cold PBS and lysed in 100 μl of sample buffer. Equal amounts of protein were separated for immunoblotting.
Immunoprecipitations.
Cos-7 cells were transfected with 1 μg of HA-RSK2, 1 μg of FLAG-ERK2, and 0, 0.05, 0.5, or 1 μg of His-PEA-15. The amount of DNA was equilibrated to 4 μg by using an empty vector. Cells were lysed in Nonidet P-40 buffer [20 mM Tris (pH 8), 137 mM NaCl, 10% glycerol, and 1% Nonidet P-40] supplemented with protease inhibitors and sodium orthovanadate. Lysates were incubated with 0.4 μg of 12CA5 antibody or anti-FLAG antibody (Sigma–Aldrich). The immune complexes were precipitated with 50 μl of protein-A Sepharose for 1 h at 4°C. The precipitates were washed with Nonidet P-40 lysis buffer supplemented with 1 M NaCl. Immune complexes were resuspended in 50 μl of sample buffer and immunoblotted for FLAG, HA, or PEA-15.
To identify PEA-15/RSK2/ERK complexes, immunoprecipitations were carried out as described earlier to precipitate FLAG ERK2 (anti-FLAG). The immune complexes were washed four times with lysis buffer and then subjected to competitive elution by using a FLAG peptide (Sigma–Aldrich) according to the manufacturer's instructions. The eluates were pooled, and HA-RSK2 was immunoprecipitated from this eluate by using an anti-HA antibody (Santa Cruz Biotechnology). The complexes were washed one time with lysis buffer supplemented with 1 M NaCl and then three times with lysis buffer without NaCl. The immune complexes were immunoblotted to detect ERK, HA-RSK2, and PEA-15.
His-Tagged Protein Precipitations.
His6-tagged PEA-15 or control empty vector was transfected into HeLa cells. His-PEA-15 was precipitated by using a Ni2+ column as described (8). The precipitated protein complexes were washed two times in lysis buffer containing 20 mM imidazole and then another two times in lysis buffer without imidazole. Control precipitations were carried out simultaneously by using the same procedure. Precipitates were immunoblotted for PEA-15, endogenous RSK2, and ERK1/2.
In Vitro Kinase Assays.
Cos-7 cells were transfected with pMT2 HA-RSK2. The cells were serum-starved for 20 h before stimulation with 10 ng/ml EGF for 10 min. Cells were washed in PBS and lysed in cold lysis buffer [20 mM Tris (pH 8), 137 mM NaCl, 10% glycerol, and 1% Nonidet P-40]. Cell debris was discarded after centrifuging at 14,576 × g for 20 min at 4°C. HA-RSK2 was immunoprecipitated from the supernatant with HA-antibody 12CA5. Immune complexes were precipitated with 50 μl of protein A-Sepharose (Amersham Biosciences) beads for 1 h at 4°C. The precipitates were washed in lysis buffer supplemented with 1 M NaCl and then resuspended in assay buffer [20 mM MOPS (pH 7.2), 25 mM β-glycerophosphate, 5 mM EGTA, 1 mM DTT] and split into two equal aliquots. One aliquot was used in a32P-radiolabled S6-peptide kinase assay according to the manufacturer's protocol (Upstate Biotechnology), in which 0, 4, 10, or 20 μg of purified, dialyzed GST-PEA-15 in kinase buffer was added to the kinase assays. The other aliquot was used to verify immunoprecipitation by blotting for RSK2 and ERK1/2. Alternatively, a nonradiolabeled kinase assay was carried out on one aliquot with increasing amounts of PEA15, and the reaction was arrested by using sample buffer. The complexes were immunoblotted for phospho-RSK2 (Ser 386, T577, Ser-369, and Thr-365) and RSK2.
Transcription Assays.
Transcription assays for RSK2 were carried out by using the Stratagene Pathdetect kit specific for CREB-mediated transcription as described (8). Cos-7 cells were cotransfected with 250 ng of pFR-Luc, 50 ng of pFA2-CREB, 100 ng each of pMT2HA-RSK2 and pCMV-ERK2, and 0, 25, 75, 250, or 500 ng of pcDNA3HA-PEA-15. DNA amounts were equalized to 1 μg per 35-mm well with empty vector. Luciferase readings were obtained with the Promega assay kit according to the manufacturer's protocol.
