Abstract
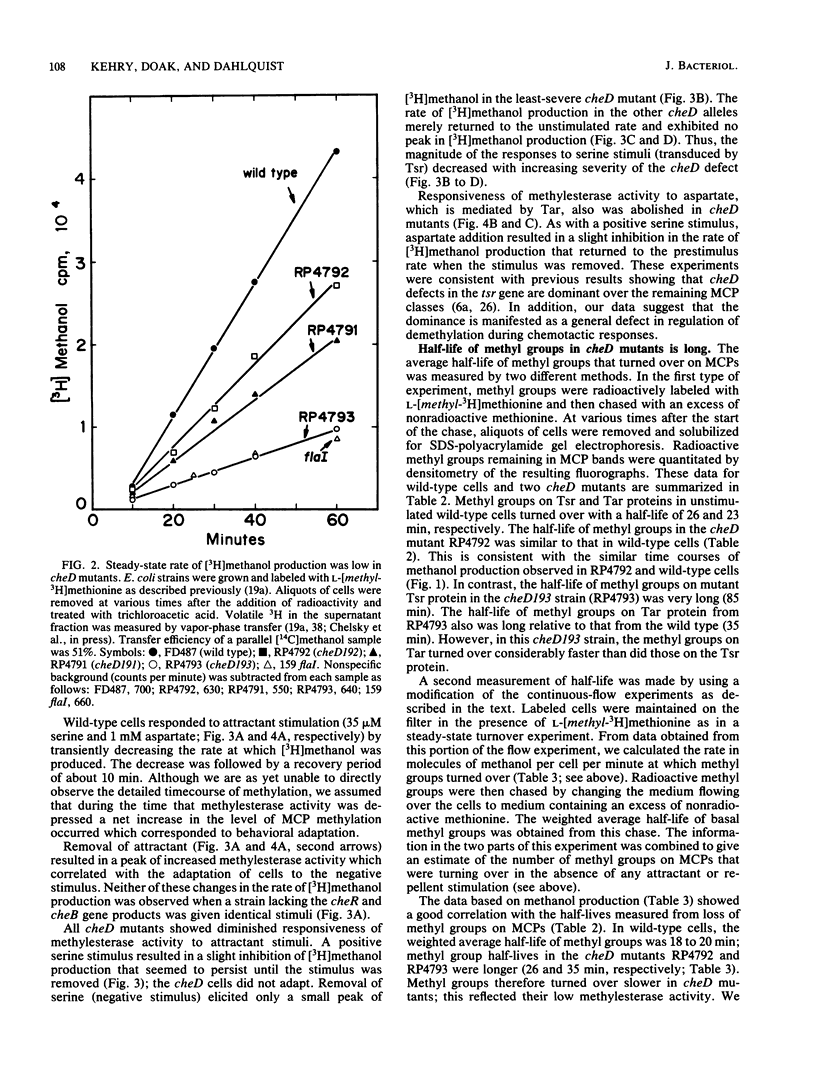
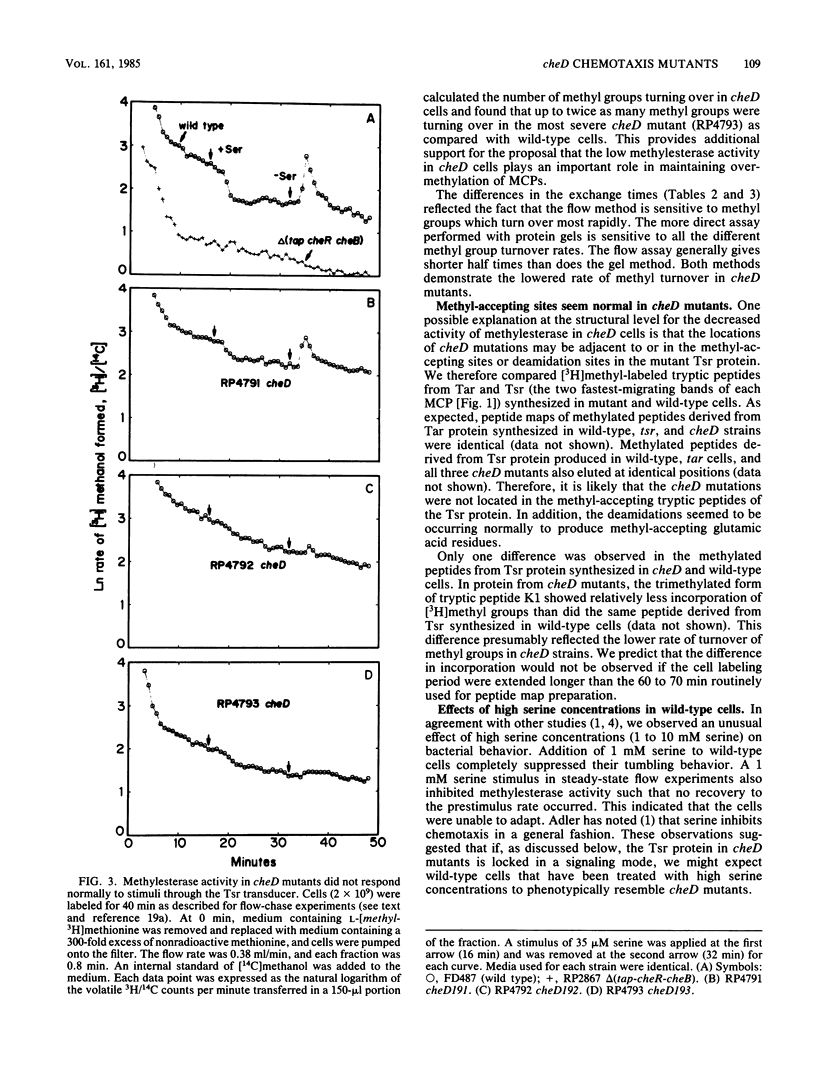
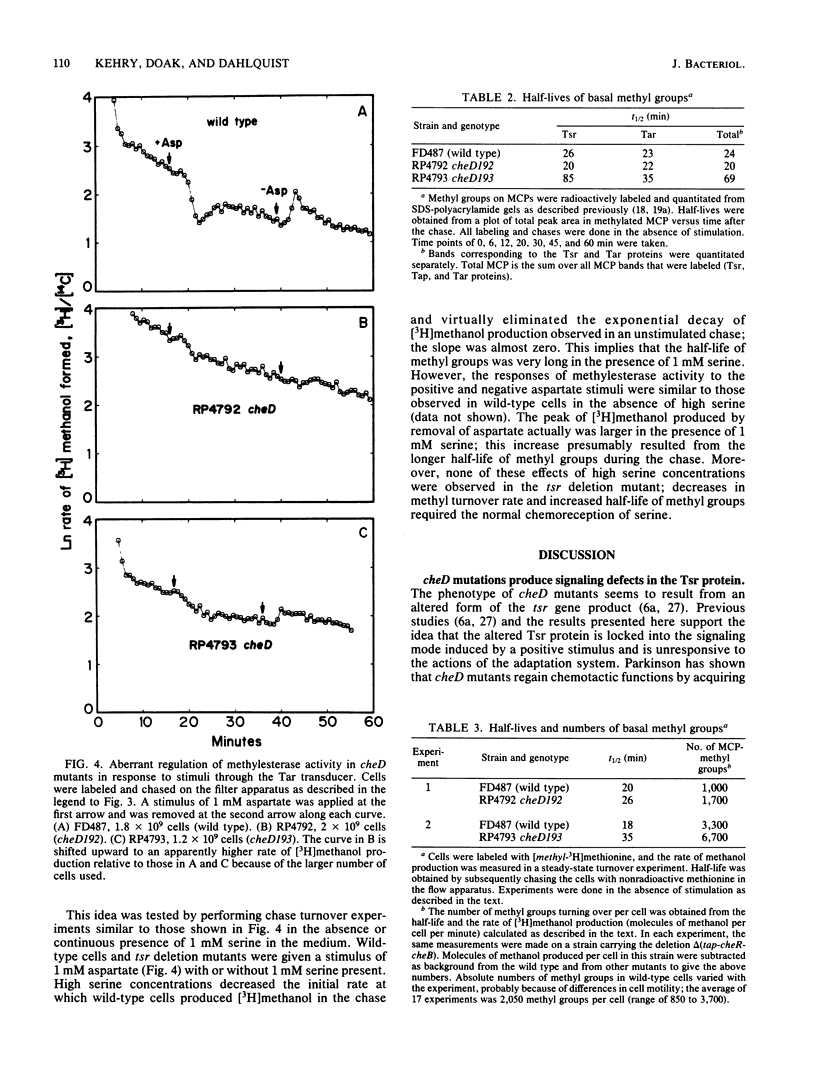
The adaptation process in several cheD chemotaxis mutants, which carry defects in tsr, the serine transducer gene, was examined. cheD mutants are smooth swimming and generally nonchemotactic; the defect is dominant to the wild-type tsr gene (J. S. Parkinson, J. Bacteriol. 142:953-961, 1980). All classes of methyl-accepting chemotaxis proteins synthesized in unstimulated cheD strains are overmethylated relative to the wild type. We found that the steady-state rate of demethylation in cheD mutants was low; this may explain their overmethylated phenotype. In addition, all cheD mutants showed diminished responsiveness of methylesterase activity to attractant and repellent stimuli transduced by either the Tsr or Tar protein, and they did not adapt. These results suggest that the dominant nature of the cheD mutations is manifested as a general defect in the regulation of demethylation. Some of these altered properties of methylesterase activity in cheD mutants were exhibited in wild-type cells that were treated with saturating concentrations of serine. The mutant Tsr protein thus seems to be locked into a signaling mode that suppresses tumbling and inhibits methylesterase activity in a global fashion. We found that the Tar and mutant Tsr proteins synthesized in cheD strains were methylated and deamidated at the correct sites and that the mutations were not located in the methylated peptides. Thus, the signaling properties of the transducers may be controlled at sites distinct from the methyl-accepting sites.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adler J. Chemoreceptors in bacteria. Science. 1969 Dec 26;166(3913):1588–1597. doi: 10.1126/science.166.3913.1588. [DOI] [PubMed] [Google Scholar]
- Adler J. Chemotaxis in bacteria. Annu Rev Biochem. 1975;44:341–356. doi: 10.1146/annurev.bi.44.070175.002013. [DOI] [PubMed] [Google Scholar]
- Berg H. C., Anderson R. A. Bacteria swim by rotating their flagellar filaments. Nature. 1973 Oct 19;245(5425):380–382. doi: 10.1038/245380a0. [DOI] [PubMed] [Google Scholar]
- Berg H. C., Brown D. A. Chemotaxis in Escherichia coli analysed by three-dimensional tracking. Nature. 1972 Oct 27;239(5374):500–504. doi: 10.1038/239500a0. [DOI] [PubMed] [Google Scholar]
- Boyd A., Krikos A., Simon M. Sensory transducers of E. coli are encoded by homologous genes. Cell. 1981 Nov;26(3 Pt 1):333–343. doi: 10.1016/0092-8674(81)90202-6. [DOI] [PubMed] [Google Scholar]
- Boyd A., Simon M. I. Multiple electrophoretic forms of methyl-accepting chemotaxis proteins generated by stimulus-elicited methylation in Escherichia coli. J Bacteriol. 1980 Aug;143(2):809–815. doi: 10.1128/jb.143.2.809-815.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan A. M., Parkinson J. S. Genetics of methyl-accepting chemotaxis proteins in Escherichia coli: cheD mutations affect the structure and function of the Tsr transducer. J Bacteriol. 1985 Jan;161(1):96–104. doi: 10.1128/jb.161.1.96-104.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelsky D., Dahlquist F. W. Chemotaxis in Escherichia coli: associations of protein components. Biochemistry. 1980 Sep 30;19(20):4633–4639. doi: 10.1021/bi00561a015. [DOI] [PubMed] [Google Scholar]
- Chelsky D., Dahlquist F. W. Structural studies of methyl-accepting chemotaxis proteins of Escherichia coli: evidence for multiple methylation sites. Proc Natl Acad Sci U S A. 1980 May;77(5):2434–2438. doi: 10.1073/pnas.77.5.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFranco A. L., Koshland D. E., Jr Multiple methylation in processing of sensory signals during bacterial chemotaxis. Proc Natl Acad Sci U S A. 1980 May;77(5):2429–2433. doi: 10.1073/pnas.77.5.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFranco A. L., Parkinson J. S., Koshland D. E., Jr Functional homology of chemotaxis genes in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1979 Jul;139(1):107–114. doi: 10.1128/jb.139.1.107-114.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström P., Hazelbauer G. L. Multiple methylation of methyl-accepting chemotaxis proteins during adaptation of E. coli to chemical stimuli. Cell. 1980 May;20(1):165–171. doi: 10.1016/0092-8674(80)90244-5. [DOI] [PubMed] [Google Scholar]
- Goldbeter A., Koshland D. E., Jr Simple molecular model for sensing and adaptation based on receptor modification with application to bacterial chemotaxis. J Mol Biol. 1982 Nov 5;161(3):395–416. doi: 10.1016/0022-2836(82)90246-7. [DOI] [PubMed] [Google Scholar]