Exogenous Expression of PEA-15 in Lymphocytes.
Lentiviral GFP-tagged PEA-15 was constructed and produced according to the manufacturer's protocol by using the Lentiviral Directional TOPO expression kit (Invitrogen). Briefly, GFP/PEA-15 cDNA was amplified and introduced into the pLenti6/V5DTOPO vector. For virus production, pLentiGFP-PEA vector was transfected into 293FT cells together with the packaging plasmids. Viral titer was evaluated on HT1080 cells and ranged between 0.8 and 2 × 107 transduction units per milliliter. For lymphocyte infection, freshly isolated splenocytes were first stimulated with 0.5 and 2 μg/ml CD3/CD28, respectively, for 48 h and then transduced by one cycle of 12-h exposure to the viral supernatant at 37°C and 5% CO2. After transduction, cells were washed and cultured for 96 h in media containing CD3/CD28 antibodies. Transduction efficiency was assessed by flow cytometry for EGFP fluorescence, and PEA-15 expression was analyzed by Western blot analysis. EGFP/PEA-15-expressing cells were stimulated with 10 μg/ml PMA for 5 min and lysed for detection of RSK(T577) and total RSK protein.
Proliferation Assay.
WT and KO thymocytes (2 × 105 cells per 200 μl complete RPMI cultured in triplicates in a 96-well plate) were stimulated with 10 ng/ml PMA and 0.5 μg/ml ionomycin for 48 h, followed by addition of 1 μCi of tritiated thymidine per well for 18 h. Cells were harvested and subjected to TCA precipitation with 600 μl of 5% TCA (cold) for 30 min at 4°C. The TCA precipitated samples were then transferred to glass fiber filter discs (GF-B; Whatman) on a vacuum aspirator flask and washed with 5 ml each of 5% TCA and 70% ethanol. The filter discs were transferred to scintillation vials, Scintiverse scintillation mixture was added, and incorporated thymidine was measured on a Beckman Coulter liquid scintillation counter.
Acknowledgments
We thank Mark Ginsberg and Hervé Chneiweiss for critical review of this manuscript and Minna Komu, Mohana Gudur-Valmiki, and Joanna Gawecka for insightful discussions. This work was supported by National Institutes of Health (NIH) Grant CA93849. M.L.M. was supported by NIH National Center for Research Resources Grant RR016453.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Schaeffer HJ, Weber MJ. Mol Cell Biol. 1999;19:2435–2444. doi: 10.1128/mcb.19.4.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marshall CJ. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 3.Pawson T, Scott JD. Science. 1997;19(278):2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 4.Pearson G, Robinson F, Beers GT, Xu BE, Karandikar M, Berman K, Cobb MH. Endocr Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 5.Nguyen A, Burack WR, Stock JL, Kortum R, Chaika OV, Afkarian M, Muller WJ, Murphy KM, Morrison DK, Lewis RE, et al. Mol Cell Biol. 2002;22:3035–3045. doi: 10.1128/MCB.22.9.3035-3045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morrison DK. J Cell Sci. 2001;114:1609–1612. doi: 10.1242/jcs.114.9.1609. [DOI] [PubMed] [Google Scholar]
- 7.Formstecher E, Ramos JW, Fauquet M, Calderwood DA, Hsieh JC, Canton B, Nguyen XT, Barnier JV, Camonis J, Ginsberg MH, et al. Dev Cell. 2001;1:239–250. doi: 10.1016/s1534-5807(01)00035-1. [DOI] [PubMed] [Google Scholar]
- 8.Vaidyanathan H, Ramos JW. J Biol Chem. 2003;278:32367–32372. doi: 10.1074/jbc.M303988200. [DOI] [PubMed] [Google Scholar]