- Goy M. F., Springer M. S., Adler J. Sensory transduction in Escherichia coli: role of a protein methylation reaction in sensory adaptation. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4964–4968. doi: 10.1073/pnas.74.11.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H., Koiwai O., Kozuka M. Studies on bacterial chemotaxis. II. Effect of cheB and cheZ mutations on the methylation of methyl-accepting chemotaxis protein of Escherichia coli. J Biochem. 1979 May;85(5):1213–1223. [PubMed] [Google Scholar]
- Hazelbauer G. L., Engström P. Multiple forms of methyl-accepting chemotaxis proteins distinguished by a factor in addition to multiple methylation. J Bacteriol. 1981 Jan;145(1):35–42. doi: 10.1128/jb.145.1.35-42.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehry M. R., Bond M. W., Hunkapiller M. W., Dahlquist F. W. Enzymatic deamidation of methyl-accepting chemotaxis proteins in Escherichia coli catalyzed by the cheB gene product. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3599–3603. doi: 10.1073/pnas.80.12.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehry M. R., Dahlquist F. W. Adaptation in bacterial chemotaxis: CheB-dependent modification permits additional methylations of sensory transducer proteins. Cell. 1982 Jul;29(3):761–772. doi: 10.1016/0092-8674(82)90438-x. [DOI] [PubMed] [Google Scholar]
- Kehry M. R., Dahlquist F. W. The methyl-accepting chemotaxis proteins of Escherichia coli. Identification of the multiple methylation sites on methyl-accepting chemotaxis protein I. J Biol Chem. 1982 Sep 10;257(17):10378–10386. [PubMed] [Google Scholar]
- Kehry M. R., Doak T. G., Dahlquist F. W. Stimulus-induced changes in methylesterase activity during chemotaxis in Escherichia coli. J Biol Chem. 1984 Oct 10;259(19):11828–11835. [PubMed] [Google Scholar]
- Kehry M. R., Engström P., Dahlquist F. W., Hazelbauer G. L. Multiple covalent modifications of Trg, a sensory transducer of Escherichia coli. J Biol Chem. 1983 Apr 25;258(8):5050–5055. [PubMed] [Google Scholar]
- Khan S., Macnab R. M., DeFranco A. L., Koshland D. E., Jr Inversion of a behavioral response in bacterial chemotaxis: explanation at the molecular level. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4150–4154. doi: 10.1073/pnas.75.9.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleene S. J., Toews M. L., Adler J. Isolation of glutamic acid methyl ester from an Escherichia coli membrane protein involved in chemotaxis. J Biol Chem. 1977 May 25;252(10):3214–3218. [PubMed] [Google Scholar]
- Kondoh H., Ball C. B., Adler J. Identification of a methyl-accepting chemotaxis protein for the ribose and galactose chemoreceptors of Escherichia coli. Proc Natl Acad Sci U S A. 1979 Jan;76(1):260–264. doi: 10.1073/pnas.76.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kort E. N., Goy M. F., Larsen S. H., Adler J. Methylation of a membrane protein involved in bacterial chemotaxis. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3939–3943. doi: 10.1073/pnas.72.10.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D. E., Jr A response regulator model in a simple sensory system. Science. 1977 Jun 3;196(4294):1055–1063. doi: 10.1126/science.870969. [DOI] [PubMed] [Google Scholar]
- Macnab R. M., Koshland D. E., Jr The gradient-sensing mechanism in bacterial chemotaxis. Proc Natl Acad Sci U S A. 1972 Sep;69(9):2509–2512. doi: 10.1073/pnas.69.9.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S. Novel mutations affecting a signaling component for chemotaxis of Escherichia coli. J Bacteriol. 1980 Jun;142(3):953–961. doi: 10.1128/jb.142.3.953-961.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S. cheA, cheB, and cheC genes of Escherichia coli and their role in chemotaxis. J Bacteriol. 1976 May;126(2):758–770. doi: 10.1128/jb.126.2.758-770.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins C., Dahlquist F. W. The methyl-accepting chemotaxis proteins of E. coli: a repellent-stimulated, covalent modification, distinct from methylation. Cell. 1981 Aug;25(2):333–340. doi: 10.1016/0092-8674(81)90051-9. [DOI] [PubMed] [Google Scholar]
- Sherris D., Parkinson J. S. Posttranslational processing of methyl-accepting chemotaxis proteins in Escherichia coli. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6051–6055. doi: 10.1073/pnas.78.10.6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M., Simon M. Chemotaxis in Escherichia coli: methylation of che gene products. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3317–3321. doi: 10.1073/pnas.74.8.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M., Simon M. Flagellar rotation and the mechanism of bacterial motility. Nature. 1974 May 3;249(452):73–74. doi: 10.1038/249073a0. [DOI] [PubMed] [Google Scholar]