- 9.Frodin M, Gammeltoft S. Mol Cell Endocrinol. 1999;151:65–77. doi: 10.1016/s0303-7207(99)00061-1. [DOI] [PubMed] [Google Scholar]
- 10.Clark DE, Errington TM, Smith JA, Frierson HF, Jr, Weber MJ, Lannigan DA. Cancer Res. 2005;65:3108–3116. doi: 10.1158/0008-5472.CAN-04-3151. [DOI] [PubMed] [Google Scholar]
- 11.Smith JA, Poteet-Smith CE, Xu Y, Errington TM, Hecht SM, Lannigan DA. Cancer Res. 2005;65:1027–1034. [PubMed] [Google Scholar]
- 12.Ramos JW, Hughes PE, Renshaw MW, Schwartz MA, Formstecher E, Chneiweiss H, Ginsberg MH. Mol Biol Cell. 2000;11:2863–2872. doi: 10.1091/mbc.11.9.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramos JW, Kojima TK, Hughes PE, Fenczik CA, Ginsberg MH. J Biol Chem. 1998;273:33897–33900. doi: 10.1074/jbc.273.51.33897. [DOI] [PubMed] [Google Scholar]
- 14.Levchenko A, Bruck J, Sternberg PW. Proc Natl Acad Sci USA. 2000;97:5818–5823. doi: 10.1073/pnas.97.11.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith JA, Poteet-Smith CE, Malarkey K, Sturgill TW. J Biol Chem. 1999;274:2893–2898. doi: 10.1074/jbc.274.5.2893. [DOI] [PubMed] [Google Scholar]
- 16.Hill JM, Vaidyanathan H, Ramos JW, Ginsberg MH, Werner MH. EMBO J. 2002;21:6494–6504. doi: 10.1093/emboj/cdf641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kubes M, Cordier J, Glowinski J, Girault JA, Chneiweiss H. J Neurochem. 1998;71:1307–1314. doi: 10.1046/j.1471-4159.1998.71031307.x. [DOI] [PubMed] [Google Scholar]
- 18.Renganathan H, Vaidyanathan H, Knapinska A, Ramos JW. Biochem J. 2005;390:729–735. doi: 10.1042/BJ20050378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krueger J, Chou FL, Glading A, Schaefer E, Ginsberg MH. Mol Biol Cell. 2005;16:3552–3561. doi: 10.1091/mbc.E04-11-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calvo V, Bierer BE, Vik TA. Eur J Immunol. 1992;22:457–462. doi: 10.1002/eji.1830220225. [DOI] [PubMed] [Google Scholar]
- 21.Schinelli S, Zanassi P, Paolillo M, Wang H, Feliciello A, Gallo V. J Neurosci. 2001;21:8842–8853. doi: 10.1523/JNEUROSCI.21-22-08842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Araujo H, Danziger N, Cordier J, Glowinski J, Chneiweiss H. J Biol Chem. 1993;268:5911–5920. [PubMed] [Google Scholar]
- 23.Trencia A, Perfetti A, Cassese A, Vigliotta G, Miele C, Oriente F, Santopietro S, Giacco F, Condorelli G, Formisano P, et al. Mol Cell Biol. 2003;23:4511–4521. doi: 10.1128/MCB.23.13.4511-4521.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joneson T, Fulton JA, Volle DJ, Chaika OV, Bar-Sagi D, Lewis RE. J Biol Chem. 1998;273:7743–7748. doi: 10.1074/jbc.273.13.7743. [DOI] [PubMed] [Google Scholar]
- 25.Denouel-Galy A, Douville EM, Warne PH, Papin C, Laugier D, Calothy G, Downward J, Eychene A. Curr Biol. 1998;8:46–55. doi: 10.1016/s0960-9822(98)70019-3. [DOI] [PubMed] [Google Scholar]
- 26.Morrison DK, Davis RJ. Annu Rev Cell Dev Biol. 2003;19:91–118. doi: 10.1146/annurev.cellbio.19.111401.091942. [DOI] [PubMed] [Google Scholar]
- 27.Yu W, Fantl WJ, Harrowe G, Williams LT. Curr Biol. 1998;8:56–64. doi: 10.1016/s0960-9822(98)70020-x. [DOI] [PubMed] [Google Scholar]
- 28.Kitsberg D, Formstecher E, Fauquet M, Kubes M, Cordier J, Canton B, Pan G, Rolli M, Glowinski J, Chneiweiss H. J Neurosci. 1999;19:8244–8251. doi: 10.1523/JNEUROSCI.19-19-08244.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]