- Silverman M., Simon M. Identification of polypeptides necessary for chemotaxis in Escherichia coli. J Bacteriol. 1977 Jun;130(3):1317–1325. doi: 10.1128/jb.130.3.1317-1325.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M. S., Goy M. F., Adler J. Protein methylation in behavioural control mechanisms and in signal transduction. Nature. 1979 Jul 26;280(5720):279–284. doi: 10.1038/280279a0. [DOI] [PubMed] [Google Scholar]
- Springer M. S., Goy M. F., Adler J. Sensory transduction in Escherichia coli: two complementary pathways of information processing that involve methylated proteins. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3312–3316. doi: 10.1073/pnas.74.8.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer W. R., Koshland D. E., Jr Identification of a protein methyltransferase as the cheR gene product in the bacterial sensing system. Proc Natl Acad Sci U S A. 1977 Feb;74(2):533–537. doi: 10.1073/pnas.74.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J. B., Koshland D. E., Jr A protein methylesterase involved in bacterial sensing. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3659–3663. doi: 10.1073/pnas.75.8.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J. B., Koshland D. E., Jr Changing reactivity of receptor carboxyl groups during bacterial sensing. J Biol Chem. 1981 Nov 10;256(21):10826–10833. [PubMed] [Google Scholar]
- Terwilliger T. C., Bogonez E., Wang E. A., Koshland D. E., Jr Sites of methyl esterification on the aspartate receptor involved in bacterial chemotaxis. J Biol Chem. 1983 Aug 25;258(16):9608–9611. [PubMed] [Google Scholar]
- Toews M. L., Adler J. Methanol formation in vivo from methylated chemotaxis proteins in Escherichia coli. J Biol Chem. 1979 Mar 25;254(6):1761–1764. [PubMed] [Google Scholar]
- Toews M. L., Goy M. F., Springer M. S., Adler J. Attractants and repellents control demethylation of methylated chemotaxis proteins in Escherichia coli. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5544–5548. doi: 10.1073/pnas.76.11.5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Werf P., Koshland D. E., Jr Identification of a gamma-glutamyl methyl ester in bacterial membrane protein involved in chemotaxis. J Biol Chem. 1977 Apr 25;252(8):2793–2795. [PubMed] [Google Scholar]